https://reasonandscience.catsboard.com/t2239-evolution-common-descent-the-tree-of-life-a-failed-hypothesis
Claim: Eugene V. Koonin (2020): The genomes of all cellular organisms encompass about a hundred universal genes that encode, almost exclusively, protein and RNA components of the translation system. The presence of this universal gene core is strong evidence for the origin of all cellular life from a last universal cellular ancestor (LUCA) 1
Reply: There are also many differences, that cannot be overlooked:
1. The DNA replication machinery is not homologous in the 3 domains of life. The bacterial core replisome enzymes do not share a common ancestor with the analogous components in eukaryotes and archaea.
2. Bacteria and Archaea differ strikingly in the chemistry of their membrane lipids. Cell membrane phospholipids are synthesized by different, unrelated enzymes in bacteria and archaea, and yield chemically distinct membranes.
3. Sequences of glycolytic enzymes differ between Archaea and Bacteria/Eukaryotes. There is no evidence of a common ancestor for any of the four glycolytic kinases or of the seven enzymes that bind nucleotides.
4. There are at least six distinct autotrophic carbon fixation pathways. If common ancestry were true, an ancestral Wood–Ljungdahl pathway should have become life's one and only principle for biomass production.
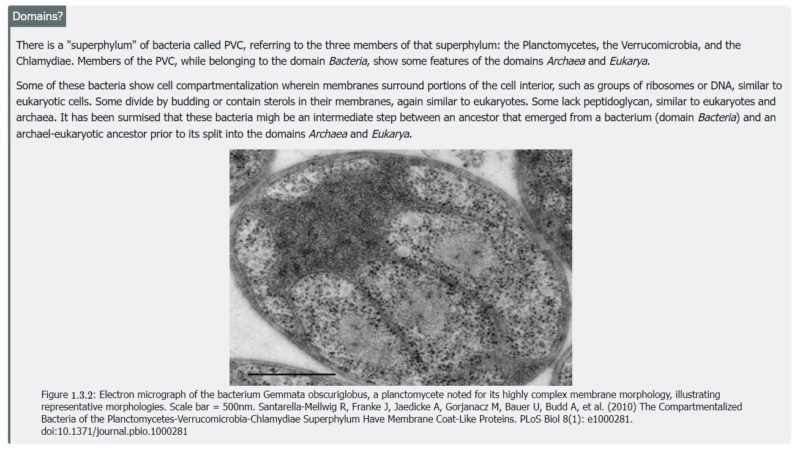
5. There is a sharp divide in the organizational complexity of the cell between eukaryotes, which have complex intracellular compartmentalization, and even the most sophisticated prokaryotes (archaea and bacteria), which do not.
6. A typical eukaryotic cell is about 1,000-fold bigger by volume than a typical bacterium or archaeon, and functions under different physical principles: free diffusion has little role in eukaryotic cells but is crucial in prokaryotes
7. Subsequent massive sequencing of numerous, complete microbial genomes have revealed novel evolutionary phenomena, the most fundamental of these being: pervasive horizontal gene transfer (HGT), in large part mediated by viruses and plasmids, that shapes the genomes of archaea and bacteria and call for a radical revision (if not abandonment) of the Tree of Life concept
8. RNA Polymerase differences: Prokaryotes only contain three different promoter elements: -10, -35 promoters, and upstream elements. Eukaryotes contain many different promoter elements
9. Ribosome and ribosome biogenesis differences: Although we could identify E. coli counterparts with comparable biochemical activity for 12 yeast ribosome biogenesis factors (RBFs), only 2 are known to participate in bacterial ribosome assembly. This indicates that the recruitment of individual proteins to this pathway has been largely independent in the bacterial and eukaryotic lineages. 22
10. Like the origins of DNA replication, the promoters of bacterial and yeast genes have different structures, are recognized by different proteins, and are not exchangeable. The absolute incompatibility between prokaryote (e.g., E. coli) and eukaryote (e.g., yeast) origins of replication and promoters, as well as DNA replication, transcription, and translation machinery, stands as a largely unrecognized challenge to the evolutionary view that the two share a common ancestor.
1. Genome sequencing of cells from the three domains of life, bacteria, archaea, and eukaryotes, reveal that the DNA replication machinery, most of the core replisome enzymes and components are not homologous. Thus, the bacterial core replisome enzymes do not share a common ancestor with the analogous components in eukaryotes and archaea.
2. Bacteria and Archaea differ strikingly in the chemistry of their membrane lipids. Cell membrane phospholipids are synthesized by different, unrelated enzymes in bacteria and archaea, and yield chemically distinct membranes. Bacteria and archaea have membranes made of water-repellent fatty molecules. Bacterial membranes are made of fatty acids bound to the phosphate group while archaeal membranes are made of isoprenes bonded to phosphate in a different way. This leads to something of a paradox: Since a supposed last universal common ancestor, LUCA already had an impermeable membrane for exploiting proton gradients, why would its descendants have independently evolved two different kinds of impermeable membrane?
3. Sequences of glycolytic enzymes differ between Archaea and Bacteria/Eukaryotes. There is no evidence of a common ancestor for any of the four glycolytic kinases or of the seven enzymes that bind nucleotides.
4. There are at least six distinct autotrophic carbon fixation pathways. Since the claim is that this is how life began fixing carbon, and the first carbon fixation pathways were anaerobic, this represents a major puzzle for proponents of common ancestry, and its proponents are led to wonder why an ancestral Wood–Ljungdahl pathway has not become life's one and only principle for biomass production. What is even more puzzling, is the fact that searches of the genomes of acetogenins for enzymes clearly homologous to those of the methanogenic C1-branch came up empty-handed with one notable exception, i.e. the initial step of CO2 reduction which is, in both cases, catalyzed by a molybdo/tungstopterin enzyme from the complex iron-sulfur molybdoenzyme (CISM) superfamily. So, partially, carbon fixation pathways share partially the same enzymes. This points clearly to a common designer choosing different routes for the same reaction but using partially convergent design. Similarities between living organisms could be because they have been designed by the same intelligence, just as we can recognize a Norman Foster building by his characteristic style, or a painting by Van Gogh. We expect to see repeated motifs and re-used techniques in different works by the same artist/designer.
