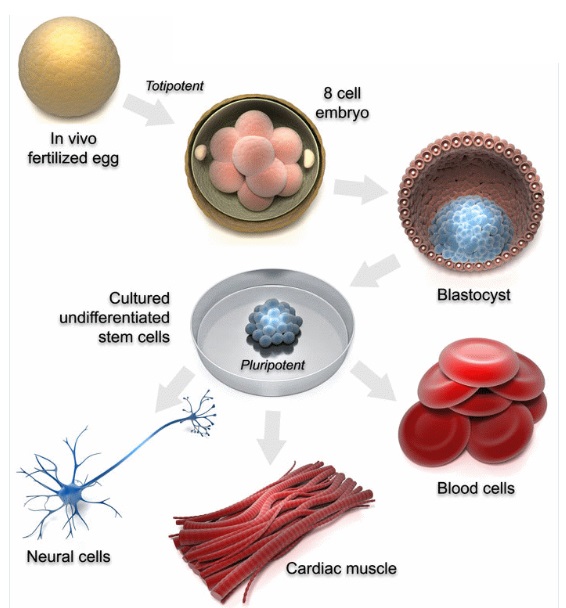
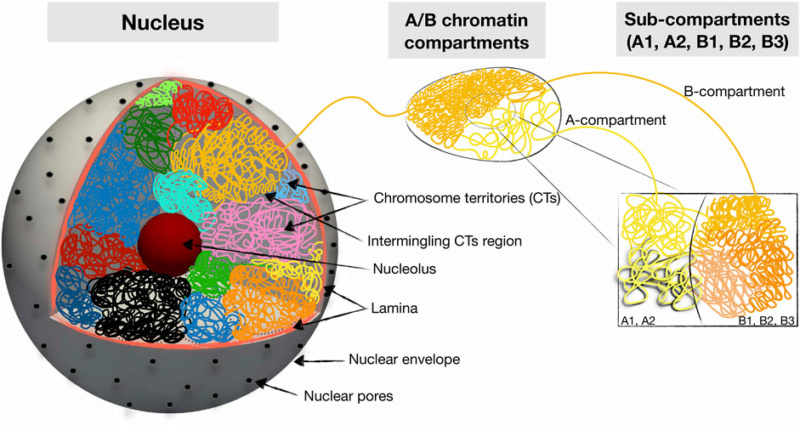
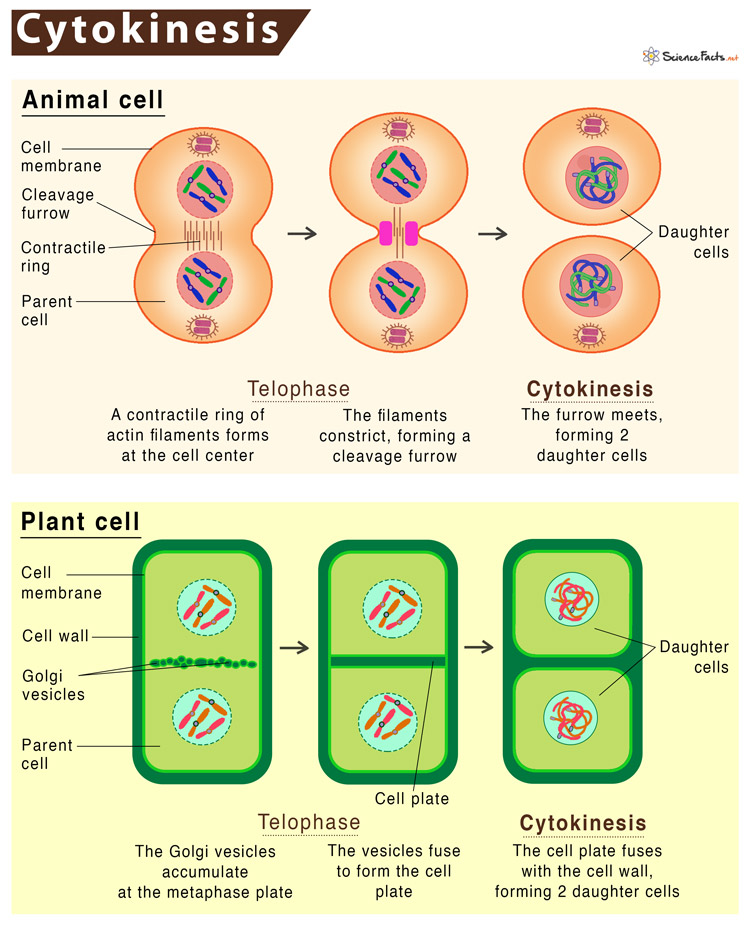
Developmental biology encompasses a wide range of processes that dictate the growth, form, and function of organisms from conception to maturity.
Cell Differentiation: This is where cells evolve and become specialized in their function.
Morphogenesis: The mechanism by which the structure of an organism develops.
Growth: Refers to the increase in cell number and size, allowing the organism to develop in size and complexity.
Developmental processes are foundational in shaping the life of organisms. They determine how cells renew and differentiate, ensuring that each organism is not only formed correctly but is also functionally adept. These processes have extensive implications in medicine, evolutionary biology, and agriculture. This extensive list represents a comprehensive overview of key developmental processes that are essential for the formation and function of an organism. The following list encompasses processes ranging from the molecular to the organ level, each vital for the proper development, structure, and function of an organism. These processes, often interlinked, collectively orchestrate the intricate dance of development from a single cell to a multicellular organism.
The dichotomy between structural and regulatory codes is a suitable tool to start the systematic study of codes. Barbieri assumes the existence of three types of organic codes, namely manufacturing, signaling, and regulatory codes.
Codes are pervasive in nature, woven into the fabric of life, ensuring that organisms function, communicate, and adapt. While the word "code" might immediately conjure images of binary digits or cryptographic sequences, the biological realm showcases its own elegant systems of information storage and processing. Let's unravel these intricate codes.
Manufacturing Codes: At the heart of life lies a manufacturing system, a protocol that defines how the blueprints of life are read and realized. Picture a vast assembly line where raw materials are meticulously transformed into complex products. This is what the manufacturing code oversees. It's a set of instructions determining how basic molecular building blocks are assembled into the machinery that powers life. Whether it's the synthesis of proteins vital for structure and function or the replication of the genetic material itself, this code ensures the flawless production of life's necessities.
Signaling Codes: Imagine a vast network where information flows seamlessly, messages sent and received with precision. This is the realm of signaling codes. Every organism, no matter how simple or complex, exists in a dynamic environment. To thrive, it must sense and respond to myriad signals, be they external cues or internal messages. The signaling code encompasses the myriad ways cells communicate, both with each other and with their surroundings, ensuring harmony and coordination in the face of constant change.
