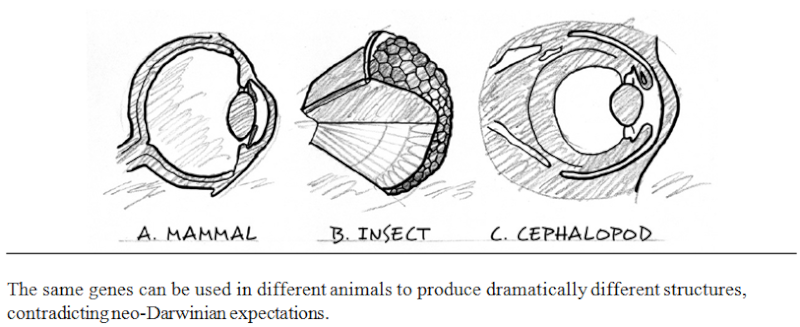
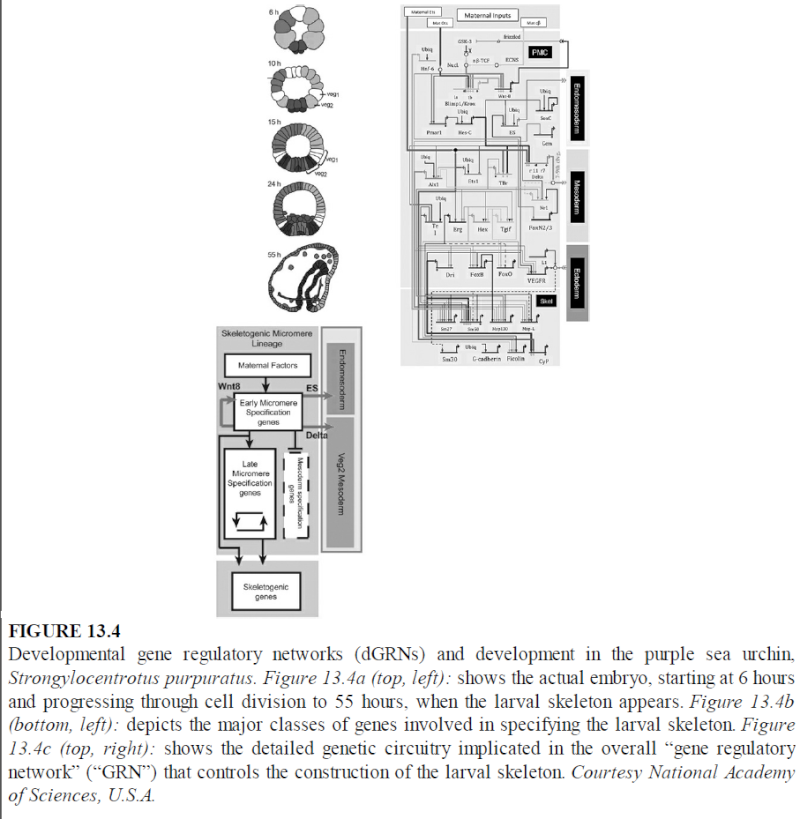
https://reasonandscience.catsboard.com/t2316-evolution-where-do-complex-organisms-come-from
How do biological multicellular complexity and a spatially organized body plan emerge?
https://reasonandscience.catsboard.com/t2990-how-does-biological-multicellular-complexity-and-a-spatially-organized-body-plan-emerge
The architecture of multicellular organisms arises from a complex interplay of at least 47 key developmental processes, 33 genetic codes, and over 223 epigenetic, manufacturing, and regulatory codes. These elements are coordinated through hundreds of signaling networks, embodying functional integration and systemic complexity. This holistic ensemble, showcasing emergent properties and synergy, hints at a level of organization often observed to require an intelligent agent for setup.
Key Developmental Processes Shaping Organismal Form and Function
https://reasonandscience.catsboard.com/t2316-evolution-where-do-complex-organisms-come-from
The different genetic codes
https://reasonandscience.catsboard.com/t2277-the-different-genetic-codes
The various codes in the cell
https://reasonandscience.catsboard.com/t2213-the-various-codes-in-the-cell
Overview on Various Signaling Pathways
https://reasonandscience.catsboard.com/t2351-the-essential-signaling-pathways-for-animal-development#10693
The mechanisms behind the complexity of organisms, anatomical features, and the diversity of life is pre-programmed and complex information encoded in multiple genetic codes—at least 33 known variations—as well as in over 223 epigenetic, manufacturing, and regulatory codes. These codes are necessary for complex communication networks that operate at a structural level, functioning in a coordinated and interconnected manner. This pre-set programming is essential for guiding growth and development, adapting to nutritional needs and environmental changes, regulating reproduction, maintaining homeostasis, managing metabolism, orchestrating defense mechanisms, and controlling cell lifecycle events. Such sophisticated, information-driven systems, given their integrated complexity, require an intelligent designer to establish them. This perspective challenges the idea that undirected, gradual evolutionary processes can fully account for such complexity, especially considering that if all necessary components are not simultaneously present, individual parts would lack function and thus could not be favored by natural selection.
1. Genetic and epigenetic information ( at least 33 variations of genetic codes, and over 230 manufacturing, signaling, and operational codes 4) and hundreds, (in prokaryotes probably thousands) of signaling networks direct the making of complex multicellular organisms, biodiversity, form, and architecture. Furthermore, there is growing evidence, that life is based on polyphyly, not monophyly. There are over 50 lines of evidence, that refute universal common descent.