5. There is a sharp divide in the organizational complexity of the cell between eukaryotes, which have complex intracellular compartmentalization, and even the most sophisticated prokaryotes (archaea and bacteria), which do not. The compartmentalization of eukaryotic cells is supported by an elaborate endomembrane system and by the actin-tubulin-based cytoskeleton. There are no direct counterparts of these organelles in archaea or bacteria. The other hallmark of the eukaryotic cell is the presence of mitochondria, which have a central role in energy transformation and perform many additional roles in eukaryotic cells, such as in signaling and cell death.
6. A typical eukaryotic cell is about 1,000-fold bigger by volume than a typical bacterium or archaeon, and functions under different physical principles: free diffusion has little role in eukaryotic cells but is crucial in prokaryotes
7. Subsequent massive sequencing of numerous, complete microbial genomes have revealed novel evolutionary phenomena, the most fundamental of these being: pervasive horizontal gene transfer (HGT), in large part mediated by viruses and plasmids, that shapes the genomes of archaea and bacteria and call for a radical revision (if not abandonment) of the Tree of Life concept 22
8. RNA Polymerase differences: Prokaryotes only contain three different promoter elements: -10, -35 promoters, and upstream elements. Eukaryotes contain many different promoter elements: TATA box, initiator elements, downstream core promoter element, CAAT box, and the GC box to name a few. Eukaryotes have three types of RNA polymerases, I, II, and III, and prokaryotes only have one type. Eukaryotes form and initiation complex with the various transcription factors that dissociate after initiation is completed. There is no such structure seen in prokaryotes. Another main difference between the two is that transcription and translation occurs simultaneously in prokaryotes and in eukaryotes the RNA is first transcribed in the nucleus and then translated in the cytoplasm. RNAs from eukaryotes undergo post-transcriptional modifications including: capping, polyadenylation, and splicing. These events do not occur in prokaryotes. mRNAs in prokaryotes tend to contain many different genes on a single mRNA meaning they are polycystronic. Eukaryotes contain mRNAs that are monocystronic. Termination in prokaryotes is done by either rho-dependent or rho-independent mechanisms. In eukaryotes transcription is terminated by two elements: a poly(A) signal and a downstream terminator sequence. 21
9. Ribosome and ribosome biogenesis differences: Although we could identify E. coli counterparts with comparable biochemical activity for 12 yeast ribosome biogenesis factors (RBFs), only 2 are known to participate in bacterial ribosome assembly. This indicates that the recruitment of individual proteins to this pathway has been largely independent in the bacterial and eukaryotic lineages. 22
The bacterial version of a universal ribosomal protein tends to be remarkably different from its archaeal equivalent, the same being true, even more dramatically, for the aminoacyl-tRNA synthetases. In both cases, in a sequence alignment, a position constant in composition in the Bacteria tends to be so in its archaeal homolog as well, but the archaeal and bacterial compositions for that position often differ from each other. Moreover, among the aminoacyl-tRNA synthetases, a total lack of homology between large (and characteristic) sections of the bacterial version of a molecule and its archaeal counterpart is common 19
10. Like the origins of DNA replication, the promoters of bacterial and yeast genes have different structures, are recognized by different proteins, and are not exchangeable. The absolute incompatibility between prokaryote (e.g., E. coli) and eukaryote (e.g., yeast) origins of replication and promoters, as well as DNA replication, transcription, and translation machineries, stands as a largely unrecognized challenge to the evolutionary view that the two share a common ancestor.