Regulatory Codes: Regulatory codes act as the grand orchestrators, ensuring that the myriad processes within a cell or organism are coordinated in time and space. Think of them as conductors of a grand symphony, ensuring each instrument plays its part at the right moment. They determine when a gene should be activated or silenced, when a cell should grow or pause, and how resources are allocated. The regulatory code ensures that each process dovetails seamlessly with others, fostering a harmonious interplay that sustains life.
Epigenetic Codes: While genetics lays down the foundational script of life, epigenetics offers a layer of finesse, allowing organisms to modulate their genetic instructions based on experience and environment. It's akin to annotations on a manuscript, guiding how the original script is to be interpreted under different circumstances. Without altering the fundamental script, the epigenetic code offers a dynamic interface, allowing organisms to adapt and respond with a level of plasticity that is both astounding and essential.
Together, these codes offer a glimpse into the intricate ballet of life, a dance of molecules choreographed with precision and purpose. Delving into each code, we uncover the myriad ways nature encodes, processes, and utilizes information, showcasing the elegance and complexity inherent in the living world. As we continue our exploration, the wonder of life's codes becomes ever more evident, reminding us of the marvels that underpin our existence.
First, I will individual questions specifically to each of them will be answered, and in the following, I will ask these questions related to each of them:
The appearance of X (1-47) in the evolutionary timeline
De Novo Genetic Information necessary to instantiate X (1-47)
Manufacturing codes and languages that would have to emerge and be employed to instantiate X (1-47)
Epigenetic Regulatory Mechanisms necessary to be instantiated for X (1-47)
Signaling Pathways necessary to create, and maintain X (1-47)
Regulatory codes necessary for maintenance and operation of X (1-47)
Is there scientific evidence supporting the idea that X (1-49) was brought about by the process of evolution?
Irreducibility and Interdependence of the systems to instantiate and operate X (1-47)
Once is instantiated and operational, what other intra and extracellular systems is X (1-47) interdependent with?
Elucidating these questions will provide the reader a comprehensive understanding of the intricacies involved in the formation, operation, and emergence of the most relevant biological processes or components (1-47) that convey biological form and architecture.
Appearance of (1-47) in the evolutionary timeline: This will map out when and in which organisms these processes or components supposedly first appeared according to the evolutionary timeline. This historical context can provide insights into the environmental, genetic, or ecological factors that is claimed to have driven their emergence.
De Novo Genetic Information necessary to instantiate (1-47): Understanding the new genetic instructions required for these processes will shed light on the complexity and uniqueness of their emergence and the kind of required innovations they represent.
Manufacturing codes and languages that would have to emerge and be employed to instantiate 1-47: This looks into the specialized 'codes' or mechanisms that these processes require. It emphasizes the complexity of biological systems and how they can't function without a precise set of instructions.
Epigenetic Regulatory Mechanisms necessary to be instantiated for X (1-47): Exploring the non-DNA sequence-based regulations emphasizes the layers of control and fine-tuning in biological systems, showing that it's not just the genes themselves but also their regulation that's crucial.
Signaling Pathways necessary to create, and maintain X (1-47): This emphasizes the interconnectedness of biological processes. Signaling pathways are like communication networks, ensuring that processes are coordinated.
Regulatory codes necessary for maintenance and operation of X (1-47): Delves into the continuous requirements for these processes to function properly over an organism's lifespan, showing the persistence of complexity beyond just the initial emergence.
Is there scientific evidence supporting the idea that X (1-47) was brought about by the process of evolution? This question seeks to tie the aforementioned complexities back to the current scientific understanding, emphasizing empirical grounding and challenging or affirming existing paradigms.
Irreducibility and Interdependence of the systems to instantiate and operate X (1-47): Exploring these concepts touches upon the evidence that some systems are so complex that they seem to require all their parts to be present and functioning from the outset, posing questions about incremental evolutionary paths.
Once X is instantiated and operational, what other intra and extracellular systems is X (1-47) interdependent with? This emphasizes the holistic nature of biology, where processes are not isolated but are parts of larger networks, influencing and being influenced by myriad other systems.
Following are the key developmental processes shaping organismal form and function with a brief description of each process, The list is in alphabetic order
1. Angiogenesis and Vasculogenesis: Formation of new blood vessels from pre-existing ones (angiogenesis) and de novo vessel formation (vasculogenesis).