2. Biological information is programmed in many cases through digital semiotic language ( syntax, semantics, and pragmatics) to get a functional outcome. Each protein, metabolic pathway, organelle, or system, each biomechanical structure and motion works based on principles that provide a specific function.
3. Information is not physical, it is conceptual, and is simply beyond the sphere of influence of any undirected physical process. To suggest that a physical process can create semiotic code is like suggesting that a rainbow can write poetry, or a blueprint, is never going to happen! Programming instructional assembly information and getting a specific functional outcome as we see in biology is always prescriptive, the result of a mind with intention, goals, and foresight, able to instantiate a distant specific goal.
4. Therefore, the origin of complex organismal architecture and biodiversity is best explained as the result of intelligent design, a deliberate creative intellectual process, rather than unguided evolution.
Premise 1. Life is embroidered with a profound amalgamation of genetic and epigenetic codes. With at least 33 distinct genetic variations and over 230 intertwined manufacturing, signaling, and operational codes, together with hundreds, and in prokaryotes probably thousands of intricate signaling networks, these codes collectively choreograph the exquisite dance of multicellular organisms, sculpting the vast biodiversity, nuanced form, and majestic architecture we observe.
Premise 2. This monumental wealth of information isn't random. It's meticulously orchestrated, often resembling the digital semiotic languages characterized by syntax, semantics, and pragmatics. Every protein, each metabolic trajectory, and every biomechanical construct operates with precision, abiding by principles finely tuned for specific functions.
Premise 3. While the foundations of life are tangible, the essence of information transcends the physical. It's conceptual, operating in realms beyond the reach of spontaneous and aimless physical phenomena. Arguing that such processes can birth semiotic codes is akin to believing a rainbow might pen a sonnet or winds could draft architectural wonders. The directive, purposeful nature inherent in biological programming suggests intention, foresight, and specific goals.
Conclusion: The staggering complexity and deliberate design seen in organismal architecture and biodiversity beckon consideration beyond mere chance. It suggests that these wonders are not just products of random evolution but the handiwork of a deliberate and intelligent design.
Instructional assembly information encoded in 33 genetic, and at least 230 epigenetic codes and languages, and hundreds of signaling networks operating on a structural level in an integrated interlocked fashion are the mechanisms explaining the organismal architecture, development, operation, and adaptation, directing/dictating the make and control of complex molecular physiological integrated structures and their development - the making of irreducible complex molecular machines, robotic molecular production lines, protein-protein interactions, metabolic signaling and transcription-regulatory networks, chemical cell factories, and multicellular organisms and organizations. There is a high degree of internal order that governs the cell’s molecular, and extracellular organization.
Having that information at hand, we can ask:
Do evolutionary, non-intelligent mechanisms explain adequately the observations? In my opinion, the answer is no. All historical, observational, testable, and repeatable examples have demonstrated that instantiating information and operational functionality dictated by prescribed information come from intelligent sources. Etiology, the science of causes, in regards to the origin of biocomplexity, the hypothesis that natural, unguided random or evolutionary mechanisms, natural selection, genetic drift, and flow, were responsible, have led to dead ends, to the constatation that these causes produce too limited results that do not explain biological form. Only an intelligent agency with intent and foresight can in principle, conceptualize and instantiate hardware and software, that directs the making of integrated operational systems with specific functions that work in an integrated manner. Finely orchestrated order and fine-tuning, instructions that convey and create purposeful function, are all positive evidence of a mind at work as opposed to blind, randomness operating in stochastic chaos. What we find is a gold nugget of truth...a mind is more than likely behind biological form and variation...therefore it is rational and reasonable to believe in an intelligent designer.
Observation:
The integrated complexity and specificity of biological systems based on a large number of genetic and epigenetic codes, complex signaling networks, and the precise orchestration of cellular and organismal functions, closely mirror the complexity of human-engineered systems. These biological systems involve not only a vast array of components but also highly detailed instructions for assembling proteins, regulating cellular activities, and ensuring survival and reproduction. Such a level of complexity and integration is typically associated with the output of intelligent design in human contexts, where complex systems are deliberately planned and executed to achieve specific goals.
Hypothesis:
If the biological structures and systems observed in nature exhibit a level of complexity, specificity, information, multilevel interdependent communications systems, and purposefulness akin to those produced by intelligent human designers, it suggests that such natural systems be the result of intelligent design. This hypothesis posits that the information and systems seen in biology are not solely the result of undirected evolutionary processes but indicate the involvement of a guiding intelligence.
Experiment:
The existence of complex information systems in biology, such as the genetic and epigenetic codes, the optimized nature of the genetic code, and the elaborate signaling networks that govern cellular and organismal functions has been unraveled. Characteristics that indicate design, such as irreducible complexity, where systems are composed of several well-matched, interacting parts that contribute to the basic function, and the system ceases to function if any one of the parts is removed, have been found. Also, specified and integrated complexity, where systems exhibit a complexity that is highly unlikely to have occurred by evolutionary mechanisms also conforms to an independently given pattern or set of rules.