1. DNA Replication Across Taxa
http://library.lol/main/F4FD87CCFF8554BAF60936F1A8BFCFFC
2. Phylogenomic Investigation of Phospholipid Synthesis in Archaea
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3533463/
3. Glycolysis Is an Energy-Conversion Pathway in Many Organisms
https://www.ncbi.nlm.nih.gov/books/NBK22593/
4. Beating the acetyl coenzyme A-pathway to the origin of life
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3685460/
5. The origin and early evolution of eukaryotes in the light of phylogenomics
https://genomebiology.biomedcentral.com/articles/10.1186/gb-2010-11-5-209
6. Energetics and genetics across the prokaryote-eukaryote divide
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3152533/
7. Evolution of microbes and viruses: a paradigm shift in evolutionary biology?
https://pubmed.ncbi.nlm.nih.gov/22993722/
8. Prokaryotic vs. Eukaryotic Trancription
https://www.chem.uwec.edu/webpapers2006/sites/demlba/folder/provseuk.html
9. The evolution of the ribosome biogenesis pathway from a yeast perspective
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3919561/
10. DNA Replication Origins
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3783049/
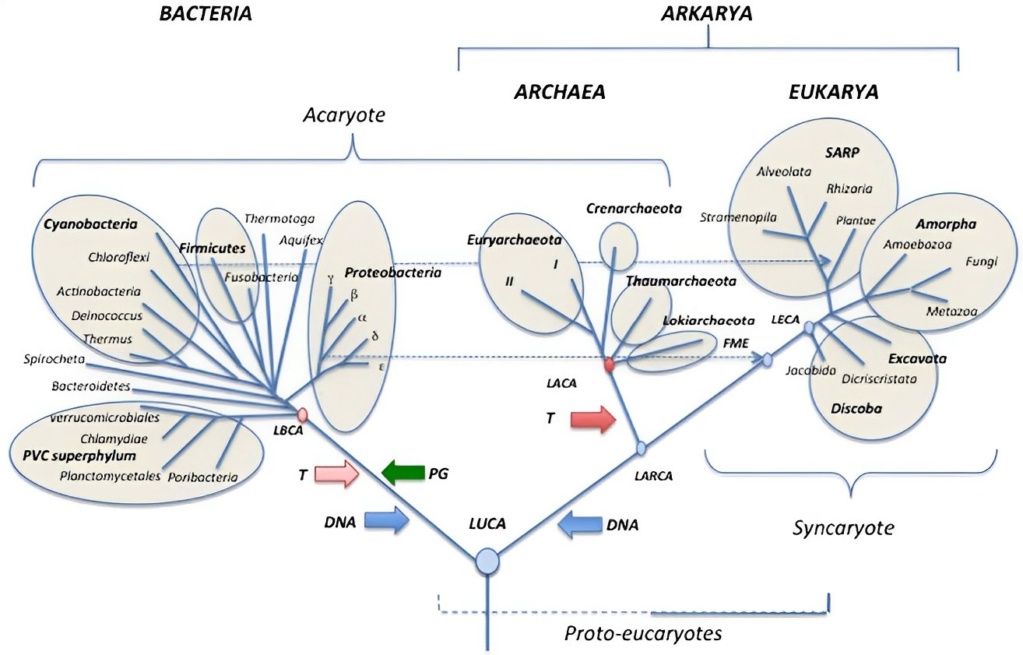
Laura A. Hug A new view of the tree of life 11 April 2016
The tree of life as we know it has dramatically expanded due to new genomic sampling of previously enigmatic or unknown microbial lineages. This depiction of the tree captures the current genomic sampling of life, illustrating the progress that has been made in the last two decades following the first published genome. What emerges from analysis of this tree is the depth of evolutionary history that is contained within the Bacteria, in part due to the CPR, which appears to subdivide the domain. Most importantly, the analysis highlights the large fraction of diversity that is currently only accessible via cultivation-independent genome-resolved approaches.
https://www.nature.com/articles/nmicrobiol201648
Following points are a clear smackdown to the claim of Common descent and Darwin's tree of life
Has the hypothesis of common ancestry merit? Behe thinks so. I asked him about it: 53:40
https://www.youtube.com/watch?v=sOz4vuge0bY&feature=youtu.be&fbclid=IwAR0wHIaq--8mKLajN26vISIQ1Xun1bpeXrB5RihoKkEDDQpnPZN0ZoLa_uw
In his book, Darwin suggests that all living organisms are related by ascendency, and therefore they are all derived from ancestral species, which migrate around the world and diversify, generating the amazing biodiversity of organisms (Darwin, 1859).
Jonathan Lambert What a Newfound Kingdom Means for the Tree of Life December 11, 2018
Many types of organisms that live in an environment, “but unless you have a larger known reference sequence, it’s very difficult to put these different things into an evolutionary framework 21
https://www.quantamagazine.org/what-a-newfound-kingdom-means-for-the-tree-of-life-20181211/
Eugene V. Koonin The Logic of Chance: The Nature and Origin of Biological Evolution 2011
Arguments for a LUCA that would be indistinguishable from a modern prokaryotic cell have been presented, along with scenarios depicting LUCA as a much more primitive entity (Glansdorff, et al., 2008).
The difficulty of the problem cannot be overestimated. Indeed, all known cells are complex and elaborately organized. The simplest known cellular life forms, the bacterial (and the only known archaeal) parasites and symbionts, clearly evolved by degradation of more complex organisms; however, even these possess several hundred genes that encode the components of a fully-fledged membrane; the replication, transcription, and translation machineries; a complex cell-division apparatus; and at least some central metabolic pathways. As we have already discussed, the simplest free-living cells are considerably more complex than this, with at least 1,300 genes.
All the difficulties and uncertainties of evolutionary reconstructions notwithstanding, parsimony analysis combined with less formal efforts on the reconstruction of the deep past of particular functional systems leaves no serious doubts that LUCA already possessed at least several hundred genes. In addition to the aforementioned “golden 100” genes involved in the expression, this diverse gene complement consists of numerous metabolic enzymes, including pathways of the central energy metabolism and the biosynthesis of amino acids, nucleotides, and some coenzymes, as well as some crucial membrane proteins, such as the subunits of the signal recognition particle (SRP) and the H+- ATPase.
DNA Replication Across Taxa
Genome sequencing of cells from the three domains of life, bacteria, archaea, and eukaryotes, reveals that most of the core replisome components evolved twice, independently. Thus, the bacterial core replisome enzymes do not share a common ancestor with the analogous components in eukaryotes and archaea, while the archaea and eukaryotic core replisome machinery share a common ancestor
Page 331:The reconstructed gene repertoire of LUCA also has gaping holes. The two most shocking ones are
(i) the absence of the key components of the DNA replication machinery, namely the polymerases that are responsible for the initiation (primases) and elongation of DNA replication and for gap-filling after primer removal, and the principal DNA helicases (Leipe, et al., 1999), and
(ii) the absence of most enzymes of lipid biosynthesis. These essential proteins fail to make it into the reconstructed gene repertoire of LUCA because the respective processes in bacteria, on one hand, and archaea, on the other hand, are catalyzed by different, unrelated enzymes and, in the case of membrane phospholipids, yield chemically distinct membranes.
Bacteria and archaea have membranes made of water-repellent fatty molecules. Simple fatty molecules tend to flip around, making the membrane leaky, so both bacteria and archaea tacked on a water-loving phosphate group to stabilize the molecules and make their membranes impermeable. They took very different routes, though. Bacterial membranes are made of fatty acids bound to the phosphate group while archaeal membranes are made of isoprenes bonded to phosphate in a different way. This suggests that their membranes evolved independently. This leads to something of a paradox: if LUCA already had an impermeable membrane for exploiting proton gradients, why would its descendants have independently evolved two different kinds of impermeable membrane? 17
https://3lib.net/book/1167956/035b71
My comment: Shocking and remarkable indeed : The DNA replication machinery is essential in all domains, and so is lipid biosynthesis for cell membranes. Its not possible that the first cells emerged without membranes and DNA replication in a LUCA, and then evolved distinguished membranes and DNA replication, each by its own.
