https://reasonandscience.catsboard.com/t2213-the-various-codes-in-the-cell
http://www.codebiology.org/database.pdf
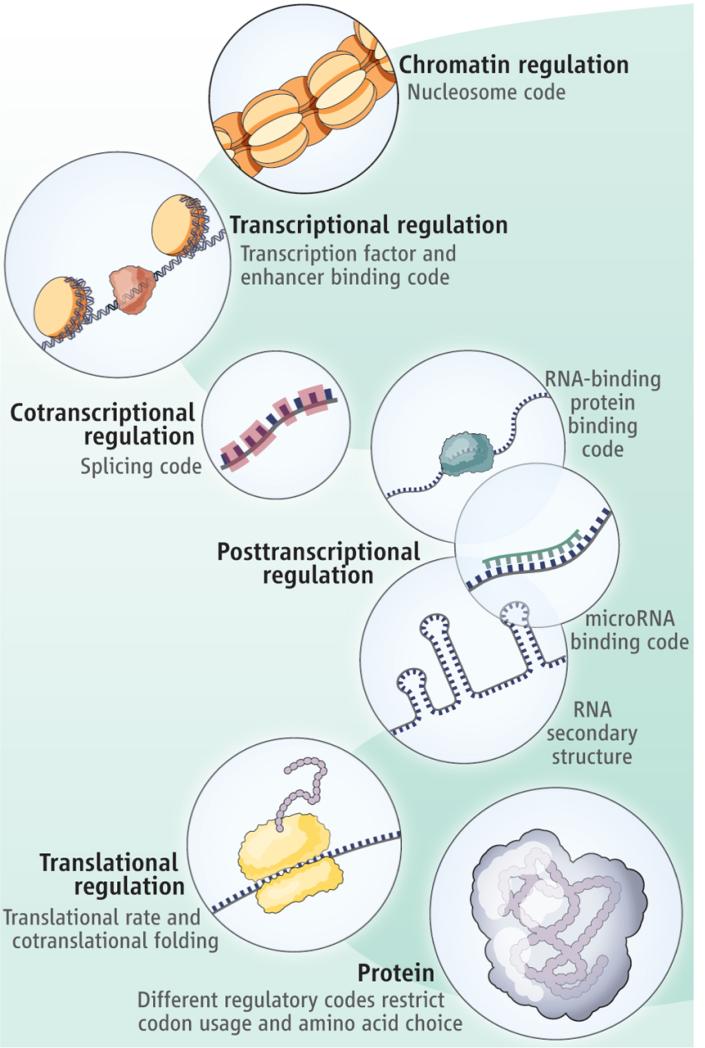
Not all codes other than the genetic code are considered epigenetic codes. Epigenetic codes specifically refer to modifications or marks that influence the accessibility, expression, and heritability of genetic information. These modifications can occur on DNA, histones, or other molecules involved in gene regulation. The term "regulatory codes" is sometimes used to describe various types of molecular information that guide cellular processes, gene expression, and interactions within a cell. Regulatory codes encompass a broader range of codes that dictate how cellular components interact, respond to signals, and carry out specific functions. While "epigenetic codes" and "regulatory codes" are terms that are often used in the context of cellular processes and gene regulation, the naming conventions can vary depending on the specific area of study and scientific literature.
The dichotomy between structural and regulatory codes is a suitable tool to start the systematic study of codes. Barbieri assumes the existence of three types of organic codes, namely manufacturing, signaling, and regulatory codes.