Conclusion:
The presence of complex information systems in biology, characterized by a multitude of interdependent codes, highly efficient and error-correcting processes, and elaborate networks for regulation and communication, challenges the explanatory power of unguided evolutionary mechanisms alone. The resemblance of these biological systems to human-engineered systems, known to result from intelligent design, suggests the plausible inference that a similar type of intelligence may underlie the origin of complex biological systems. This argument does not necessarily identify the nature of the designing intelligence but posits that the best explanation for the observed complexity and specificity in biological systems is an intelligent cause, rather than undirected natural evolutionary processes.

Paul C. W. Davies: The algorithmic origins of life 2013 Feb 6
The algorithm for building an organism is not only stored in a linear digital sequence (tape), but also in the current state of the entire system (e.g. epigenetic factors such as the level of gene expression, post-translational modifications of proteins, methylation patterns, chromatin architecture, nucleosome distribution, cellular phenotype and environmental context). The algorithm itself is therefore highly delocalized, distributed inextricably throughout the very physical system whose dynamics it encodes.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3565706/
Gerd B. Müller: Why an extended evolutionary synthesis is necessary 18 August 2017
These examples of conceptual change in various domains of evolutionary biology represent only a condensed segment of the advances made since the inception of the MS theory some 80 years ago. Relatively minor attention has been paid to the fact that many of these concepts, which are in full use today, sometimes contradict or expand central tenets of the MS theory. Given proper attention, these conceptual expansions force us to consider what they mean for our present understanding of evolution. Obviously, several of the cornerstones of the traditional evolutionary framework need to be revised and new components incorporated into a common theoretical structure.
Although today's organismal systems biology is mostly rooted in biophysics and biological function, its endeavors are profoundly integrative, aiming at multiscale and multilevel explanations of organismal properties and their evolution. Instead of chance variation in DNA composition, evolving developmental interactions account for the specificities of phenotypic construction.
https://royalsocietypublishing.org/doi/10.1098/rsfs.2017.0015
Laland, Uller, et al (2014) state In our view, this ‘gene-centric’ focus fails to capture the full gamut of processes that direct evolution. Missing pieces include how physical development influences the generation of variation (developmental bias); how the environment directly shapes organisms’ traits (plasticity); how organisms modify environments (niche construction); and how organisms transmit more than genes across generations (extra-genetic inheritance).
How do biological multicellular complexity and a spatially organized body plan emerge?
https://reasonandscience.catsboard.com/t2990-how-does-biological-multicellular-complexity-and-a-spatially-organized-body-plan-emerge
The findings show a clear Lamarckian epigenetic contribution to gene network evolution and the classic Darwinian interpretation of evolution alone cannot explain our observations. “The findings support the idea that both genetic and epigenetic mechanisms need to be combined in a ‘grand unified theory of evolution,’”
https://news.yale.edu/2020/10/27/yeast-study-yields-insights-longstanding-evolution-debate
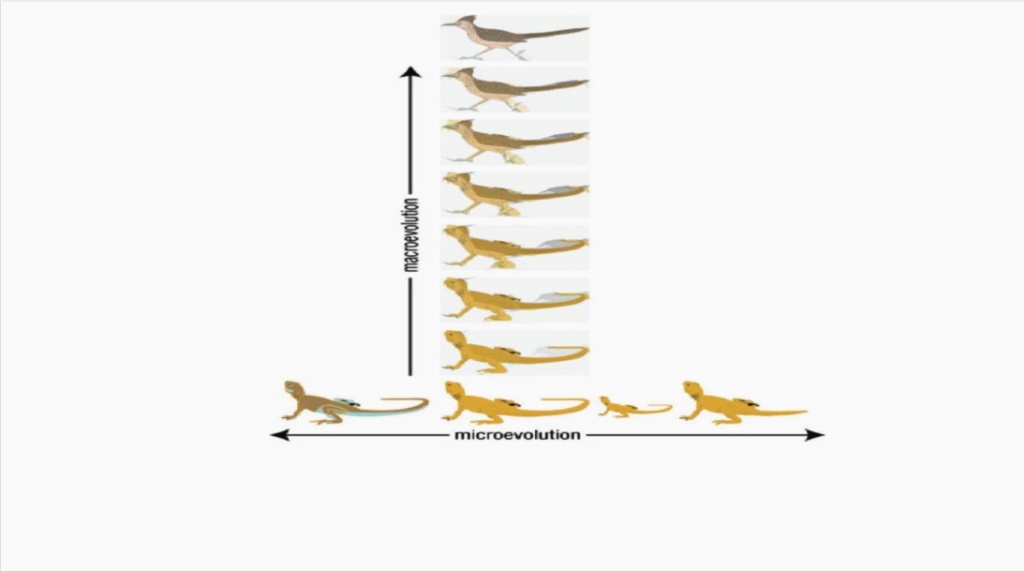
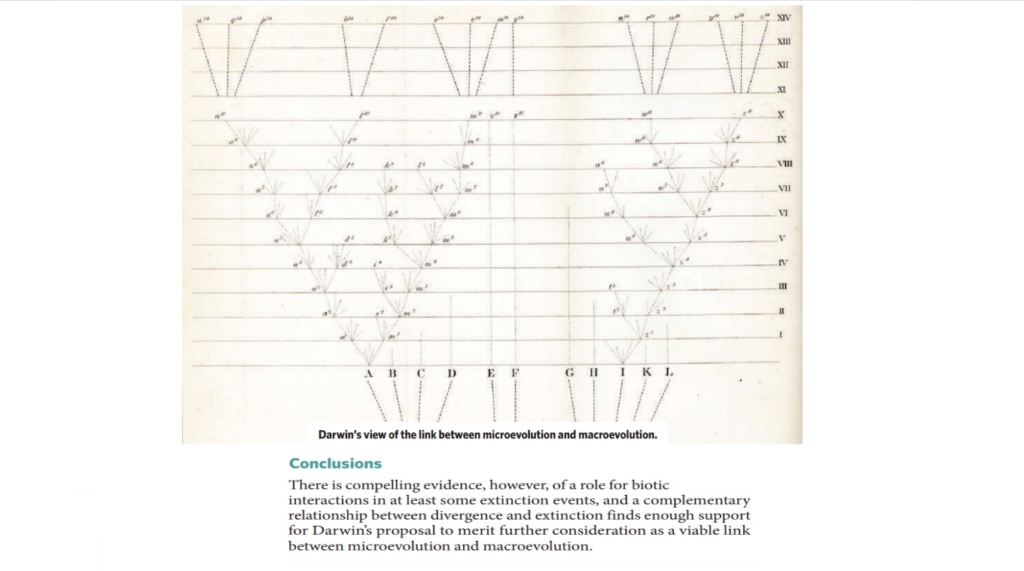
Macroevolution has also been defined by Professor Jerry Coyne as “large changes in body form or the evolution of one type of plant or animal from another type”
MICHAEL J. BEHE Experimental Support for the Design Inference DECEMBER 27, 1987
In order to say that some function is understood, every relevant step in the process must be elucidated. The relevant steps in biological processes occur ultimately at the molecular level, so a satisfactory explanation of a biological phenomenon such as sight, or digestion, or immunity, must include a molecular explanation. It is no longer sufficient, now that the black box of vision has been opened, for an ‘evolutionary explanation’ of that power to invoke only the anatomical structures of whole eyes, as Darwin did in the 19th century and as most popularizers of evolution continue to do today. Anatomy is, quite simply, irrelevant. So is the fossil record. It does not matter whether or not the fossil record is consistent with evolutionary theory, any more than it mattered in physics that Newton’s theory was consistent with everyday experience. The fossil record has nothing to tell us about, say, whether or how the interactions of 11-cis-retinal with rhodopsin, transducin, and phosphodiesterase could have developed step-by-step. Neither do the patterns of biogeography matter, or of population genetics, or the explanations that evolutionary theory has given for rudimentary organs or species abundance.
