The trajectory from a prebiotic synthesis of the basic building blocks of life, to the sophisticated synthesis by cell factories: an unsolved riddle
https://reasonandscience.catsboard.com/t2894-prevital-unguided-origin-of-the-four-basic-building-blocks-of-life-impossible#7650402 individual reactions synthesize amino acids, nucleotides and cofactors from H2, CO2, NH3, H2S and phosphate in modern cells
https://www.frontiersin.org/articles/10.3389/fmicb.2021.793664/full%20?utm_source=fweb&utm_medium=nblog&utm_campaign=ba-sci-fmicb-luca-hydrothermal-vents-core-energy
We identified a set of roughly 400 ancient reactions that cells use to synthesize the 20 amino acids of proteins, the four bases of genes, and the 18 vitamins that cells need to make those 400 reactions work. Those core chemical reactions are conserved across all microbial lineages, hence they were present in the last universal common ancestor of all life, LUCA. They provide a window into the source of building blocks, but also the source of energy at origins.
https://blog.frontiersin.org/2022/01/19/frontiers-microbiology-origin-of-life-energy-hydrothermal-vents/
Marian Breuer Essential metabolism for a minimal cell Jan 18, 2019
Our model features 338 reactions organized in nine subsystems involving 304 metabolites, catalyzed by gene products of 155 genes, thus covering a third of the genes of JCVI-syn3A.
https://elifesciences.org/articles/36842
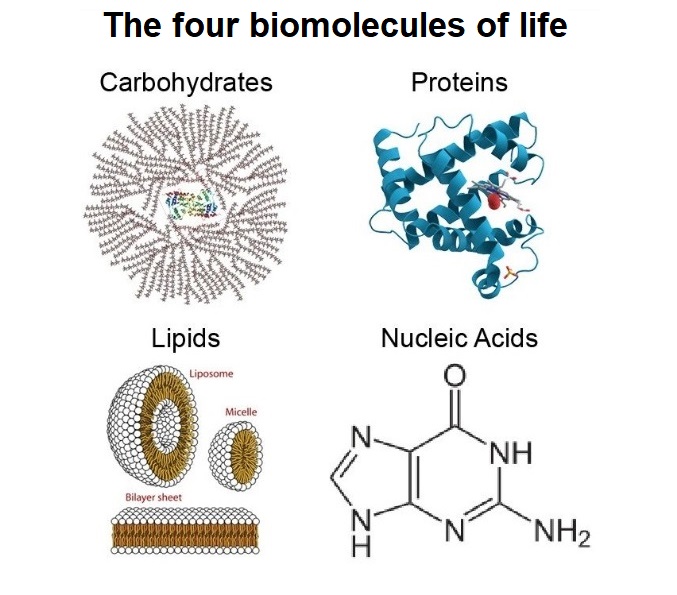
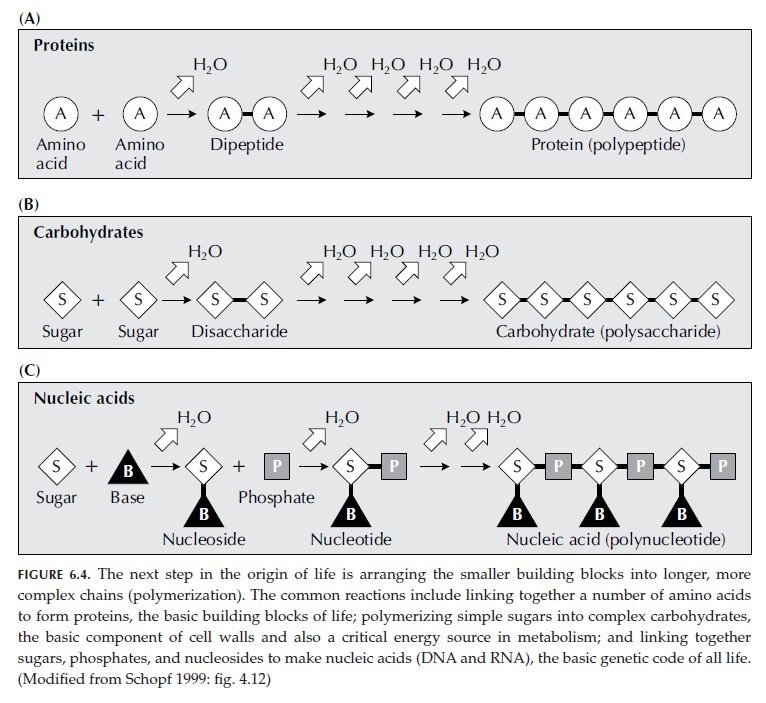
Modern cells use vast resources of information, basic elements of matter, and energy, to synthesize the four basic building blocks: 1. Amino acids 2. Nucleotides 3. Carbohydrates 4. phospholipids. These individual components are themselves complex, and very specific, and the cell machinery uses several highly sophisticated and controlled
metabolic pathways to synthesize them, where both, anabolism, and catabolism play a role, that is, de-novo synthesis, and recycling. This requires a minimal genome, proteome, and metabolome. Sophisticated signaling networks are constantly in use to detect the required quantities based on the external availability of food and to maintain intracellular homeostasis. Obviously, this ultrasophisticated chemical factory was not extant on the early earth.