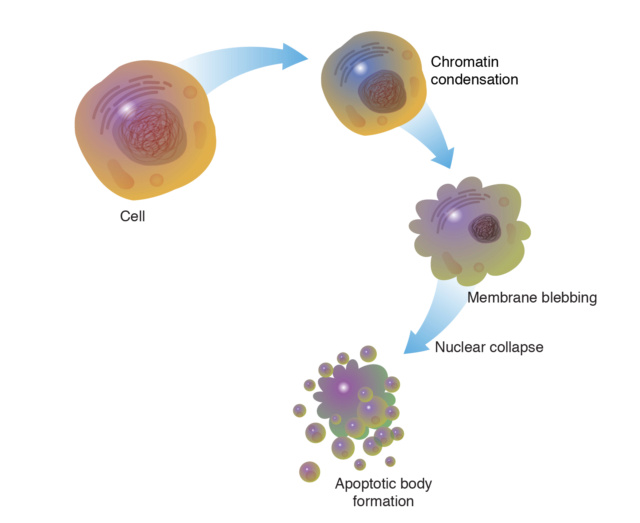
2. Apoptosis: Programmed cell death essential for removing unwanted cells.
3. Cell-Cycle Regulation: Controls the progression of cells through the stages of growth and division.
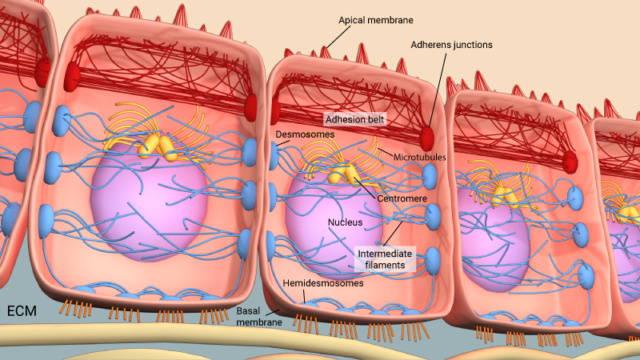
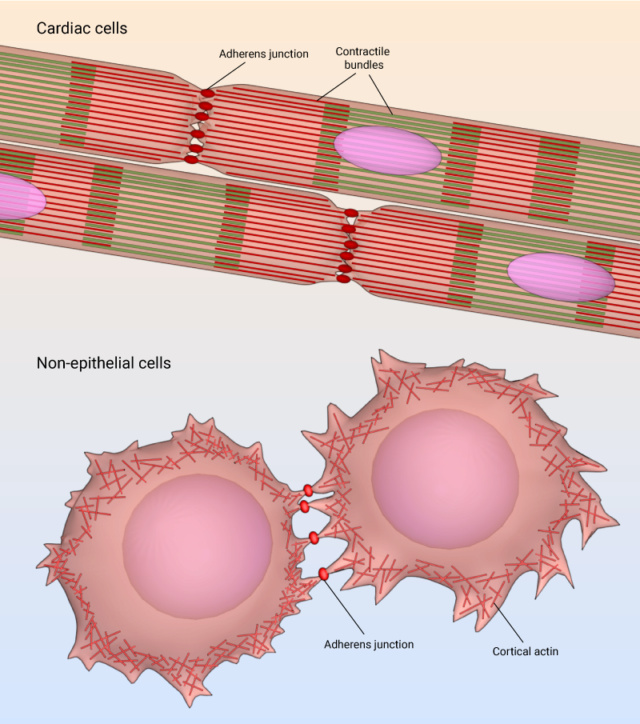
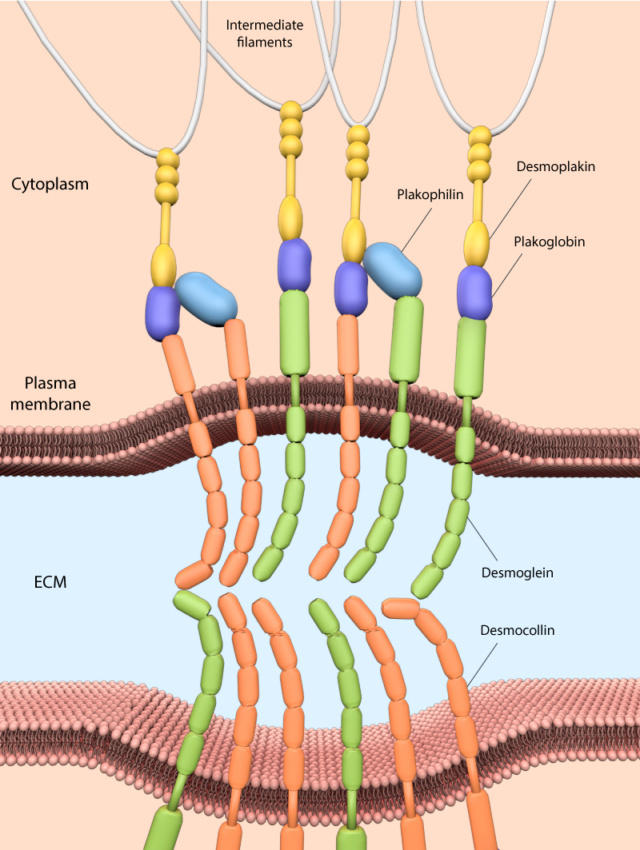
4. Cell-cell adhesion and the ECM: Refers to how cells stick to each other and to the extracellular matrix, essential for tissue formation.
5. Cell-Cell Communication: Cells communicate to coordinate their actions.
6. Cell Fate Determination and Lineage Specification (Cell differentiation): Process by which cells become specialized in their function.
7. Cell Migration and Chemotaxis: Movement of cells, guided by certain chemical gradients.
8. Cell Polarity and Asymmetry: Defines distinct cellular 'sides' or 'ends', crucial for many cell functions.
9. Cellular Pluripotency: Cells can give rise to multiple cell types.
10. Cellular Senescence: State of stable cell cycle arrest.
11. Centrosomes: Organize microtubules and provide structure to cells.
12. Chromatin Dynamics: How DNA and proteins are organized in the nucleus.
13. Cytokinesis: Physical process of cell division.
14. Cytoskeletal Arrays: Framework of the cell, involved in cell shape, movement, and division.
15. DNA Methylation: Addition of methyl groups to DNA, often involved in gene silencing.
16. Egg-Polarity Genes: Determine the axes of the egg and subsequently the organism.
17. Epigenetic Codes: Changes in gene function without changing DNA sequence.
18. Gene Regulation Network: Interactions between genes, controlling when and where genes are expressed.
19. Germ Cell Formation and Migration: Development and movement of reproductive cells.
20. Germ Layer Formation (Gastrulation): Development of primary tissue layers in embryos.
21. Histone PTMs: Modifications to histone proteins affecting DNA accessibility.
22. Homeobox and Hox Genes: Control the body plan of an embryo along the head-tail axis.
23. Hormones: Chemical messengers coordinating bodily functions.
24. Immune System Development: Formation and maturation of immune cells.
25. Ion Channels and Electromagnetic Fields: Channels allowing ions to flow in/out of cells; electromagnetic fields can influence development.
26. Membrane Targets: Processes focusing on cell membrane components.
27. MicroRNA Regulation: Small RNAs regulating gene expression post-transcriptionally.
28. Morphogen Gradients: Concentration gradients of substances determining tissue development.
29. Neural Crest Cells Migration: Movement of cells contributing to diverse structures, including peripheral nerves.
30. Neural plate folding and convergence: Formation of the neural tube in early development.
31. Neuronal Pruning and Synaptogenesis: Refinement of neural connections and formation of synapses.
32. Neurulation and Neural Tube Formation: Development of the neural tube, precursor to the CNS.
33. Noncoding RNA from Junk DNA: RNA molecules not coding for protein but having various functions.
34. Oogenesis: Egg cell (oocyte) formation.
35. Oocyte Maturation and Fertilization: Development of mature egg and its fusion with sperm.
36. Pattern Formation: Processes determining organized spatial arrangement of cells/tissues.
37. Photoreceptor development: Formation of cells detecting light in the eye.
38. Regional specification: Defining distinct regions within developing tissues.
39. Segmentation and Somitogenesis: Division of body into segments and formation of somites in embryos.
40. Signaling Pathways: Series of molecular events relaying extracellular signals to intracellular targets.
41. Spatiotemporal gene expression: Time and place-specific gene expression.
42. Spermatogenesis: The process of sperm cell formation and maturation.
43. Stem Cell Regulation and Differentiation: Control of stem cell fate and their development into specialized cells.
44. Symbiotic Relationships and Microbiota Influence: Interactions with microbial partners and their influence on host development.
45. Syncytium formation: Multinucleated cell formation, especially important in muscle tissues.
46. Transposons and Retrotransposons: Mobile genetic elements, sometimes influencing gene regulation.
47. Tissue Induction and Organogenesis: Formation of tissues and organs from undifferentiated cells.
The processes and systems listed are essential to the complex orchestration of development and physiology in multicellular organisms. Many of these are interwoven and interdependent to ensure accurate and timely development, function, and maintenance.
Angiogenesis and Vasculogenesis are closely linked to Tissue Induction and Organogenesis. As tissues and organs form, they require a blood supply for nutrient and oxygen delivery.
Apoptosis works in tandem with Cell-Cycle Regulation. As cells progress through the cycle, faulty or unwanted cells undergo programmed death to maintain tissue homeostasis.
Cell-Cell Adhesion and the ECM are fundamental for Cell Migration and Chemotaxis, allowing cells to navigate the 3D environment of the body.
Cell Fate Determination and Lineage Specification are influenced by Cell-Cell Communication and Morphogen Gradients, which provide cues for cells to differentiate into specific lineages.