https://www.discovery.org/a/54/
To understand novelty in evolution, we need to understand organisms down to their individual building blocks, down to the workings of their deepest components, for these are what undergo change.
Dr. Marc W. Kirschner: The Plausibility of Life: Resolving Darwin's Dilemma 2005
For a complete understanding of biological processes that define the intricate development of body architecture with striking precision, the orchestration of organismal development, cell and tissue shape, organization, and body form, it is necessary to understand as many integrative elements of biological systems as possible. Complex pattern formation involves numerous highly intricate biomolecular mechanisms that lead to the superb formation of tissue structures. That includes providing information that gives mechanical cues directing intra and extracellular shape changes and movements on the level of individual cells, but also tissue substratum as a whole. Answering the questions about how cells, tissues, and organisms masterfully develop and form, precedes the question IF evolutionary claims are compelling answers, explaining IF the evolutionary changes permit a purely blind primary macroevolutionary transition zone, morphogenesis of an entire organism moving and morph from one species to another on a first-degree speciation level, where novel features arise, like wings, eyes, ears, legs, arms, and so forth. The fact and truth are, that science is still far and away from having a complete answer to that question. But what we do know, permits to make informed conclusions.
Biodiversity and complex organismal architecture is explained by trillions of bits. Incredible amounts of data far beyond our imagination. Instructions, complex codified specifications, INFORMATION. Algorithms masterfully encoded in various genetic and sophisticated epigenetic languages and communication channels and networks. Neurotransmitters, through nanotubes between cells, communication through vesicles and amazingly, even light photons. Genes, but as well and especially various striking epigenetic signaling and bioelectric codes through various signaling networks provide cues to molecules and macromolecule complexes, and ingenious scaffold networks interpret and react in a variety of ways upon decoding and data processing of those instructions. Since signaling pathways work in an extraordinarily precise, in a synergetic integrated manner with the transcriptional regulatory network and complex short and long-range cross-talk between cells, these crucial instructions, crucial for advanced life forms, could not be the result of a random gradual increase of information. These superb information networks only operate and work in an integrated fashion, and had to be "born", and fully set up right from the beginning. Conveying codes, a system of rules to convert information, such as letters and words, into another form, and translation ciphers of one language to another are always sourced back to intelligent set-up. What we see in biochemistry is incredibly complex instructional codified information being stored through the genetic code ( codons) in a masterful information-storage molecule (DNA), encoded & transcribed ( RNA polymerase), sent (mRNA), and decoded & translated ( Ribosome), as well as epigenetic codes and languages, and several signaling pathways. The morphogenesis of organismal structure and shape is classified into two groups: The various instructional codes and languages using molecules that provide complex instructional cues of action based on information through signaling and secondly by force-generating molecules that are precisely directed through those signals, which are responsible for fantastic cell morphogenesis. Blueprints, instructional information, and master plans, which permit the striking autonomous self-organization and control of complex machines ( molecular machines) and exquisite factory parks ( cells) upon these are both always tracked back to an intelligent source which made both for purposeful, specific goals. That brings us unambiguously to intelligent design. To the origin by an intelligent designer.