That means, the at least several hundred genes possessed in all three domains of life would have had to emerge in a convergent manner ( that is separately they would have come into existence with the same genome, proteome, and metabolome except lipid biosynthesis and DNA replication which were the two only distinct parts that diverged each from the other domains. This is a hard sell when evoking evolution. Even more when only unguided random mechanisms were at hand, that is chance and luck. If the emergency of one cell type would have been exceedingly improbable, imagine the same feat tree separate times.
Stephen J. Gould wrote in Wonderful Life: The Burgess Shale and the Nature of History 1990
“…No finale can be specified at the start, none would ever occur a second time in the same way, because any pathway proceeds through thousands of improbable stages. Alter any early event, ever so slightly, and without apparent importance at the time, and evolution cascades into a radically different channel.
https://3lib.net/book/677772/5f1e6d
Neither a LUCA is credible, nor naturally emerging tree separate domains of life through partially convergent manner. The only rational explanation is a designer creating the three domains of life separately, and using the same toolkit where required, and a separate divergent toolkit for other parts.
Should that not be evidence that a LUCA never existed, and that the three domains of life had to emerge separately through an intelligent designer?
Markus A Keller The widespread role of non-enzymatic reactions in cellular metabolism 22nd January 2015
Sequences of glycolytic enzymes differ between Archaea and Bacteria/Eukaryotes
https://sci-hub.ren/10.1016/j.copbio.2014.12.020
Franklin M. Harold In Search of Cell History: The Evolution of Life's Building Blocks page 96
Membranes also pose one of the most stubborn puzzles in all of cell evolution. Shortly after the discovery of the Archaea, it was realized that these organisms differ strikingly from the Bacteria in the chemistry of their membrane lipids. Archaea make their plasma membranes of isoprenoid subunits, linked by ether bonds to glycerol-1-phosphate; by contrast, Bacteria and Eukarya employ fatty acids linked by ester bonds to glycerol-3-phosphate. There are a few partial exceptions to the rule. Archaeal membranes often contain fatty acids, and some deeply branching Bacteria, such as Thermotoga, favor isoprenoid ether lipids (but even they couple the ethers to glycerol-3-phosphate). This pattern of lipid composition, which groups Bacteria and Eukarya together on one side and Archaea on the other, stands in glaring contrast to what would be expected from the universal tree, which puts Eukarya with the Archaea
https://3lib.net/book/2463874/2f4dd0
Keith A. Webster Evolution of the coordinate regulation of glycolytic enzyme genes by hypoxia 01 SEPTEMBER 2003
There is no evidence of a common ancestor for any of the four glycolytic kinases or of the seven enzymes that bind nucleotides
Genetic, protein and DNA analysis, together with major differences in the biochemistry and molecular biology of between all three domains – Bacteria, Archaea and Eukaryota – suggest that the three fundamental cell types are distinct and evolved separately (i.e. Bacteria are not actually pro-precursors of the eukaryotes, which have sequence similarities in particular parts of their biochemistry between both Bacteria or Archaea). Only a relatively small percentage of genes in Archaea have sequence similarity to genes in Bacteria or Eukaryota. Furthermore, most of the cellular events triggered by intracellular Ca2+ in eukaryotes do not occur in either Bacteria or Archaea.
https://journals.biologists.com/jeb/article/206/17/2911/13849/Evolution-of-the-coordinate-regulation-of
Jay Wile This Could Be One of the Most Important Scientific Papers of the Decade July 23, 2018
More than eight years ago (have I really been blogging that long?), I was excited to see the appearance of a new peer-reviewed journal, BIO-Complexity. I thought it was going to have a lot of impact on the science of biology, but so far, its impact has been minimal. A few good studies (like this one and this one) have been published in it, but overall, it has not published the ground-breaking research I had hoped it would.
That might have changed. I just devoured the most recent study published in the journal, and I have to say, it is both innovative and impressive. It represents truly original thinking in the field of biology, and if further research confirms the results of the paper, we might very well be on the precipice of an important advancement in the field of biological taxonomy (the science of classifying living organisms).
http://blog.drwile.com/this-could-be-one-of-the-most-important-scientific-papers-of-the-decade/
Winston Ewert The Dependency Graph of Life 2018
The hierarchical classification of life has been claimed as compelling evidence for universal common ancestry. However, research has uncovered much data which is not congruent with the hierarchical pattern. Nevertheless, biological data resembles a nested hierarchy sufficiently well to require an explanation. While many defenders of intelligent design dispute common descent, no alternative account of the approximate nested hierarchy pattern has been widely adopted. We present the dependency graph hypothesis as an alternative explanation, based on the technique used by software developers to reuse code among different software projects. This hypothesis postulates that different biological species share modules related by a dependency graph. We evaluate several predictions made by this model about both biological and synthetic data, finding them to be fulfilled.
http://bio-complexity.org/ojs/index.php/main/article/view/BIO-C.2018.3
Eric Bapteste Prokaryotic evolution and the tree of life are two different things 2009 Sep 29
The concept of a tree of life is prevalent in the evolutionary literature. It stems from attempting to obtain a grand unified natural system that reflects a recurrent process of species and lineage splittings for all forms of life. Traditionally, the discipline of systematics operates in a similar hierarchy of bifurcating (sometimes multifurcating) categories. The assumption of a universal tree of life hinges upon the process of evolution being tree-like throughout all forms of life and all of biological time. In prokaryotes, they do not. Prokaryotic evolution and the tree of life are two different things, and we need to treat them as such, rather than extrapolating from macroscopic life to prokaryotes. In the following we will consider this circumstance from philosophical, scientific, and epistemological perspectives, surmising that phylogeny opted for a single model as a holdover from the Modern Synthesis of evolution.
