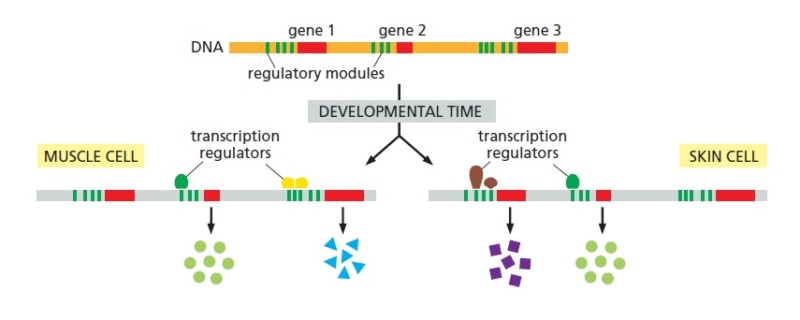
https://reasonandscience.catsboard.com/t2318-gene-regulatory-networks-controlling-body-plan-development
Animal evolution has been permitted or driven by gene regulatory evolution. 1
EVOLUTIONARY BIOSCIENCE AS REGULATORY SYSTEMS BIOLOGY 1
Never in the modern history of evolutionary bioscience have such essentially different ideas about how to understand evolution of the animal body plan been simultaneously current. The first is the classic neo-Darwinian concept that evolution of animal morphology occurs by means of small continuous changes in primary protein sequence which in general require homozygosity to effect phenotype. The second paradigm holds that evolution at all levels can be illuminated by detailed analysis of cis-regulatory changes in genes that are direct targets of sequence level selection, in that they control variation of immediate adaptive significance. An entirely different way of thinking is that the evolution of animal body plans is a system level property of the developmental gene regulatory networks (dGRNs) which control ontogeny of the body plan.
Just as development is a system property of the regulatory genome, causal explanation of evolutionary change in developmental process must be considered at a system level.
Never in the modern history of evolutionary bioscience have such essentially different ideas about how to understand evolution of the animal body plan been simultaneously current.
The first is the classic neo-Darwinian concept that evolution of animal morphology occurs by means of small continuous changes in primary protein sequence which in general require homozygosity to effect phenotype.
The second paradigm holds that evolution at all levels can be illuminated by detailed analysis of cis-regulatory changes in genes that are direct targets of sequence level selection, in that they control variation of immediate adaptive significance.
Both approaches often focus on changes at single gene loci, and both are framed within the concepts of population genetics.
An entirely different way of thinking is that the evolution of animal body plans is a system level property of the developmental gene regulatory networks (dGRNs) which control ontogeny of the body plan. It follows that gross morphological novelty required dramatic alterations in dGRN architecture, always involving multiple regulatory genes, and typically affecting the deployment of whole network subcircuits.
Because dGRNs are deeply hierarchical, and it is the upper levels of these GRNs that control major morphological features in development, a question dealt with below in this essay arises: how can we think about selection in respect to dGRN organization? The answers lie in the architecture of dGRNs and the developmental logic they generate at the system level, far from micro-evolutionary mechanism. While adaptive evolutionary variation occurs constantly in modern animals at the periphery of dGRNs, the stability over geological epochs of the developmental properties that define the major attributes of their body plans requires special explanations rooted deep in the structure/function relations of dGRNs.
Neo-Darwinian evolution is uniformitarian in that it assumes that all process works the same way, so that evolution of enzymes or flower colors can be used as current proxies for study of evolution of the body plan. It erroneously assumes that change in protein coding sequence is the basic cause of change in developmental program; and it erroneously assumes that evolutionary change in body plan morphology occurs by a continuous process. All of these assumptions are basically counterfactual. This cannot be surprising, since the neo-Darwinian synthesis from which these ideas stem was a pre-molecular biology concoction focused on population genetics and adaptation natural history, neither of which have any direct mechanistic import for the genomic regulatory systems that drive embryonic development of the body plan.