What are the REAL mechanisms of biodiversity, replacing macroevolution?
https://www.youtube.com/watch?v=_IGrzrk6iBEre=youtu.be
Why Darwin's theory of evolution does not explain biodiversity
https://reasonandscience.catsboard.com/t2623-why-darwins-theory-of-evolution-does-not-explain-biodiversity
1. Biological sciences have come to discover in the last decades that major morphological innovation, development and body form are based on at least 47 different, but integrative mechanisms.
2. These varied mechanisms orchestrate gene expression, generate Cell types and patterns, perform various tasks essential to cell structure and development, are responsible for important tasks of organismal development, affect gene transcription, switch protein-coding genes on or off, determine the shape of the body, regulate genes, provide critical structural information and spatial coordinates for embryological development, influence the form of a developing organism and the arrangement of different cell types during embryological development, organize the axes, and act as chemical messengers for development
3. Neo-Darwinism and Modern Synthesis have proposed traditionally a gene-centric view, a scientific metabiological proposal going back to Darwin's " On the Origin of species ", where first natural selection was proposed as the mechanism of biodiversity, and later, gene variation defining how bodies are built and organized. Not even recently proposed alternatives, like the third way, neutral theory, inclusive fitness theory, Saltationism, Saltatory ontogeny, mutationism, Genetic drift, or combined theories, do full justice by taking into account all organizational physiological hierarchy and complexity which empirical science has come to discover.
4. Only a holistic view, namely structuralism and systems biology, takes into account all influences that form cell form and size, body development, and growth, providing adequate descriptions of the scientific evidence.
1. Genetic and epigenetic information directs the making of complex multicellular organisms, biodiversity, form, and architecture
2. This information is preprogrammed and prescribed to get a purposeful outcome. Each protein, metabolic pathway, organelle or system, each biomechanical structure and motion works based on principles that provide a specific function.
3. Preprogramming and prescribing a specific outcome is always the result of intention with foresight, able to instantiate a distant specific goal.
4. Foresight comes always from an intelligent agent. Therefore, biodiversity is the result of intelligent design, rather than unguided evolution.
They are apt to communicate, crosstalk, signal, regulate, govern, control, recruit, interpret, recognize, orchestrate, elaborate strategies, guide and so forth. All codes, blueprints, and languages are inventions by intelligence. Therefore, the genetic and epigenetic codes and signaling networks and the instructions to build cells and complex biological organisms were most likely created by an intelligent agency.
Biological cells and organisms are characterized by Irreducible complexity and hierarchical top-down systems interdependence, which is understood as irreducible functional systems complexity. And such is specified by genetic and epigenetic informational codes and signals used to set up and create front-loaded instructional blueprints, which direct how bodies are built, but also how life can self-correct, adapt to the environment and evolve. It is perfectly comparable to how a blueprint instructs to make machines and factories, and industries. Such things come undoubtedly from preexisting intelligence.
To understand the major trends in animal diversity and if the various kinds of morphology are due to evolution, we must first understand how animal form is generated. As science has unraveled, the make of body form, phenotype, and organismal architecture is due to several genetic and principally, epigenetic interlocked and interconnected mechanisms. The modern, extended evolutionary synthesis does not take into consideration all relevant factors. Structuralism proposes that complex structure emerges holistically from the dynamic interaction of all parts of an organism. It denies that biological complexity can be reduced to natural selection, gene drift and gene flow, and argues that pattern formation is driven principally by multilevel processes that involve various functional units, working in an interdependent manner, pre-programmed to respond to ecological and environmental cues and conditions, food resource availability, and development programs. Various genetic and epigenetic Codes, an integrated understanding of the structural and functional aspects of epigenetics and several signalling pathways, nuclear architecture during differentiation, chromatin organization, morphogenetic fields, amongst many other mechanisms.
The Gene regulation network
Epigenetics refers to heritable changes in gene expression that occur without alteration in DNA sequence. These changes may be induced spontaneously, induced by environmental factors or as a consequence of specific mutations. There are two primary and interconnected epigenetic mechanisms: DNA methylation and covalent modification of histones. 42 . In addition, it has become apparent that non-coding RNA is also intimately involved in this process. The different mechanisms that control epigenetic changes do not stand alone, and there are a clear interconnection and interdependency between:
Genes involved in Cell-Cell communication and transcriptional control are especially important for animal development
What are the genes that animals share with one another but not with other kingdoms of life? These would be expected to include genes required specifically for animal development but not needed for unicellular existence. Comparison of animal genomes with the genome of budding yeast—a unicellular eukaryote— suggests that three classes of genes are especially important for multicellular organization.
The first class includes genes that encode proteins used for cell–cell adhesion and cell signaling; hundreds of human genes encode signal proteins, cell-surface receptors, cell adhesion proteins, or ion channels that are either not present in yeast or present in much smaller numbers.
The second class includes genes encoding proteins that regulate transcription and chromatin structure: more than 1000 human genes encode transcription regulators, but only about 250 yeast genes do so. The development of animals is dominated by cell–cell interactions and by differential gene expression.