In eukaryotes, plasma membrane consists of sterols and carbohydrates.In prokaryotes, plasma membrane does not contain carbohydrates or sterols. Prokarotic membranes have only a few types of phospholipids while eukaryotic membranes have can have over 6 different phospholipids as well as other types of lipids. Prokaryotic membranes do not commonly have cholesterol inside the hydrophobic core whereas eukaryotic membranes use chloresterol to regulate their fluidity. Eukaryotic cell membrane is basically trilamellar with double layer of phospholipid. It is asymmetrical. It has intrinsic and extrinsic proteins that also help in transport across membrane. It has other components like cholesterol to maintain fluidity of membrane. Where as prokaryotic or bacterial cell membrane is composed of peptidoglycan that is cross chain of N acetyl glycosamine and muramic acid.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2761302/
Eugene V Koonin The origin and early evolution of eukaryotes in the light of phylogenomics 05 May 2010
The origin of eukaryotes is a huge enigma and a major challenge for evolutionary biology. There is a sharp divide in the organizational complexity of the cell between eukaryotes, which have complex intracellular compartmentalization, and even the most sophisticated prokaryotes (archaea and bacteria), which do not. A typical eukaryotic cell is about 1,000-fold bigger by volume than a typical bacterium or archaeon, and functions under different physical principles: free diffusion has little role in eukaryotic cells, but is crucial in prokaryotes. The compartmentalization of eukaryotic cells is supported by an elaborate endomembrane system and by the actin-tubulin-based cytoskeleton. There are no direct counterparts of these organelles in archaea or bacteria. The other hallmark of the eukaryotic cell is the presence of mitochondria, which have a central role in energy transformation and perform many additional roles in eukaryotic cells, such as in signaling and cell death.
https://genomebiology.biomedcentral.com/articles/10.1186/gb-2010-11-5-209
Mark A. Ragan The network of life: genome beginnings and evolution 2009 Aug 12
The rapid growth of genome-sequence data since the mid-1990s is now providing unprecedented detail on the genetic basis of life, and not surprisingly is catalysing the most fundamental re-evaluation of origins and evolution since Darwin’s day. Several papers in this theme issue argue that Darwin’s tree of life is now best seen as an approximation—one quite adequate as a description of some parts of the living world (e.g. morphologically complex eukaryotes), but less helpful elsewhere (e.g. viruses and many prokaryotes); indeed, one of our authors goes farther, proclaiming the “demise” of Darwin’s tree as a hypothesis on the diversity and seeming naturalness of hierarchical arrangements of groups of living organisms.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2874017/
W. ford doolittle Uprooting the Tree of Life february 2000
Charles Darwin contended more than a century ago that all modern species diverged from a more limited set of ancestral groups, which themselves evolved from still fewer progenitors and so on back to the beginning of life. In principle, then, the relationships among all living and extinct organisms could be represented as a single genealogical tree.Most contemporary researchers agree. Many would even argue that the general features of this tree are already known, all the way down to the root—a solitary cell, termed life’s last universal common ancestor, that lived roughly 3.5 to 3.8 billion years ago. The consensus view did not come easily but has been widely accepted for more than a decade. Yet ill winds are blowing. To everyone’s surprise, discoveries made in the past few years have begun to cast serious doubt on some aspects of the tree, especially on the depiction of the relationships near the root.
http://labs.icb.ufmg.br/lbem/aulas/grad/evol/treeoflife-complexcells.pdf
Konstantin Khalturin Newly Discovered 'Orphan Genes' Defy Evolution 2009 Sep;25
An important category of "rogue" genetic data that utterly defies evolutionary predictions is the common occurrence of taxonomically restricted genes, otherwise known as "orphan genes." These are now being discovered in the sequencing of all genomes. Many multicellular animals share similar sets of genes that produce proteins that perform related biochemical functions. This is a common feature of purposefully engineered systems. In addition to these standard genes, all organisms thus far tested also have unique sets of genes specific to that type of creature.
The authors of a recent review paper, published in Trends in Genetics, on the subject of orphan genes stated, "Comparative genome analyses indicate that every taxonomic group so far studied contains 10–20% of genes that lack recognizable homologs [similar counterparts] in other species."1
These orphan genes are also being found to be particularly important for specific biological adaptations that correspond with ecological niches in relation to the creature's interaction with its environment.2 The problem for the evolutionary model of animal origins is the fact that these DNA sequences appear suddenly and fully functional without any trace of evolutionary ancestry (DNA sequence precursors in other seemingly related organisms). And several new studies in both fish and insect genomes are now highlighting this important fact.
https://pubmed.ncbi.nlm.nih.gov/19716618/
W. Ford Doolittle What Is the Tree of Life? April 14, 2016
A universal Tree of Life (TOL) has long been a goal of molecular phylogeneticists, but reticulation at the level of genes and possibly at the levels of cells and species renders any simple interpretation of such a TOL, especially as applied to prokaryotes, problematic. 12 One of the several ways in which microbiology puts the neo-Darwinian synthesis in jeopardy is by the threatening to “uproot the Tree of Life (TOL)” [1]. Lateral gene transfer (LGT) is much more frequent than most biologists would have imagined up until about 20 years ago, so phylogenetic trees based on sequences of different prokaryotic genes are often different. How to tease out from such conflicting data something that might correspond to a single, universal Tree of Life becomes problematic. Moreover, since many important evolutionary transitions involve lineage fusions at one level or another, the aptness of a tree (a pattern of successive bifurcations) as a summary of life’s history is uncertain [2–4].
https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1005912
Rob Waugh Octopuses ‘are aliens’, scientists decide after DNA study 12 Aug 2015
Not to freak you out or anything, but scientists have just revealed that octopuses are so weird they’re basically aliens.
The first full genome sequence shows of that octopuses (NOT octopi) are totally different from all other animals – and their genome shows a striking level of complexity with 33,000 protein-coding genes identified, more than in a human.
There we were thinking it was quite freaky enough when they learned how to open jam jars.
US researcher Dr Clifton Ragsdale, from the University of Chicago, said: ;The octopus appears to be utterly different from all other animals, even other molluscs, with its eight prehensile arms, its large brain and its clever problem-solving abilities.
‘The late British zoologist Martin Wells said the octopus is an alien. In this sense, then, our paper describes the first sequenced genome from an alien.’