Cell Polarity and Asymmetry are critical for processes like Cytokinesis and work closely with the Cytoskeletal Arrays to ensure accurate cell division.
Cellular Pluripotency ties into Stem Cell Regulation and Differentiation, with pluripotent cells being a subset of stem cells that can give rise to almost all cell types.
Chromatin Dynamics, particularly Histone PTMs and DNA Methylation, play roles in Epigenetic Codes and influence Gene Regulation Network.
Egg-Polarity Genes are involved in Oogenesis and also influence the Oocyte Maturation and Fertilization processes.
Homeobox and Hox Genes work in synchrony with Pattern Formation and Segmentation and Somitogenesis to define body plans.
Hormones can impact various processes, including Immune System Development and Oogenesis.
MicroRNA Regulation, Noncoding RNA from Junk DNA, and Transposons and Retrotransposons provide additional layers of post-transcriptional control on the Gene Regulation Network.
Neural Crest Cells Migration is a part of Neurulation and Neural Tube Formation and directly affects Neural plate folding and convergence.
Neuronal Pruning and Synaptogenesis are essential aspects of Photoreceptor development and general neural development.
Spermatogenesis and Oogenesis interplay during Oocyte Maturation and Fertilization to initiate the next generation's development.
Signaling Pathways are pervasive and influence many of the aforementioned processes, from Cell-Cycle Regulation to Pattern Formation to Stem Cell Regulation and Differentiation.
Symbiotic Relationships and Microbiota Influence can indirectly impact processes like Immune System Development.
It's worth noting that this list only scratches the surface. The interconnectedness of these systems is immensely complex, with each one potentially influencing or being influenced by multiple others. They collectively underscore the intricacy and delicate choreography inherent to biology.
In biology, at least 47 complex systems, and over 230 structural and regulatory codes, genetic and epigenetic languages play key roles in developmental processes shaping organismal form and function. They are so complex and intertwined, that they seem to require all their parts to be present and functioning from the outset, posing big question marks about incremental evolutionary paths being an adequate explanation of their origins. In Biology, it is not only the complex biomolecular machines, systems, and languages that are irreducibly complex, and work in an interdependent manner together, but once instantiated, they are often part of a larger, higher-order system. Irreducibly complex and interdependent systems are often required to convey and produce a complex function-bearing product, like a machine. Often, that machine alone bears no function on its own, unless it is interconnected with other machines to convey a final higher-order system with a specific function. An example is a car, that has an engine, which is interconnected and interdependent with the Transmission System, Exhaust System, cooling System, etc.
Premise 1. Life is embroidered with a profound amalgamation of genetic and epigenetic codes. With at least 33 distinct genetic variations and over 230 intertwined manufacturing, signaling, and operational codes, together with hundreds, and in prokaryotes probably thousands of intricate signaling networks, these codes collectively choreograph the exquisite dance of multicellular organisms, sculpting the vast biodiversity, nuanced form, and majestic architecture we observe.
Premise 2. This monumental wealth of information isn't random. It's meticulously orchestrated, often resembling the digital semiotic languages characterized by syntax, semantics, and pragmatics. Every protein, each metabolic trajectory, and every biomechanical construct operates with precision, abiding by principles finely tuned for specific functions.
Premise 3. While the foundations of life are tangible, the essence of information transcends the physical. It's conceptual, operating in realms beyond the reach of spontaneous and aimless physical phenomena. Arguing that such processes can birth semiotic codes is akin to believing a rainbow might pen a sonnet or winds could draft architectural wonders. The directive, purposeful nature inherent in biological programming suggests intention, foresight, and specific goals.
Conclusion: The staggering complexity and deliberate design seen in organismal architecture and biodiversity beckon consideration beyond mere chance. It suggests that these wonders are not just products of random evolution but the handiwork of a deliberate and intelligent design.
Interdependence between Intrinsic and Extrinsic Irreducibly Complex Systems
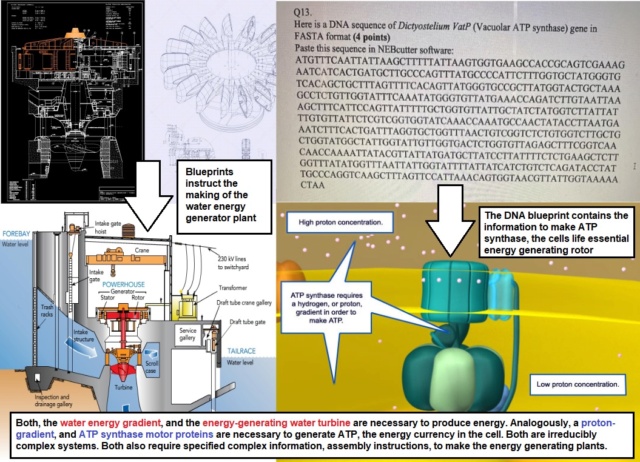
The concept of irreducible complexity posits that certain systems are so intricate in their interdependent components that removing any one of them would cause the system to cease functioning. This principle can be applied to both intrinsic components (those that are an inherent part of the system) and extrinsic components (external factors or conditions that the system relies upon). The principle of interdependence is evident in many systems, both artificial and biological. Let me give an example, examining and comparing both hydroelectric turbines and ATP synthase ( molecular turbines, that generate ATP, the energy currency in the cell:
Hydroelectric energy production
The hydroelectric turbine is intrinsically irreducibly complex
As water flows over the blades of a turbine, it causes the turbine to turn, converting the water's kinetic energy into mechanical energy. It requires several intrinsic components that make the turbine functional:
Turbine Blades (Runner): These are the components that capture the kinetic energy of water. They are designed in specific shapes and configurations to optimize the conversion of water's kinetic energy into rotational mechanical energy. Without the blades, water would simply flow through without generating rotation.
Shaft: The shaft is connected to the turbine blades and translates the rotational movement of the blades into the generator. Without the shaft, the rotation of the blades couldn't be transferred.
Generator: This is where the mechanical energy (from the rotating shaft) is converted into electrical energy. The generator contains magnets and coils of wire. As the shaft rotates, it induces a flow of electric current in the wires.
Wicket Gates: These are adjustable gates that control the flow of water onto the turbine blades. Without effective wicket gates, the turbine could be overwhelmed by too much water or underutilized with too little.
But the turbine alone will bear no function. It requires a set of extrinsic parts, that together, in a joint venture, will achieve the final goal, which is to generate usable energy. These parts are:
Extrinsic irreducible complexity
Dam: A dam holds back water, creating a reservoir or a lake. This stored water has potential energy due to its height. Without the dam, there's no potential energy from water height.
Water: The moving water's kinetic energy, or the stored potential energy in the dam's reservoir, is what gets converted into mechanical energy by the turbines. Without water, there's no kinetic energy.
Penstock: This is a conduit that brings water from the reservoir to the turbine. It plays a role in regulating the flow and pressure of the water hitting the turbine blades. Without the penstock, there's no controlled flow of water.