Modern abiogenesis research focusses on attempting to elucidate how these basic molecules could have emerged by chemical evolution on the early earth, prior to the emergence of the modern chemical cell factories that all life forms use. In almost 70 years, rather than making progress to solve the riddle, general frustration is the case.
Steve Benner, one of the world’s leading authorities on abiogenesis, summed it up:We are now 60 years into the modern era of prebiotic chemistry. That era has produced tens of thousands of papers attempting to define processes by which “molecules that look like biology” might arise from “molecules that do not look like biology” …. For the most part, these papers report “success” in the sense that those papers define the term…. And yet, the problem remains unsolvedIn the following, I am outlining a MINIMAL number of enzymes involved in synthesizing these building blocks. Not taken into consideration, is all the machinery, that is involved synthesizing the co-factors, metal clusters, metal import channels, chaperones, etc. that are required to get functional proteins. NucleotidesFolate is necessary for the production and maintenance of new cells, for
DNA synthesis and
RNA synthesis through
methylation. The synthesis of NADPH requires six enzymes. 5 Six proteins are required in the folate pathway. 4 RNA Nucleotides are building blocks for DNA and RNA. Three Pyrimidines and Two Purines Are Commonly Found in Cells.The pyrimidine synthesis pathway requires six regulated steps, seven enzymes, and energy in the form of ATP.The starting material for purine biosynthesis is Ribose 5-phosphate, a product of the highly complex pentose phosphate pathway, which uses 12 enzymes. 1 De novo purine synthesis pathway requires ten regulated steps, eleven enzymes, and energy in the form of ATP. DNAThe replacement of RNA as the repository of genetic information by its more stable cousin, DNA, provides a more reliable way of transmitting information. DNA uses thymine (T) as one of its four informational bases, whereas RNA uses uracil (U). At the C2' position of ribose, an oxygen atom is removed. The remarkable enzymes that do this are named Ribonucleotide reductases (RNR). The iron-dependent enzyme is essential for DNA synthesis, and most essential enzymes of life 32, and it has one of the most sophisticated allosteric regulations known today. 50 The thymine-uracil exchange constitutes one of the major chemical differences between DNA and RNA. Before being incorporated into the chromosomes, this essential modification takes place. Uracil bases in RNA are transformed into thymine bases in DNA. The synthesis of thymine requires seven enzymes. De novo biosynthesis of thymine is an intricate and energetically expensive process. All in all, not considering the metabolic pathways and enzymes required to make the precursors to start RNA and DNA synthesis requires at least
26 enzymes. Amino AcidsSeveral classes of reactions play special roles in the biosynthesis of amino acids and nucleotides
(1) transamination reactions and other rearrangements promoted by enzymes containing
pyridoxal phosphate (PLP), which requires
eight enzymes to be synthesized.
3(2) transfer of one-carbon groups, with either
tetrahydrofolate or
S-adenosylmethionine as a cofactor;
tetrahydrofolate is derived from the folate pathway,
(3) transfer of amino groups derived from the amide nitrogen of glutamine.
As implied by the root of the word (amine), the key atom in amino acid composition is nitrogen. All organisms contain the enzymes
glutamate dehydrogenase and
glutamine synthetase, which convert ammonia to glutamate and glutamine, respectively.