Octopuses: What even ARE they?
They inhabit every ocean at almost all depths and possess a range of features that call to mind sci-fi aliens.
These include prehensile sucker-lined tentacles, highly mobile, camera-like eyes sensitive to polarised light, sophisticated camouflage systems that alter skin colour and patterns, jet-propulsion, three hearts, and the ability to regenerate severed limbs.
The scientists estimate that the two-spot octopus genome contains 2.7 billion base pairs – the chemical units of DNA – with long stretches of repeated sequences.
https://metro.co.uk/2015/08/12/octopuses-are-aliens-scientists-decide-after-dna-study-5339123/
Samantha Mathewson Octopus Have Been Found to have Unique Genes Aug 20, 2015
hundreds of other genes that are common in cephalopods, but unknown in other animals, were found.
http://www.natureworldnews.com/articles/16161/20150820/octopus-found-unique-genes.htm
EurekAlert! Decoding the genome of an alien 12-AUG-2015
Besides recognizable genes, vast swathes of the genome consist of regulatory networks that control how genes are expressed in cells. In the octopus, nearly half of the genome was found to be composed of mobile elements called transposons, one of the highest proportions in the animal kingdom. Transposons replicate and move around with a life of their own, disrupting or enhancing gene expression and facilitating reshufflings of gene order. The researchers found many of them to be particularly active in the octopus nervous system. The "Hox" genes, involved in embryonic development in all animals, are a particularly dramatic example. Although clustered together in most animals, including other mollusks, they are scattered in snippets in the octopus, presumably enabling the evolution of the versatile cephalopod body plan.
https://www.eurekalert.org/pub_releases/2015-08/oios-dtg081215.php
My comment: Presumably. Yes. Or, in other words, guesswork as always...... The architecture of a body plan must be right from the beginning. Everything goes, or nothing goes. The question is, where does the information of this reshuffling of genes come from? In my view, the only rational explanation is intentional design.
JEFFREY P. TOMKINS, PH.D Are Rotifers Gene Stealers or Uniquely Engineered? DECEMBER 03, 2012
The tools of DNA sequencing are becoming cheaper to use and more productive than ever, and the deluge of DNA comparison results between organisms coming forth are becoming a quagmire for the evolutionary paradigm. To prop it up, biologists resort to ever more absurd explanations for discrepancies. A prime example of this trickery is in a recent DNA sequencing project performed in a microscopic aquatic multi-cellular animal called a rotifer.1
In this effort, the researchers targeted those gene sequences that are expressed as proteins for DNA sequencing because the genome was too large and complex to sequence and assemble all of its DNA. They recorded over 61,000 gene sequences that were expressed from rotifers grown in stressed and non-stressed conditions. Of these, they could only find sequence similarities between rotifers and other creatures for 28,922 sequences (less than half). The researchers tossed the unknown DNA sequences out of their analysis since the non-similar genes were novel, apparently specific to rotifer, and essentially difficult for evolution to explain.
Of the 28,922 sequences for which they could obtain a match in a public database of other creature's DNA and protein sequences, a significant proportion (more than in any other creature sequenced) did not fit evolutionary expectations of common descent. Further complicating this picture, the rotifer gene sequences were found in a diverse number of non-rotifer creatures! Some of the creatures that had gene matches to rotifers included a variety of plants, other multicellular animals, protists (complex single celled animals), archaea, bacteria, and fungi. Evolutionists have two options in which to categorize these unusual gene matches based on their naturalistic presuppositions. First, they can say that these genes evolved independently in separate creatures in a hypothetical process called "convergent evolution." However, in cases where there are literally hundreds of these DNA sequences popping up in multiple organisms, this scenario becomes so unlikely that even evolutionists have too much difficulty imagining it. The second option is called "horizontal gene transfer," or HGT. This involves the transfer of genes, perhaps via some sort of microbial host vector such as a bacterium.
https://www.icr.org/article/are-rotifers-gene-stealers-or-uniquely
What a bust against common ancestry from a mainstream scientist :
Eugene V Koonin The Biological Big Bang model for the major transitions in evolution 2007 Aug 20
Major transitions in biological evolution show the same pattern of sudden emergence of diverse forms at a new level of complexity. The relationships between major groups within an emergent new class of biological entities are hard to decipher and do not seem to fit the tree pattern that, following Darwin's original proposal, remains the dominant description of biological evolution. The cases in point include the origin of complex RNA molecules and protein folds; major groups of viruses; archaea and bacteria, and the principal lineages within each of these prokaryotic domains; eukaryotic supergroups; and animal phyla. In each of these pivotal nexuses in life's history, the principal "types" seem to appear rapidly and fully equipped with the signature features of the respective new level of biological organization. No intermediate "grades" or intermediate forms between different types are detectable. Usually, this pattern is attributed to cladogenesis compressed in time, combined with the inevitable erosion of the phylogenetic signal.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1973067/
Wells, Jonathan The Politically Incorrect Guide to Darwinism And Intelligent Design 2006
At about the same time, Dalhousie University evolutionary biologist W. Ford Doolittle concluded that lateral gene transfer among ancient organisms meant that molecular phylogeny might never be able to discover the “true tree” of life, not because it is using the wrong methods or the wrong genes, “but because the history of life cannot properly be represented as a tree.” He concluded: “Perhaps it would be easier, and in the long run more productive, to abandon the attempt to force” the molecular data “into the mold provided by Darwin.” Instead of a tree, Doolittle proposed “a web- or net-like pattern.” 10
The controversy over the universal tree of life continues. In 2002, Woese suggested that biology should go beyond Darwin’s doctrine of common descent. In 2004, he wrote: “The root of the universal tree is an artifact resulting from forcing the evolutionary course into a tree representation when that representation is inappropriate.” In 2004, Doolittle and his colleagues proposed replacing the tree of life with a net-like “synthesis of life,” and in 2005 they recommended that “representations other than a tree should be investigated.” Meanwhile, other scientists continue to defend the hypothesis that the universal ancestor existed but was complex rather than simple 11
“DR ROSE SAID: ‘THE TREE OF LIFE IS BEING POLITELY BURIED – WE ALL KNOW THAT. WHAT’S LESS ACCEPTED IS OUR WHOLE FUNDAMENTAL VIEW OF BIOLOGY NEEDS
TO CHANGE.’ HE SAYS BIOLOGY IS VASTLY MORE COMPLEX THAN WE THOUGHT AND FACING UP TO THIS COMPLEXITY WILL BE AS SCARY AS THE CONCEPTUAL UPHEAVALS PHYSICISTS HAD TO TAKE ON BOARD IN THE EARLY 20TH CENTURY.”