The third class of noncoding RNAs has a more uncertain status: it includes genes that encode microRNAs (miRNAs); there are at least 500 of these in humans. Along with the regulatory proteins, they play a significant part in controlling gene expression during animal development, but the full extent of their importance is still unclear.
Chromatin dance in the nucleus through extensile motors
Transcription and gene regulation Genome topology has emerged as a key player in all genome functions. Although a contribution of local genome looping in transcription has long been appreciated, recent observations have revealed the importance of long-range interactions, and genome-wide studies have uncovered the universal nature of such regulatory genome topology interactions in gene regulation. Several types of chromosomal interactions, either in the form of loops between sequences on the same chromosomes or interchromosomal interactions, have emerged as key mechanisms in gene regulation.
The nature of genome topology is very precise, and so its regulatory functions in gene expression and genome maintenance. The emerging picture is one of extensive self-enforcing feedback between activity and spatial organization of the genome, suggestive of a self-organizing and self-perpetuating system that uses epigenetic dynamics to regulate genome function in response to regulatory cues and to propagate cell-fate memory
Post-transcriptional modifications (PTMs) of histones affect gene transcription
According to the histone code hypothesis, the pattern of histone modification is recognized by proteins much like a language or code. One pattern of histone modification may attract proteins that inhibit transcription. Alternatively, a different combination of histone modifications may attract proteins, such as ATP-dependent chromatin-remodeling complexes, that promote gene transcription. In this way, the histone code plays a key role in accessing the information within the genomes of eukaryotic species. Post-translational modifications (PTMs) of histones provide a fine-tuned mechanism for regulating chromatin structure and dynamics. In addition to combinatorial PTMs that function together both synergistically and antagonistically, there is now an appreciation for PTM asymmetry within individual nucleosomes, novel types of PTMs with unique functions, nucleosomes bearing histone variants, and nuclear compartmentalization events that are all contributing to the final output of chromatin organization and function
The DNA methylation code and language
One of the best known epigenetic mechanisms is DNA methylation in which a small molecule (a methyl group) is added to the DNA macromolecule at particular locations. Like a barcode or marker, the methyl group indicates, for instance, which genes in the DNA are to be turned on. This DNA methylation is accomplished via the action of a protein machine that adds the methyl group at precisely the right location in the DNA strand. Methylation of DNA occurs at certain target sites along the DNA sequence where specific short DNA sequences appear. These sequences are found by special proteins as they move along the DNA. The special proteins search for these sequences and add a methyl group to the adenine base that appears within the sequence. The protein binds to the DNA, twists the helix so the adenine base rotates into a precisely shaped pocket in the protein, and the protein then facilitates the transfer of the methyl group from a short donor molecule for example to adenine.
Homeobox and Hox Genes
This family of related genes determines the shape of the body. It subdivides the embryo along the head-to-tail axis into fields of cells that eventually become limbs and other structures. Starting as a fertilized egg with a homogeneous appearance, an embryo made of skin, muscles, nerves and other tissues gradually arises through the division of cells. Long before most cells in the emerging body begin to specialize, however, a plan that designates major regions of the body-the head, the trunk, the tail and so on is established. This plan helps seemingly identical combinations of tissues arrange themselves into distinctly different anatomical structures, such as arms and legs. Individual genes mediate some of the developmental decisions involved in establishing the embryonic body plan.
" Junk DNA "
MicroRNAs--"Once Dismissed as Junk"--Confirmed To Have Important Gene Regulatory Function In 2008 Scientific American noted that microRNAs were "once dismissed as junk" and said the following:
Tiny snippets of the genome known as microRNA were long thought to be genomic refuse because they were transcribed from so-called "junk DNA," sections of the genome that do not carry information for making proteins responsible for various cellular functions. Evidence has been building since 1993, however, that microRNA is anything but genetic bric-a-brac. Quite the contrary, scientists say that it actually plays a crucial role in switching protein-coding genes on or off and regulating the amount of protein those genes produce.
Transposons and Retrotransposons
striking evidence has accumulated indicating that some proviral sequences and HERV proteins might even serve the needs of the host and are therefore under positive selection. The remarkable progress in the analysis of host genomes has brought to light the significant impact of HERVs and other retroelements on genetic variation, genome evolution, and gene regulation.
Centrosomes
Centrosomes play a central role in development: a frog egg can be induced to develop into a frog merely by injecting a sperm centrosome—no sperm DNA is needed. Another non-genetic factor involved in development is the membrane pattern of the egg cell.
Cytoskeletal arrays
The precise arrangement of microtubules in the cytoskeleton constitutes a form of critical structural information. neither the tubulin subunits nor the genes that produce them account for the differences in the shape of the microtubule arrays that distinguish different kinds of embryos and developmental pathways. Instead, the structure of the microtubule array itself is, once again, determined by the location and arrangement of its subunits, not the properties of the subunits themselves. Jonathan Wells explains it this way: “What matters in [embryological] development is the shape and location of microtubule arrays, and the shape and location of a microtubule array is not determined by its units.” Directed transport involves the cytoskeleton, but it also depends on spatially localized targets in the membrane that are in place before transport occurs. Developmental biologists have shown that these membrane patterns play a crucial role in the embryological development of fruit flies.