Number of enzymes required to synthesize the life-essential amino acids:
1. Glycine requires
five enzymes 6, 72. Alanine can be obtained by three different synthesis pathways, and the number of enzymes is speculative. At least a transamination reaction is required, so i will put
one enzyme. 8
3. Valine is derived from Pyruvate. Pyruvic acid can be made from glucose through glycolysis.
2 Nine enzymes are required to go from Glucose to pyruvate. A further pathway required
three enzymes is required. 9
4. Leucine The biosynthesis pathways of the branched-chain amino acids (valine, isoleucine and leucine) all begin with the same precursors (pyruvate or pyruvate and 2-ketobutyrate). Subsequently, four enzymes are required. 10 5. Isoleucine The isoleucine pathway is almost the same as the valine biosynthesis pathway. 116. Methionine requires
4 enzymes, plus
Vitamin B12 thirty enzymes are necessary for one of nature’s largest characterized biosynthetic pathways. 127. Proline: three enzymes are required in the pathway
8. Phenylalanine:
one enzyme is needed. 13
9. Tryptophan biosynthesis is a biologically expensive, complicated process. In fact, the products of four other pathways are essential contributors of carbon or nitrogen during tryptophan formation. Thirteen enzymes are used
1410. Serine requires
three enzymes in its biosynthesis pathway 15
11. Threonine requires
five enzymes in its biosynthesis pathway 16
12. Asparagine requires
two enzymes in its biosynthesis pathway 17
13. Glutamine requires
two enzymes in its biosynthesis pathway 18
14. Tyrosine requires
two enzymes in its biosynthesis pathway 19
15. Cysteine requires
one enzyme in its biosynthesis pathway 20
16. Lysine requires
two enzymes in its biosynthesis pathway 21
17. Methionine requires
two enzymes in its biosynthesis pathway 22
18. Threonine requires
one enzyme in its biosynthesis pathway 23
19. Arginine requires
eight enzymes in its biosynthesis pathway
2420. Aspartate requires
two enzymes in its biosynthesis pathway 25
21. Glutamate requires
one enzyme in its biosynthesis pathway 26
22. Histidine requires
eight enzymes in its biosynthesis pathway 27
A minimum of
112 enzymes are required to synthesize the 20 (+2) amino acids used in proteins
CarbohydratesLUCA used the simplest and most ancient of the six known pathways of CO2 fixation, called the acetyl–CoA (or Wood–Ljungdahl) pathway. It uses
nine enzymes. MembranesAt least
74 enzymes are required for phospholipid synthesis in prokaryotes
29The origin of life: what we know, what we can know and what we will never knowA detailed understanding of that process will have to wait until ongoing studies in systems chemistry reveal both the classes of chemical materials and the kinds of chemical pathways that simple replicating systems are able to follow in their drive towards greater complexity and replicative stability.
Paul Davies, the fifth miracle page 53:Pluck the DNA from a living cell and it would be stranded, unable to carry out its familiar role. Only within the context of a highly specific molecular milieu will a given molecule play its role in life. To function properly, DNA must be part of a large team, with each molecule executing its assigned task alongside the others in a cooperative manner. Acknowledging the interdependability of the component molecules within a living organism immediately presents us with a stark philosophical puzzle. If everything needs everything else, how did the community of molecules ever arise in the first place? Since most large molecules needed for life are produced only by living organisms, and are not found outside the cell, how did they come to exist originally, without the help of a meddling scientist? Could we seriously expect a Miller-Urey type of soup to make them all at once, given the hit-and-miss nature of its chemistry?On the Development Towards the Modern World: A Plausible Role of Uncoded Peptides in the RNA World 52009Arguably one of the most outstanding problems in understanding the progress of early life is the transition from the RNA world to the modern protein-based world. One of the main requirements of this transition is the emergence of mechanism to produce functionally meaningful peptides and later, proteins. What could have served as the driving force for the production of peptides and what would have been their properties and purpose in the RNA world?My endnote: I have simplified the illustration, just to give an idea of the problem. In reality, a minimal, fully operational cell requires FAR many more enzymes than just what was mentioned above. But it shows the unbridgeable gap.1. On the one side, we have the putative prebiotic soup with the random chaotic floating around of the basic building blocks of life, and on the other side, the first living self-replicating cell ( LUCA ), a supposed fully operational minimal self-replicating cell, using the
highly specific and sophisticated molecular milieu with a large team of enzymes which catalyze the reactions to produce the four basic building blocks of life in a cooperative manner, and furthermore, able to maintain intracellular homeostasis, reproduce, obtaining energy and converting it into a usable form, getting rid of toxic waste, protecting itself from dangers of the environment, doing the cellular repair, and communicate.