https://3lib.net/book/3318290/4fabee
Herve´ Philippe The Rooting of the Universal Tree of Life Is Not Reliable 19999
Several composite universal trees connected by an ancestral gene duplication have been used to root the universal tree of life. In all cases, this root turned out to be in the eubacterial branch. However, the validity of results obtained from comparative sequence analysis has recently been questioned, in particular, in the case of ancient phylogenies. For example, it has been shown that several eukaryotic groups are misplaced in ribosomal RNA or elongation factor trees because of unequal rates of evolution and mutational saturation. Furthermore, the addition of new sequences to data sets has often turned apparently reasonable phylogenies into confused ones. We have thus revisited all composite protein trees that have been used to root the universal tree of life up to now (elongation factors, ATPases, tRNA synthetases, carbamoyl phosphate synthetases, signal recognition particle proteins) with updated data sets. In general, the two prokaryotic domains were not monophyletic with several aberrant groupings at different levels of the tree. Furthermore, the respective phylogenies contradicted each others, so that various ad hoc scenarios (paralogy or lateral gene transfer) must be proposed in order to obtain the traditional Archaebacteria–Eukaryota sisterhood. More importantly, all of the markers are heavily saturated with respect to amino acid substitutions. As phylogenies inferred from saturated data sets are extremely sensitive to differences in evolutionary rates, present phylogenies used to root the universal tree of life could be biased by the phenomenon of long branch attraction. Since the eubacterial branch was always the longest one, the eubacterial rooting could be explained by an attraction between this branch and the long branch of the outgroup. Finally, we suggested that an eukaryotic rooting could be a more fruitful working hypothesis, as it provides, for example, a simple explanation to the high genetic similarity of Archaebacteria and Eubacteria inferred from complete genome analysis.
http://www.somosbacteriasyvirus.com/rooting.pdf
William F Martin Early evolution without a tree of life 30 June 2011
There is more to evolution than will fit on any tree. For understanding major transitions in early evolution, we might not need a tree of life at all. But we need to keep our ideas testable with data from genomes or other independent data so as to keep our nose pinned to the grindstone of observations. The very early evolution of life is mostly written in the language of chemistry, some of which is (arguably) still operating today in modern metabolism if we look at the right groups . The environments and starting material that the Earth had to offer to fuel early chemistry are variables that only geochemists can reasonably constrain . One can make a case that acetogens (clostridial firmicutes) and hydrogenotrophic methanogens (euryarchareotes) harbour the ancestral states of microbial physiology in the eubacteria and archaebacteria respectively , and some trees are compatible with that view , as is the distribution of primitive energy-conserving mechanisms . But given a transition from the elements on early Earth to replicating cells, the course of prokaryote evolution does not appear to play out along the branches of a phylogenetic tree. For example, Whitman surveyed the biology and diversity of prokaryotes, showing an rRNA tree to discuss matters of classification; but branching orders in that tree play no role in his discussion of diversity or underlying evolutionary processes. If that is the direction we are headed , it is not all bad. But having the eukaryotes sitting on one branch in the rRNA tree of life rather than on two, as they should be (or three in the case of plants with their plastids), is far enough off the mark that we should be striving for a better representation of the relationship of eukaryotes to the two kinds of prokaryotes from which they stem.
Eukaryotes are genetic chimaeras and the role of mitochondria in the origin of that chimaerism is apparent . Eukaryotes are complex and the pivotal role of mitochondria in the origin of that complexity (as opposed to a pivotal role of phagocytosis) seems increasingly difficult to dispute, for energetic reasons . That leaves little reasonable alternative to the view that the host for the origin of mitochondria was a prokaryote, in the simplest of competing alternatives an archaebacterium . The antiquity of anaerobic energy metabolism and sulfide metabolism among eukaryotes meshes well with newer views of Proterozoic ocean chemistry . A challenge remains in computing networks of genomes that include lateral gene transfers among prokaryotes and the origin of eukaryotes in the same graph. Tracking early evolution without a tree of life affords far more freedom to explore ideas than thinking with a tree in hand. The ideas need to generate predictions and be testable, though, otherwise they are not science. If we check our thoughts too quickly against a tree whose truth nobody can determine anyway, the tree begins to decide which thoughts we may or may not have and which words we may or may not use. Should a tree of life police our thoughts? Working without one is an option.
https://biologydirect.biomedcentral.com/articles/10.1186/1745-6150-6-36
http://youngearth.com/marine-worm-infects-trunk-darwins-tree-be-felled-soon
marine worms are more closely related to humans than are mollusks and insects
https://www.nature.com/articles/470161a/box/1
Douglas L. Theobald: A formal test of the theory of universal common ancestry 2010
In all cases tried, with a wide variety of evolutionary models (from the simplest to the most parameter rich), the multiple-ancestry models for shuffled data sets are preferred by a large margin over common ancestry models (LLR on the order of a thousand), even with the large internal branches.
https://sci-hub.ren/10.1038/nature09014
Notes:
1. Vertebrate-type intron-rich genes in the marine annelid Platynereis dumerilii F. Raible, K. Tessmar-Raible, K. Osoegawa, P. Wincker, C. Jubin, G. Balavoine, D. Ferrier, V. Benes, P. de Jong, J. Weissenbach, P. Bork and D. Arendt.