Generator: The turbine is connected to a generator. As the turbine spins, so does the generator, converting mechanical energy into electrical energy. Without the generator, there is electrical energy.
ATP energy production in the cell
ATP synthase is intrinsically irreducibly complex
ATP synthase is an intricate molecular machine that plays a pivotal role in cellular respiration, generating ATP (adenosine triphosphate) from ADP (adenosine diphosphate) and inorganic phosphate. It harnesses the energy stored in a proton gradient, usually across the inner mitochondrial membrane in eukaryotes, to catalyze this reaction. For this complex enzyme to function, several critical components must be in place.
F1 Subunit (Catalytic Core)
α and β subunits: These subunits form the catalytic core where ATP synthesis or hydrolysis takes place. The β subunit is where ATP is synthesized from ADP and inorganic phosphate.
γ, δ, and ε subunits: These form the central stalk that rotates within the α/β core. The rotation of the γ subunit drives the conformational changes in the β subunit required for ATP synthesis.
Fo Subunit (Proton Channel)
c-Ring (c subunits): This ring is in the membrane and rotates as protons flow through the Fo portion of ATP synthase. The rotation of the c-ring is directly linked to the rotation of the γ subunit in the F1 portion.
a Subunit: This subunit forms a channel allowing protons to flow through the Fo complex. It interacts with the c-ring, enabling the translocation of protons to drive the rotation of the c-ring.
b and δ subunits: These form a peripheral stalk, holding the α/β core stationary while the central stalk and c-ring rotate.
Stator (Peripheral Stalk)
This part of the complex ensures that while the central stalk and c-ring rotate, the catalytic α/β core remains stationary. It is vital for the enzyme's ability to synthesize ATP efficiently.
Proton Channel
The Fo portion, specifically the interaction between the a and c subunits, allows for the passage of protons. This flow of protons is what drives the rotation of the c-ring and, subsequently, the γ subunit inside the F1 portion.
For ATP synthase to function effectively, all of these components must be present and interact in a coordinated manner. The absence or malfunction of any of these parts would disrupt the enzyme's ability to synthesize ATP, making ATP synthase a prime example of a molecular system exhibiting irreducible complexity.
Extrinsic irreducible complexity
ATP Synthase Enzyme: This complex enzyme has a rotating component. As protons flow through the enzyme, they cause this component to rotate.
Proton Gradient: ATP synthase operates in cell membranes, especially the inner mitochondrial membrane. Here, there's a gradient of protons (H+ ions) across the membrane, known as the proton motive force.
Flow of Protons: As protons flow back across the membrane, they pass through ATP synthase.
Just like a dam creates potential energy in water for turbines, the proton gradient sets up potential energy for ATP synthase. The flow of protons through ATP synthase can be likened to the flow of water through turbines. Without the proton gradient, ATP synthase wouldn't function, just as without water, a turbine won't spin. Both systems exemplify how specific conditions and components are crucial for energy conversion processes. The structures, be it a dam or a cellular membrane, and the flow (of water or protons) are not just beneficial but essential for the proper functioning of the respective systems. This interconnectedness underscores the intricacy of both engineered and biological systems and emphasizes the importance of each component in the overall process. While intrinsic irreducible complexity deals with the essentiality of components within a system, extrinsic irreducible complexity pertains to the vital external conditions or components the system relies upon. Both concepts highlight the exquisite level of coordination and specialization present in biological systems, emphasizing their delicate balance and interdependence.
Evidence of Design in Irreducibility and Interdependence
Both hydroelectric turbines and ATP synthase exemplify systems that wouldn't function without their respective intrinsic and extrinsic components. The seamless interplay between these components suggests a high level of precision and specificity. Just as an engineer carefully calibrates the components of a hydroelectric turbine to maximize efficiency, the components of ATP synthase are finely tuned to optimize ATP production. Each part has a unique and specific function, which, if altered, can compromise the system's overall effectiveness. In the hydroelectric turbine, the wicket gates control water flow, while in ATP synthase, the proton gradient drives rotation. These mechanisms don't just randomly occur; they reflect a level of intelligence in their design and function. The interdependence of intrinsic and extrinsic components in both systems indicates that for these systems to function, multiple conditions must be met simultaneously. For instance, ATP synthase would be pointless without a proton gradient, and similarly, a hydroelectric turbine would be of no use without water flow. The existence of both components and conditions together speaks to foresight in their design. Just as a watch requires all its parts to tell time or a car needs every component to drive, the hydroelectric turbine and ATP synthase showcase the principle of interdependence. Such interdependence is a hallmark of design, where every component and condition is essential for the system's function. While science continues to explore and uncover the intricacies of natural systems, the principles of intrinsic and extrinsic irreducible complexity, coupled with the interdependence of these systems, lend compelling support to the idea of intentional design using advanced engineering. Such meticulous calibration and specificity, mechanisms to ensure optimal performance, and simultaneous occurrence and harmonious integration hint both at foreplanning and design.
Major Premise: Systems that exhibit both intrinsic and extrinsic irreducible complexity, with interdependence among their parts, demonstrate characteristics that are typically associated with deliberate design and engineering.
Minor Premise: ATP synthase and hydroelectric turbines are systems that exhibit both intrinsic and extrinsic irreducible complexity with a high degree of interdependence among their parts.
Conclusion: Therefore, ATP synthase and hydroelectric turbines demonstrate characteristics that are typically associated with deliberate design and engineering.
Manufacturing, signaling, and regulatory codes
The concept of "organic codes" provides a framework for understanding the many ways biological systems store, transmit and interpret information. Organic codes can be broadly categorized into manufacturing, signaling, and regulatory codes.
R.Prinz (2023): Barbieri assumes the existence of three types of organic codes, namely manufacturing, signaling, and regulatory codes (Barbieri 2015) 1
Manufacturing Codes
These are direct transcription-translation systems where a sequence of units in a polymer (like DNA or RNA) is converted into another sequence in a different polymer (like protein).
The canonical example is the genetic code, where triplets of nucleotides (codons) in mRNA are translated into specific amino acids in proteins.
It's a 'manufacturing' process because a physical entity (a protein) is being produced based on the information contained in another entity (the mRNA).
Signaling Codes
These codes are involved in the transmission of signals between cells or within cells. The codes here translate specific molecular signals into cellular responses. This translation often happens through signaling pathways.
Examples include hormone-receptor interactions. When a hormone binds to its specific receptor on a cell, it triggers a series of intracellular events, leading to a particular cellular response.
Regulatory Codes
These codes are responsible for controlling and coordinating the activities of manufacturing and signaling codes. An example would be the binding of transcription factors to DNA. The sequence specificity of this binding determines whether a particular gene will be turned on or off. Regulatory codes often interface with feedback mechanisms to adjust the activities of cells and maintain homeostasis.
Where do epigenetic codes fit in?