Sequence level changes in cis-regulatory modules controlling expression of these genes are demonstrated to be the cause of these variations, and in general they operate by altering the response of the cis-regulatory module to the pleisiomorphic spatial landscape of regulatory states. Evolutionary change in a cis-regulatory module controlling downstream gene expression is of course far less pleiotropically dangerous to the whole system than if either the coding region of the gene had been mutated or if the upstream regulatory landscape had been altered (Prud'homme et al., 2007).
The arguments are that essentially all evolutionary changes in morphology are at root cis-regulatory, which is indeed basically true; and that intra-modular mechanisms of cis-regulatory evolution will operate on similar principles wherever it occurs, also true. But these assumptions do not suffice to support the uniformitarian conclusion about body plan evolution: when the properties of the gene regulatory networks that actually generate body plans and body parts are taken into account, it can be seen that many entirely new and different mechanistic factors come into play. The result is that just as the paleontological record of evolutionary change in animal morphology is the opposite of uniformitarian (see the paper of D. Erwin in this collection), so, for very good reasons that are embedded in their structure/function relations, are the mechanisms of dGRN evolution.
This rather obvious argument gives rise to additional specific consequences, which taken together provide a new set of principles that apply to the mechanisms of body plan evolution (Britten and Davidson, 1971,Davidson and Erwin, 2006 and Peter and Davidson, in press). They are new in that none are specifically predicted by classical evolutionary theory.
No observations on single genes can ever illuminate the overall mechanisms of the development of the body plan or of body parts except at the minute and always partial, if not wholly illusory, level of the worm's eye view.
A distinguishing feature of dGRNs is their deep hierarchy, which essentially stems from the long sequence of successive spatial regulatory states required to be installed in building first the axial embryonic/larval body plan, and then constructing individual body parts
the universe of possible responses is vastly constrained by dGRN hierarchy at each level transition, inevitably resulting in what was classically termed “canalization” of the developmental process
For example, a frequently encountered type of subcircuit in upstream regions of dGRNs consists of two or three genes locked together by feedback inputs (Davidson, 2010). These feedback structures act to stabilize regulatory states, and there is a high penalty to change, in that interference with the dynamic expression of any one of the genes causes the collapse of expression of all, and the total loss from the system of their contributions to the regulatory state.
the development of an embryo is extremely canonical even though, as in sea urchins, the exact size of the egg, the temperature, or the amounts of many regulatory gene transcripts ( Materna et al., 2010) may vary considerably.
Whatever continuous variation occurs at individual cis-regulatory sequences, the dGRN circuit output preserves its Boolean morphogenetic character.
Therefore the action of selection differs across dGRN structure. Selection does not operate to produce continuous adaptive change except at the dGRN periphery.
the system level output is very impervious to change, except for catastrophic loss of the body part or loss of viability altogether. As long realized and much discussed in a non-mechanistic way in advance of actual knowledge of dGRN structure and function (for review see Gibson and Wagner, 2000), this imperviousness has something to do with whatever processes generate canalization and/or “buffering” of the genetic control system. We can now begin to understand canalization mechanistically in terms of dGRN hierarchy and subcircuit structure, as above, but in so far as “buffering” is taken to mean protection against “environmental fluctuations” as in many evolutionary mathematical models, it is irrelevant to animal embryonic processes, since in the main these depend not at all upon environmental inputs.
the fundamental role of upper level dGRNs is to set up in embryonic space a progressive series of regulatory states, which functionally define first the regions of the body with respect to its axes; then the location of the progenitor fields of the body parts; then the subparts of each body part.
In embryonic development the transcriptional processes mediated by dGRNs are intrinsically insensitive to varying cis-regulatory input levels.
1. https://www.pnas.org/content/115/38/E8909
Last edited by Otangelo on Wed Sep 07, 2022 8:52 am; edited 11 times in total