2. intron - Part of a gene whose sequence is transcribed but not present in a mature mRNA after splicing.
3. Mali B, Frank U. Hydroid TNF-receptor-associated factor (TRAF) and its splice variant: a role in development.Mol Immunol. (2004) 41:377-84
4. Hughes, J.F. et al. 2010. Chimpanzee and human Y chromosomes are remarkably divergent in structure gene content. Nature. 463 (7280): 536-539.
The dramatic divergence of bacteriophage genomes is an obstacle that frequently prevents the detection of homology between proteins and, thus, the determination of phylogenetic links between phages. 1
“DR ROSE SAID: ‘THE TREE OF LIFE IS BEING POLITELY BURIED – WE ALL KNOW THAT. WHAT’S LESS ACCEPTED IS OUR WHOLE FUNDAMENTAL VIEW OF BIOLOGY NEEDS TO CHANGE.’ HE SAYS BIOLOGY IS VASTLY MORE COMPLEX THAN WE THOUGHT AND FACING UP TO THIS COMPLEXITY WILL BE AS SCARY AS THE CONCEPTUAL UPHEAVALS PHYSICISTS HAD TO TAKE ON BOARD IN THE EARLY 20TH CENTURY.”
A science forum was held at Arizona State University in February 2011, where the following dialogue between Dawkins and Venter was reported:
Dr. Craig Venter Denies Common Descent in front of Richard Dawkins!
https://www.youtube.com/watch?v=MXrYhINutuI
Venter: I'm not so sanguine as some of my colleagues here that there's only one life form on this planet we have a lot of different types of metabolism different organisms I wouldn't call you the same life-form as the one we have that lives in pH12 base that would dissolve your skin if we drop you at it. The same genetic it will have a common anything well you don't have the same genetic code in fact the micoplasmas use a different genetic code and it would not work in yourself so there are a lot of variations on the unit
Dawkins: But you're not saying it belongs to a different tree of life from me
Venter: I well I think the Tree of Life is an artifact of some early scientific studies that aren't really holding up so the tree you know there may be a bush of life. Bush I don't like that word written but that's only I can see that so there's not a tree of life and in fact from our deep sequencing of organisms in the ocean out of now we have about 60 million different unique gene sets we found 12 that looked like a very very deep branching perhaps fourth domain of life that obviously is extremely rare that it only shows up out of those few sequences but it's still DNA based but you know the diversity we have in the DNA world I'm not so saying what in wedding ready to throw out the DNA world.
Evolutionnews: Venter vs. Dawkins on the Tree of Life — and Another Dawkins Whopper March 9, 2011
Since at least the publication of The Blind Watchmaker (1986), Richard Dawkins has claimed that the genetic code is universal across all organisms on earth. This is “near-conclusive proof,” he writes, that every living thing on this planet “descended from a single common ancestor” (1986, p. 270)
David Posada: Testing for Universal Common Ancestry 2014 Aug 12
We did not explore many possible combinations of trees, branch lengths, sequence sizes, and evolutionary models for instance—they show that there are many cases not unlike real data sets where the UCA test fails. Our general impression is that the original UCA test would not reject a common origin for any but obviously unrelated set of sequences.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5215821/
1) http://www.evolutionnews.org/2007/04/francix_x_clines_an_excellent003528.html
2) http://www.ncbi.nlm.nih.gov/pubmed/19716618
3) http://www.truthinscience.org.uk/tis2/index.php/component/content/article/143.html
4) http://www.icr.org/article/common-dna-sequences-evidence-evolution/
5) http://www.somosbacteriasyvirus.com/rooting.pdf
6) http://www.biologydirect.com/content/6/1/36
7) http://www.pnas.org/content/104/7/2043.full.pdf
8 )http://mmbr.asm.org/content/75/3/423.full.pdf
9) http://www.ncbi.nlm.nih.gov/pmc/articles/PMC135240/
10) W. Ford Doolittle, “Phylogenetic Classification and the Universal Tree,” Science 284 (1999): 2124–28. W. Ford Doolittle, “Lateral Genomics,” Trends in Biochemical Sciences 24 (1999): M5– M8. W. Ford Doolittle, “Uprooting the Tree of Life,” Scientific American 282 (February, 2000): 90– 95.
11) Carl Woese, “On the evolution of cells,” Proceedings of the National Academy of Sciences USA 99 (2002): 8742–47. Carl R. Woese, “A New Biology for a New Century,” Microbiology and Molecular Biology Reviews 68 (2004): 173–86. Eric Bapteste, Yan Boucher, Jessica Leigh, and W. Ford Doolittle, “Phylogenetic Reconstruction and Lateral Gene Transfer,” Trends in Microbiology 12 (2004), 406–11. E. Bapteste, E. Susko, J. Leigh, D. MacLeod, R. L. Charlebois, and W. F. Doolittle, “Do Orthologous Gene Phylogenies Really Support Tree-Thinking?” Biomed Central Evolutionary Biology 5 (2005), 33. Available online (June 2006) at: http://www.biomedcentral.com/content/pdf/1471-2148-5-33.pdf. S. L. Baldauf, “The Deep Roots of Eukaryotes,” Science 300 (2003), 1703–06.
12) http://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1005912
13) http://fire.biol.wwu.edu/cmoyer/zztemp_fire/biol345_F13/papers/Lombard_membranes_3domains_natrevmicro12.pdf
14) http://www.sciencedirect.com/science/article/pii/S0958166914002353
15) http://jeb.biologists.org/content/206/17/2911
16. Intracellular Calcium, page 577
17. ORIGIN, EVOLUTION, EXTINCTION, THE EPIC STORY OF LIFE ON EARTH, page 10
18. In Search of Cell History The Evolution of Life’s Building Blocks, page 96
Why Darwin was wrong about the tree of life
[url=https://www.sott.net/article/173647-Why-Darwin-was-wrong-about-the-tree
https://www.onezoom.org/
Last edited by Otangelo on Thu Oct 13, 2022 7:38 am; edited 91 times in total