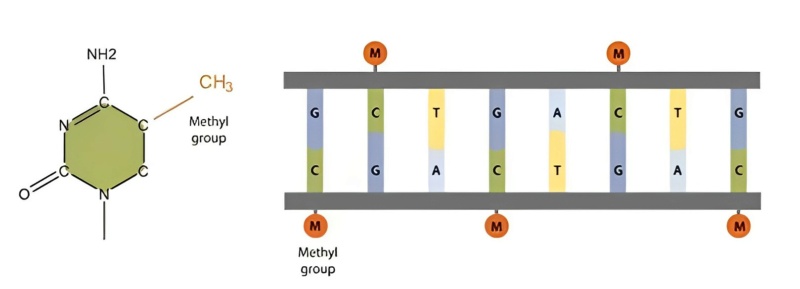
Epigenetic codes deal with heritable changes in gene function that don't involve changes in the DNA sequence itself. Instead, they involve chemical modifications to the DNA or to histone proteins with which DNA is associated. These modifications can influence gene expression by changing the structure of chromatin – the complex of DNA and proteins – and by regulating the accessibility of genes to the cellular machinery. Epigenetic codes primarily fall under regulatory codes. This is because epigenetic modifications like DNA methylation, histone acetylation, and histone methylation play crucial roles in regulating gene expression. For example, methylated DNA often corresponds to gene silencing, while acetylated histones usually correlate with gene activation. However, there's also a signaling aspect to epigenetics. Certain stimuli or signals can lead to changes in the epigenetic landscape of a cell. For instance, external stressors or developmental cues might trigger a cascade of intracellular events leading to specific epigenetic modifications. In this context, epigenetic codes can also interface with signaling codes. The categorization of organic codes into manufacturing, signaling, and regulatory codes provides a structured way to understand the myriad informational processes in biology. Epigenetic codes, given their role in regulating gene expression in response to both intrinsic and extrinsic signals, predominantly belong to the regulatory category but also intersect with signaling codes.
1. Angiogenesis and Vasculogenesis
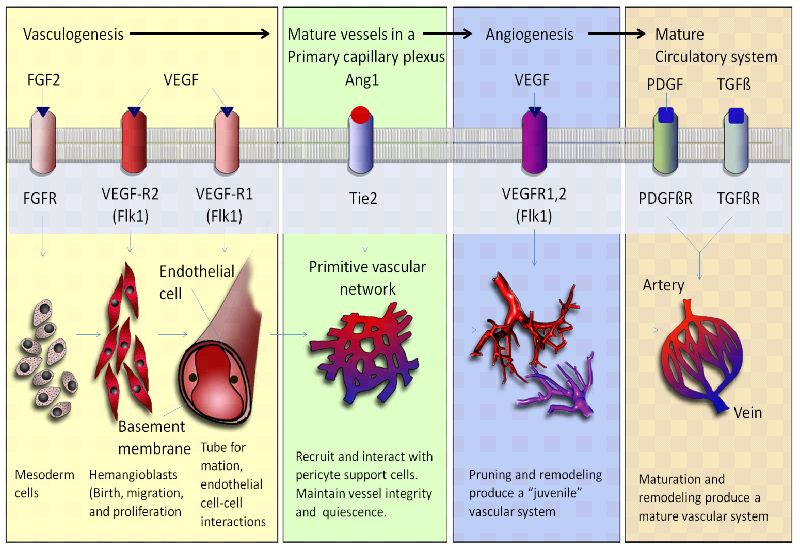
Angiogenesis and vasculogenesis are essential processes involved in the formation and maintenance of blood vessels within biological systems. Angiogenesis refers to the formation of new blood vessels from preexisting ones. It plays a crucial role in various biological contexts, such as wound healing, tissue regeneration, and development. Angiogenesis produces a network of blood vessels that supply nutrients and oxygen to tissues, remove waste products, and facilitate the exchange of molecules between the bloodstream and surrounding cells. This process is particularly important in embryonic development, tissue repair, and growth, as well as in conditions like cancer where new blood vessels support tumor growth and metastasis. Vasculogenesis is the process by which new blood vessels are formed de novo from endothelial progenitor cells. It is particularly significant during embryonic development when the cardiovascular system is initially established. Vasculogenesis contributes to the formation of the primary vascular plexus, which serves as a scaffold for further vascular remodeling and the eventual development of a mature vascular network. Defects in vasculogenesis can lead to severe developmental abnormalities. Angiogenesis and vasculogenesis are fundamental for the survival and function of complex multicellular organisms. They enable efficient transport of nutrients, oxygen, hormones, and immune cells throughout the body. These processes are vital for tissue growth, repair, and regeneration, as well as for maintaining proper physiological functions. Additionally, angiogenesis plays a role in various diseases, such as cancer, where excessive or aberrant blood vessel formation supports tumor growth. Understanding the molecular mechanisms underlying angiogenesis and vasculogenesis has implications for developing therapeutic strategies for conditions involving abnormal blood vessel formation or inadequate blood supply to tissues.
How do angiogenesis and vasculogenesis contribute to the establishment of blood vessel networks during embryonic development?
Angiogenesis and vasculogenesis are essential processes that contribute to the establishment of blood vessel networks during embryonic development. These processes involve the formation, expansion, and remodeling of blood vessels, which are crucial for supplying nutrients, oxygen, and other essential molecules to developing tissues and organs. Here's how angiogenesis and vasculogenesis work together to create functional vascular systems:
Vasculogenesis
Formation of Blood Islands: Vasculogenesis begins with the formation of blood islands, which are clusters of angioblasts (precursor cells) that differentiate into endothelial cells, the building blocks of blood vessels.
Angioblast Migration and Aggregation: Angioblasts migrate to specific areas in the developing embryo, guided by chemical signals and gradients. They aggregate to form endothelial cords or tubes, which serve as the initial structures of blood vessels.
Vascular Lumen Formation: The endothelial cords undergo lumenization, during which they develop a central channel, or lumen. This lumen becomes the pathway for blood flow.
Hemangioblast Differentiation: Some angioblasts differentiate into hemangioblasts, which give rise to both endothelial cells and blood cells. This connection between blood vessel formation and blood cell production is crucial for the functional development of the circulatory system.
Angiogenesis
Sprouting Angiogenesis: In regions where tissues require increased blood supply, endothelial cells from existing vessels start to sprout out in response to pro-angiogenic signals, such as growth factors like VEGF (vascular endothelial growth factor).
Migration and Proliferation: Sprouting endothelial cells migrate towards the source of pro-angiogenic signals and proliferate to form new endothelial sprouts.
Lumen Formation: Similar to vasculogenesis, the endothelial sprouts undergo lumenization to form functional tubular structures.
Anastomosis: The newly formed sprouts elongate and connect with adjacent sprouts, leading to the establishment of interconnected networks of blood vessels. This anastomosis process creates a functional circulation system.
Stabilization: Pericytes and smooth muscle cells are recruited to the developing vessels to provide structural support and stability to the new blood vessels.