2. The science paper: Structural analyses of a hypothetical minimal metabolism proposes a minimal number of 50 enzymatic steps catalyzed by the associated encoded proteins. They don't, however, include the steps to synthesize the 20 amino acids required in life. Including those, the minimal metabolome would consist of 221 enzymes & proteins. A large number of molecular machines, co-factors, scaffold proteins, and chaperones are not included, required to build this highly sophisticated chemical factory.
3. There simply no feasible viable prebiotic route to go from a random prebiotic soup to this minimal proteome to kick-start metabolism by unguided means. This is not a conclusion by ignorance & incredulity, but it is reasonable to be skeptic, that this
irreducibly complex biological system, entire factory complexes composed of myriads of interconnected highly optimized production lines, full of computers and robots could emerge naturally defying known and reasonable principles of the limited range of random unguided events and physical necessity. Comparing the two competing hypotheses, chance vs intelligent design, the second is simply by far the more case-adequate & reasonable explanation. 1.
https://reasonandscience.catsboard.com/t2172-the-pentose-phosphate-pathway2.
https://reasonandscience.catsboard.com/t1796-glycolysis3.
https://reasonandscience.catsboard.com/t2590p25-origins-what-cause-explains-best-our-existence-and-why#59384.
https://reasonandscience.catsboard.com/t2590p25-origins-what-cause-explains-best-our-existence-and-why#59425.
https://reasonandscience.catsboard.com/t2708-nicotinamide-adenine-dinucleotide-nad-in-origin-of-life-scenarios6. https://en.wikipedia.org/wiki/Template:Amino_acid_metabolism_enzymes
7. https://en.wikipedia.org/wiki/Glycine#Biosynthesis
8. https://en.wikipedia.org/wiki/Alanine#Biosynthesis
9. https://en.wikipedia.org/wiki/Valine#Synthesis
10. https://en.wikipedia.org/wiki/Leucine#Synthesis_in_non-human_organisms
11. https://en.wikipedia.org/wiki/Isoleucine#Biosynthesis
12. https://en.wikipedia.org/wiki/Methionine#Biosynthesis
13. https://en.wikipedia.org/wiki/Phenylalanine
14. https://en.wikipedia.org/wiki/Tryptophan#Biosynthesis_and_industrial_production
15. https://en.wikipedia.org/wiki/Serine#Synthesis_and_industrial_production
16. https://en.wikipedia.org/wiki/Threonine#Biosynthesis
17. https://en.wikipedia.org/wiki/Asparagine#Biosynthesis
18. https://en.wikipedia.org/wiki/Glutamine
19. https://en.wikipedia.org/wiki/Tyrosine#Biosynthesis
20. https://en.wikipedia.org/wiki/Cysteine
21. https://en.wikipedia.org/wiki/Lysine
22. https://en.wikipedia.org/wiki/Methionine#Biosynthesis
23. https://en.wikipedia.org/wiki/Threonine#Metabolism
24.
https://reasonandscience.catsboard.com/t1740-amino-acids-origin-of-the-canonical-twenty-amino-acids-required-for-life#606325. https://en.wikipedia.org/wiki/Aspartic_acid#Biosynthesis
26. https://en.wikipedia.org/wiki/Glutamate_(neurotransmitter)#Biosynthesis
27. https://en.wikipedia.org/wiki/Histidine#Biosynthesis
28.
https://reasonandscience.catsboard.com/t2967-fatty-acid-and-phospholipid-biosynthesis-in-prokaryotes29.
https://reasonandscience.catsboard.com/t2967-fatty-acid-and-phospholipid-biosynthesis-in-prokaryoteshttps://manet.illinois.edu/pathways.php