Signaling between cells orients the mitotic spindle
In multicellular animals, cell communication sometimes serves to orient the direction in which cells divide. Control of division orientation has been proposed to be critical for partitioning developmental determinants and for maintaining epithelial architecture. Surprisingly, there are few cases where we understand the mechanisms by which external cues, transmitted by intercellular signalling, specify the division orientation of animal cells. One would predict that cytosolic molecules or complexes exist that are capable of interpreting extrinsic cues, translating the positions of these cues into forces on microtubules of the mitotic spindle. In recent years, a key intracellular complex has been identified that is required for pulling forces on mitotic spindles in Drosophila, C. elegans and vertebrate systems. One member of this complex, a protein with tetratricopeptide repeat (TPR) and GoLoco (Gα-binding) domains, has been found localized in positions that coincide with the positions of spindle-orienting extracellular cues. Do TPR-GoLoco proteins function as conserved, spatially-regulated mediators of spindle orientation by intercellular signalling? Here, we review the relevant evidence among cases from diverse animal systems where this protein complex has been found to localize to specific cell-cell contacts and to be involved in orienting mitotic spindles.
Membrane targets
Preexisting membrane targets, already positioned on the inside surface of the egg cell, determine where these molecules will attach and how they will function. These membrane targets provide crucial information—spatial coordinates—for embryological development.
Ion Channels and Electromagnetic Fields
Experiments have shown that electromagnetic fields have “morphogenetic” effects—in other words, effects that influence the form of a developing organism. In particular, some experiments have shown that the targeted disturbance of these electric fields disrupts normal development in ways that suggest the fields are controlling morphogenesis.2 Artificially applied electric fields can induce and guide cell migration. There is also evidence that direct current can affect gene expression, meaning internally generated electric fields can provide spatial coordinates that guide embryogenesis.3 Although the ion channels that generate the fields consist of proteins that may be encoded by DNA (just as microtubules consist of subunits encoded by DNA), their pattern in the membrane is not. Thus, in addition to the information in DNA that encodes morphogenetic proteins, the spatial arrangement and distribution of these ion channels influence the development of the animal.
The Sugar Code
These sequence-specific information-rich structures influence the arrangement of different cell types during embryological development. Thus, some cell biologists now refer to the arrangements of sugar molecules as the “sugar code” and compare these sequences to the digitally encoded information stored in DNA. As biochemist Hans-Joachim Gabius notes, sugars provide a system with “high-density coding” that is “essential to allow cells to communicate efficiently and swiftly through complex surface interactions.” According to Gabius, “These [sugar] molecules surpass amino acids and nucleotides by far in information-storing capacity.” So the precisely arranged sugar molecules on the surface of cells clearly represent another source of information independent of that stored in DNA base sequences. These cascades are, along with the cell event itself, associated with the “coding information” on a cell surface, or, using another terminology, are realized due to an instruction for the cell from the morphogenetic field of an organism. The concrete signal transduction pathways connecting the "coding information" on a cell surface and the expression of the given sets of genes need to be elucidated.
Above and beyond: epigenetic information
genes alone do not determine the three-dimensional form and structure of an animal. Developmental biologists, in particular, are now discovering more and more ways that crucial information for building body plans is imparted by the form and structure of embryonic cells, including information from both the unfertilized and fertilized egg. DNA helps direct protein synthesis. Parts of the DNA molecule also help to regulate the timing and expression of genetic information and the synthesis of various proteins within cells. Yet once proteins are synthesized, they must be arranged into higher-level systems of proteins and structures. The three-dimensional structure or spatial architecture of embryonic cells plays important roles in determining body-plan formation during embryogenesis. Developmental biologists have identified several sources of epigenetic information in these cells.
Stephen C. Meyer, Darwin's doubt: Neo darwinism and the challenge of epigenetic information
These different sources of epigenetic information in embryonic cells pose an enormous challenge to the sufficiency of the neo-Darwinian mechanism. According to neo-Darwinism, new information, form, and structure arise from natural selection acting on random mutations arising at a very low level within the biological hierarchy—within the genetic text. Yet both body-plan formation during embryological development and major morphological innovation during the history of life depend upon a specificity of arrangement at a much higher level of the organizational hierarchy, a level that DNA alone does not determine. If DNA isn’t wholly responsible for the way an embryo develops— for body-plan morphogenesis—then DNA sequences can mutate indefinitely and still not produce a new body plan, regardless of the amount of time and the number of mutational trials available to the evolutionary process. Genetic mutations are simply the wrong tool for the job at hand. Even in a best-case scenario—one that ignores the immense improbability of generating new genes by mutation and selection—mutations in DNA sequence would merely produce new genetic information. But building a new body plan requires more than just genetic information. It requires both genetic and epigenetic information—information by definition that is not stored in DNA and thus cannot be generated by mutations to the DNA. It follows that the mechanism of natural selection acting on random mutations in DNA cannot by itself generate novel body plans, such as those that first arose in the Cambrian explosion.
" Junk DNA "
MicroRNAs--"Once Dismissed as Junk"--Confirmed To Have Important Gene Regulatory Function
In 2008 Scientific American noted that microRNAs were "once dismissed as junk" and said the following:
Tiny snippets of the genome known as microRNA were long thought to be genomic refuse because they were transcribed from so-called "junk DNA," sections of the genome that do not carry information for making proteins responsible for various cellular functions. Evidence has been building since 1993, however, that microRNA is anything but genetic bric-a-brac. Quite the contrary, scientists say that it actually plays a crucial role in switching protein-coding genes on or off and regulating the amount of protein those genes produce.