Appearance of angiogenesis and vasculogenesis in the evolutionary timeline
The appearance of angiogenesis and vasculogenesis in the evolutionary timeline is a complex topic that involves speculation and ongoing research. While the exact timing is not definitively known, scientists have proposed hypotheses about when these processes might have emerged based on comparative studies and the fossil record. Angiogenesis and vasculogenesis are fundamental processes for the development and maintenance of blood vessels in organisms. Angiogenesis involves the formation of new blood vessels from preexisting ones, while vasculogenesis refers to the de novo formation of blood vessels from endothelial precursor cells.
Early Life Forms (Prokaryotes): These simple organisms did not possess complex vascular systems or mechanisms like angiogenesis and vasculogenesis. Their nutrient exchange and waste removal were likely facilitated by direct diffusion through cell membranes due to their small size and relatively simple structure.
Simple Eukaryotes: As organisms supposedly evolved into more complex eukaryotic forms, some multicellular organisms would have started to develop basic mechanisms to transport nutrients and waste products. However, true vascular systems had not yet emerged, and mechanisms like diffusion and simple tissue organization served these early organisms.
Invertebrates: The appearance of more complex invertebrates would have marked the transition toward more developed circulatory systems. Many invertebrates have open circulatory systems where blood is pumped into body cavities, allowing for nutrient exchange. These systems would have involved rudimentary precursor processes that could be considered primitive forms of angiogenesis.
Vertebrates: The emergence of vertebrates would have brought about more sophisticated circulatory systems, including closed systems with dedicated blood vessels. While the exact point at which angiogenesis and vasculogenesis evolved in vertebrate history is unclear, they would have played essential roles in the development of more advanced vascular systems.
Early Vertebrates: Over time, vertebrates would have evolved into increasingly complex cardiovascular systems. The emergence of angiogenesis and vasculogenesis would have accompanied the need for more efficient nutrient delivery, waste removal, and tissue repair. These processes would have allowed vertebrates to sustain larger and more metabolically active bodies.
De Novo Genetic Information, necessary to instantiate angiogenesis and vasculogenesis
The hypothetical emergence of angiogenesis, the process of forming new blood vessels from scratch in the development of organisms that didn't previously possess a vascular system, would have required the addition of specific genetic information to enable this complex process. While the exact genetic changes are speculative, here are some potential additions or modifications to genetic information that could have been involved in the evolution of angiogenesis:
Angiogenic Factors: The development of angiogenesis from scratch would have necessitated the evolution of genes encoding angiogenic factors, such as rudimentary versions of vascular endothelial growth factors (VEGFs). These factors would act as signaling molecules to initiate vessel formation.
Receptor Proteins: The emergence of angiogenesis would have required the development of new receptor proteins on hypothetical endothelial-like precursor cells. These receptors would allow cells to detect and respond to angiogenic factors, initiating the signaling cascades necessary for vessel formation.
Cell Adhesion Molecules: As cells begin to organize into blood vessels, new genetic information could have been necessary to generate primitive cell adhesion molecules. These molecules would facilitate cell-cell interactions and the formation of vessel-like structures.
Matrix Remodeling Enzymes: The initial formation of blood vessels from scratch would involve breaking down and restructuring extracellular matrix. Genes encoding matrix remodeling enzymes could have evolved to allow cells to create pathways for vessel growth.
Transcription Factors: The evolution of angiogenesis would require new transcription factors that activate gene expression programs specific to vessel formation. These factors would regulate the expression of genes involved in cell migration, proliferation, and differentiation.
Guidance Proteins: To direct the movement of primitive endothelial-like cells toward the formation of blood vessels, hypothetical guidance proteins could have evolved to provide directional cues.
Signaling Molecules and Pathways: New genetic information would be needed to establish the initial signaling pathways that trigger cellular responses, such as migration, proliferation, and differentiation, during angiogenesis.
Cytoskeletal Regulators: The development of angiogenesis from scratch would require genes that control cytoskeletal dynamics, enabling cell movement, migration, and organization into vessel-like structures.
Apoptosis and Survival Regulators: As cells form blood vessels, mechanisms for balancing cell survival and cell death would be crucial. Genes involved in apoptosis and survival pathways might have been required.
Differentiation and Specification Genes: The evolution of angiogenesis would involve the development of genes that specify and differentiate precursor cells into endothelial-like cells with distinct roles in vessel formation.
Cell-Cell Communication Molecules: New genetic information could have been necessary to allow cells to communicate and coordinate their behaviors during the formation of blood vessels.
Cell Signaling Pathways for Coordination: To ensure proper coordination among endothelial-like cells, genetic information might have evolved to create rudimentary cell signaling pathways for communication and synchronization.
Epigenetic Regulatory Mechanisms necessary to be instantiated
The hypothetical emergence of angiogenesis "from scratch," meaning the evolution of blood vessel formation in organisms that previously lacked such a system, would have required the establishment of new epigenetic regulations to coordinate the complex cellular changes necessary for vessel development.
Epigenetic Priming for Vascular Precursors: Establishment of epigenetic marks to prime cells for vascular differentiation, creating a permissive environment for angiogenesis-related genes.
Histone Modifications for Vessel Formation: Specific histone modifications, such as acetylation, to activate genes involved in endothelial differentiation and tube formation.
DNA Methylation Dynamics: Establishment of specific DNA methylation patterns to regulate the expression of angiogenesis-essential genes, adjusting their activation during vessel formation.
Non-Coding RNA-Mediated Regulation: Emergence of non-coding RNAs to interact with chromatin-modifying complexes, regulating genes associated with vessel development.
Imprinting and Allelic Regulation: Establishment of allele-specific epigenetic marks to guide cellular roles during vessel formation.
Epigenetic Inheritance of Vascular Patterns: Ability to inherit epigenetic information related to vessel formation, passing regulatory marks necessary for angiogenesis to subsequent generations.
Temporal Epigenetic Regulations: Evolution of epigenetic mechanisms acting as molecular "clocks" for the precise timing of vessel-related processes.
Suppression of Anti-Angiogenic Factors: New epigenetic regulations to suppress genes encoding anti-angiogenic factors, ensuring uninterrupted vessel development.
Chromatin Remodeling for Vessel Assembly: Epigenetic mechanisms regulating chromatin remodeling to facilitate cell-cell interactions and tube formation during vessel assembly.
Epigenetic Regulation of Vascular Maturation: Epigenetic marks guiding the maturation and stabilization of newly formed vessels, including recruitment of support cells.
Epigenetic Sensing of Environmental Cues: Epigenetic mechanisms enabling cells to sense and respond to environmental signals for adaptive vessel formation.
Signaling Pathways necessary to create, and maintain angiogenesis and vasculogenesis
If we're considering the emergence of angiogenesis "from scratch," i.e., the initial development of the process in organisms that didn't previously have any vascular system, potential signaling pathways that would have had to be involved in the hypothetical evolution of angiogenesis: are:
Basic Growth Factor and Receptor Pathways: Emergence of fundamental growth factor pathways and receptor systems for cell proliferation, migration, and differentiation, involving simple signaling cascades.
Chemotaxis Pathways: Development of rudimentary chemotactic signaling to direct primitive endothelial-like cells towards factor gradients promoting movement.
Adhesion Pathways: Formation of signaling pathways related to cell adhesion molecules to support endothelial cell movement and organization into early blood vessels.
Cytoskeletal Remodeling Pathways: Creation of signaling pathways for cytoskeletal alterations, crucial for cell shape and movement.
Cell-Cell Communication Pathways: Development of basic pathways for communication between primitive endothelial cells during early vessel formation.
Apoptosis and Survival Pathways: Establishment of pathways to regulate the balance between cell death and survival, vital for vessel formation.
Initial Extracellular Matrix Signaling: Emergence of signaling pathways to respond to the extracellular matrix, guiding the formation of vessel-like structures.
Evolution of Ligand-Receptor Pairs: Development of new ligand-receptor pairs for sensing and responding to vessel formation cues.
The emergence of angiogenesis, the process of forming new blood vessels from preexisting ones, would have involved the establishment and modification of specific signaling pathways to coordinate the complex cellular changes required for vessel formation. While the exact signaling pathways are speculative, here are some potential pathways that could have been involved in the emergence of angiogenesis:
VEGF Signaling Pathway: Vascular endothelial growth factor (VEGF) is a central regulator of angiogenesis. The VEGF signaling pathway involves the binding of VEGF to its receptors (VEGFRs) on endothelial cells. This activates downstream signaling cascades that promote endothelial cell proliferation, migration, and tube formation.
FGF Signaling Pathway: Fibroblast growth factors (FGFs) also play a role in angiogenesis. The FGF signaling pathway, similar to the VEGF pathway, involves FGF ligands binding to their receptors, which triggers signaling events that contribute to endothelial cell proliferation and migration.
Notch Signaling Pathway: The Notch pathway is involved in cell-cell communication and could have played a role in coordinating endothelial cell differentiation and tip cell selection during vessel sprouting.
Wnt Signaling Pathway: The Wnt pathway has diverse roles in development, and its components could have been involved in angiogenesis by influencing endothelial cell behavior and vessel branching patterns.
TGF-β Signaling Pathway: Transforming growth factor-beta (TGF-β) family members could have been implicated in angiogenesis by regulating endothelial cell differentiation and extracellular matrix remodeling.
PDGF Signaling Pathway: Platelet-derived growth factor (PDGF) signaling might have been involved in recruiting pericytes and smooth muscle cells to stabilize and mature newly formed vessels.
ECM Signaling Pathways: Extracellular matrix (ECM) components, including integrins and focal adhesion kinase (FAK), could have participated in transmitting signals that guide endothelial cell migration and vessel assembly.
MAPK Signaling Pathway: Mitogen-activated protein kinase (MAPK) pathways could have been essential for transmitting signals that regulate cell proliferation, survival, and migration during angiogenesis.
PI3K/AKT Signaling Pathway: The phosphatidylinositol 3-kinase (PI3K)/AKT pathway could have been involved in promoting endothelial cell survival, migration, and angiogenesis-related cellular responses.
Rho GTPase Signaling Pathway: Rho GTPases, such as Rho, Rac, and Cdc42, could have participated in regulating cytoskeletal dynamics and cell migration during angiogenesis.
Chemokine Signaling: Chemokines and their receptors might have guided endothelial cell migration and positioning during vessel sprouting.
Hedgehog Signaling Pathway: Hedgehog signaling could have been implicated in regulating vascular patterning and endothelial cell differentiation.
Endothelial-Specific Signaling Pathways: Signaling pathways specifically active in endothelial cells could have evolved to control angiogenesis-related processes like proliferation, migration, and tube formation.
Regulatory codes necessary for maintenance and operation
The hypothetical emergence of blood vessels and angiogenesis would have likely involved the establishment of regulatory codes and languages to coordinate the development, maintenance, and operation of the vascular system. While the exact details are speculative, here are potential regulatory codes and languages that could have been instantiated:
Transcriptional Regulatory Code: The evolution of blood vessels would require a transcriptional regulatory code involving specific DNA sequences, transcription factors, and regulatory elements that control the expression of genes involved in vessel development, maintenance, and function.
Cis-Regulatory Elements: Enhancers, promoters, and other cis-regulatory elements would need to evolve to ensure proper spatiotemporal expression of angiogenesis-related genes.
Epigenetic Regulatory Language: Epigenetic modifications such as DNA methylation and histone modifications could form an epigenetic regulatory language that guides the activation and repression of genes essential for vascular development and maintenance.
Signaling Pathway Crosstalk: Complex signaling pathways involved in angiogenesis and vascular function would need to communicate and coordinate their activities through a regulatory language that ensures proper cellular responses.
Cell-Cell Communication Codes: As blood vessels involve multiple cell types, a communication code involving cell surface receptors, ligands, and their interactions would be necessary to coordinate cellular behaviors and functions.
Extracellular Matrix (ECM) Signaling: A code involving interactions between cells and the extracellular matrix would regulate processes such as cell adhesion, migration, and signaling.
Vascular Patterning Code: The establishment of hierarchical vessel networks would require a code that guides the formation and branching patterns of blood vessels to ensure efficient distribution of nutrients and oxygen.
Stability and Maturation Code: Blood vessel stabilization and maturation would require a regulatory code involving communication between endothelial cells and support cells (pericytes and smooth muscle cells) to ensure structural integrity.
Oxygen and Nutrient Sensing Code: Blood vessels need to adapt to changing oxygen and nutrient levels. A regulatory code might govern the response of vessels to these fluctuations, ensuring appropriate vessel dilation and constriction.
Immune-Endothelial Communication: Blood vessels interact with the immune system. A code would be needed to regulate the communication between endothelial cells and immune cells, enabling immune surveillance and inflammation regulation.
Inflammatory Response Code: Inflammatory responses and repair processes would require a regulatory code to activate and control specific genes involved in tissue repair and vessel remodeling.
Vascular Tone and Homeostasis Code: Blood pressure and vessel tone need to be tightly regulated. A code would be necessary to balance vasoconstriction and vasodilation to maintain blood flow and homeostasis.
Angiogenic Switch Code: The transition from quiescent vessels to angiogenesis activation would require a code that senses environmental cues and triggers angiogenic responses.
Vascular Regression Code: Vessels need to regress when not needed. A regulatory code would be necessary to initiate vessel regression and tissue remodeling.
Wound Healing and Regeneration Code: Blood vessels play a role in tissue repair. A code would be involved in coordinating vessel-related processes during wound healing and tissue regeneration.
These regulatory codes and languages would have had to emerge to ensure the development, maintenance, and operation of blood vessels. The precise details would depend on the specific context and the genetic and molecular mechanisms that contributed to the emergence of angiogenesis and vascular systems.
Last edited by Otangelo on Sat Feb 24, 2024 9:27 am; edited 66 times in total