Transposons and Retrotransposons
striking evidence has accumulated indicating that some proviral sequences and HERV proteins might even serve the needs of the host and are therefore under positive selection. The remarkable progress in the analysis of host genomes has brought to light the significant impact of HERVs and other retroelements on genetic variation, genome evolution, and gene regulation.
Morphogen Gradients and Pattern Formation
Compartment boundaries are the sources of morphogens. Morphogens are signaling molecules that are produced from a localized source forming long-range concentration gradients that pattern a field of cells
Principal Meanings of Evolution in Biology Textbooks
What is fact :
1. Change over time; history of nature; any sequence of events in nature
2. Changes in the frequencies of alleles in the gene pool of a population
3. Limited common descent: the idea that particular groups of organisms have descended from
a common ancestor.
4. The mechanisms responsible for the change required to produce limited descent with modification; chiefly natural selection acting on random variations or mutations
What is not fact:
5. Universal common descent: the idea that all organisms have descended from a single common ancestor.
6. Blind watchmaker thesis: the idea that all organisms have descended from common ancestors through unguided, unintelligent, purposeless, material processes such as natural
selection acting on random variations or mutations; the idea that the Darwinian mechanism of natural selection acting on random variation, and other similarly naturalistic mechanisms, completely suffice to explain the origin of novel biological forms and the appearance of design in complex organisms.
1. Neo-Darwinism and the Modern Synthesis propose a gene-centric view, a scientific metabiological proposal going back to Darwin's landmark book " On the origin of species " in 1859, where first natural selection was proposed as the mechanism of biodiversity, and later, gene variation defining how bodies are built and organized.
2. Science researchers have discovered, that robust networks of interactions and biological function, major morphological innovation, development and body form are based on integrative mechanisms, the interplay of genes with the gene regulatory network, Transposons and Retrotransposons, so-called Junk DNA, splicing, and over a dozen epigenetic codes, Membrane targets and patterns, Cytoskeletal arrays, Centrosomes, Ion channels, Sugar molecules on the exterior of cells (the sugar code), that are not specified by nuclear DNA - that is, inheritance is not defined through DNA sequences alone.
3. Science is coming to recognize, that none of the recently proposed alternatives, like the third way, Saltationism, Saltatory ontogeny, mutationism, Genetic drift, or combined theories, do full justice by taking into account all organizational biophysiological hierarchy and complexity which empirical science has come to discover. As such, only a holistic view, namely structuralism, takes into consideration all influences that form cell form and size, body development and growth, doing justice to the scientific evidence.
4. Scrutinizing which causes ultimately respond for the complexity discovered in life is only satisfying, once the epistemologically flawed foundation of methodological naturalism is taken out of the box, and replaced by a new paradigm, where all possible mechanisms and causal influences are permitted to be scrutinized, investigated, and scientifically tested, including the interaction and creative force of an external intelligent, mental agency outside the known physical world, which through its transcendent power creates, forms and builds all physiobiological lifeforms in all its astounding diversity.
If the mechanisms as proposed by the modern extended evolutionary synthesis are insufficient, then it has to be figured out what OTHER mechanisms define body architecture, grow, differentiation, phenotype, and body form, and THEN it can be evaluated, how the origin of these mechanisms are best explained. As it comes out, the BIG contributor to explain life and its complexity is INFORMATION. Instructional, complex, specified, codified information that acts like a blueprint ( genes ), but also information that is PRE-PROGRAMMED and stored in epigenetic cell features on a structural level, which is pre-instructed to respond to environmental cues, development, and nutrition demands, and they are apt to communicate, crosstalk, signal, regulate, govern, control, recruit, interpret, recognize, orchestrate, elaborate strategies, guide and so forth.
1. Cells use sophisticated information transmission and amplification systems (signalling pathways), information interpretation, combination and selection ( the Gene regulatory network ) encoding and transcription ( DNA & RNA polymerase machines ) transmission (mRNA), and decoding ( Ribosome ) systems.
2. Setup of information transmission systems, aka. transmission, amplification, interpretation, combination, selection, encoding, transmission, and decoding are always a deliberate act of intelligence
3. The existence of the genetic information transmission system is best explained by the implementation of an intelligent designer.
Meyer, Darwins doubt, page 212:
According to neo-Darwinism, new information, form, and structure arise from natural selection acting on random mutations arising at a very low level within the biological hierarchy—within the genetic text. Yet both body-plan formation during embryological development and major morphological innovation during the history of life depend upon a specificity of arrangement at a much higher level of the organizational hierarchy, a level that DNA alone does not determine. If DNA isn’t wholly responsible for the way an embryo develops— for body-plan morphogenesis—then DNA sequences can mutate indefinitely and still not produce a new body plan, regardless of the amount of time and the number of mutational trials available to the evolutionary process. Genetic mutations are simply the wrong tool for the job at hand.

https://reasonandscience.catsboard.com/t2316-where-do-complex-organisms-come-from
1. http://www.jodkowski.pl/ke/Meanings2000.pdf
2.https://reasonandscience.catsboard.com/t1390-macroevolution#1982
3. https://royalsocietypublishing.org/doi/10.1098/rsif.2020.0154
4. Prinz, R. (2023). Biological Codes: A Field Guide for Code Hunters. Biological Theory.
Last edited by Otangelo on Thu Mar 21, 2024 8:33 am; edited 197 times in total