M.Barbieri: The genetic code is at the centre of life but it is not the only code that exists in living systems. Any organic code is a mapping between two independent worlds and requires molecular structures that act like adaptors, i.e., that perform two independent recognition processes. The adaptors are required because there is no necessary connection between two independent worlds, and a set of rules is required in order to guarantee the specificity of the connection. The adaptors, in short, are essential in all organic codes. They are the molecular fingerprints of the codes, and their presence in a biological process is a sure sign that that process is based on a code. In splicing and in signal transduction, for example, I have shown (in 2003) that there are true adaptors at work, and that means that those processes are based on splicing codes and on signal transduction codes. 2
Code Biology: the study of all Codes of Life
Biological cells depend not only on the physical matter but essentially on pre-programmed information and at least 234 informational code and language systems which are used to host and store that information, in order to arrange and produce the complex cellular structures essential for life, and keep the life-essential functions, that is the reproduction, metabolism. food uptake, intracellular organizational arrangement, growth and development, permanence, change, and adaptation. Living cells host multiple kinds of informational code systems that are used to store complex instructional/specifying information ( CSI ). All code systems, languages, information, and translation systems can be traced back to an intelligent origin. Evolution is not a driving force to explain the origin of cells and their language systems and programmed information content. Nor does physicochemical attraction explain the arrangement of nucleotides, molecules, and amino acids resulting in the formation of complex molecular machines and intracellular molecular production lines. The only alternative to intelligence is random self-assembly by unguided lucky events. Random chaotic events are however too unspecific to explain the extremely organized, controlled, error check and repair mechanisms, and factory-like production systems that cells host. Therefore, biological cells, cell code systems and coded information ( CSI ) have most probably a mind as the causal origin.
1. Regulation, governing, controlling, recruiting, interpreting, recognition, orchestrating, elaborating strategies, guiding, and instructing are all tasks of the gene regulatory network.
2. Such activity can only be exercised if no intelligence is present if the correct actions were pre-programmed by intelligence.
3. Therefore, most probably, the gene regulatory network was programmed by an intelligent agency.
The outstanding implication of the existence of organic codes in Nature comes from the fact that any code involves meaning and we need therefore to introduce in biology, with the standard methods of science, not only the concept of biological information but also that of biological meaning. The study on the organic codes, in conclusion, is bringing to light new mechanisms that operated in the history of life and new fundamental concepts. It is an entirely new field of research, the exploration of a vast and still largely unexplored dimension of the living world, the real new frontier of biology.
The irreducible interdependence of information generation and transmission systems
1. Codified information transmission system depends on:
a) A language where a symbol, letters, words, waves or frequency variations, sounds, pulses, or a combination of those are assigned to something else. Assigning meaning of characters through a code system requires a common agreement of meaning. Statistics, Semantics, Synthax, and Pragmatics are used according to combinatorial, context-dependent, and content-coherent rules.
b) Information encoded through that code,
c) An information storage system,
d) An information transmission system, that is encoding, transmitting, and decoding.
e) Eventually translation ( the assignment of the meaning of one language to another )
f) Eventually conversion ( digital-analog conversion, modulators, amplifiers)
g) Eventually transduction converting the nonelectrical signals into electrical signals
2. In living cells, information is encoded through at least 30 genetic, and almost 30 epigenetic codes that form various sets of rules and languages. They are transmitted through a variety of means, that is the cell cilia as the center of communication, microRNA's influencing cell function, the nervous system, the system synaptic transmission, neuromuscular transmission, transmission b/w nerves & body cells, axons as wires, the transmission of electrical impulses by nerves between brain & receptor/target cells, vesicles, exosomes, platelets, hormones, biophotons, biomagnetism, cytokines and chemokines, elaborate communication channels related to the defense of microbe attacks, nuclei as modulators-amplifiers. These information transmission systems are essential for keeping all biological functions, that is organismal growth and development, metabolism, regulating nutrition demands, controlling reproduction, homeostasis, constructing biological architecture, complexity, form, controlling organismal adaptation, change, regeneration/repair, and promoting survival.
3. The origin of such complex communication systems is best explained by an intelligent designer. Since no humans were involved in creating these complex computing systems, a suprahuman super-intelligent agency must have been the creator of the communication systems used in life.
The concept of cross-talk between different cellular codes suggests a high level of coordination and purpose in the design of biological systems. The intricate interactions and communication between these codes indicate a sophisticated and integrated design that allows cells to respond to environmental cues, maintain homeostasis, and carry out their functions effectively. The complexity and specificity of these codes, and their ability to interact with each other, cannot be adequately explained by a step-wise, gradual, and unguided evolutionary process. The examples of cross-talk between the characterized codes highlight how different cellular processes are intertwined, and changes in one code can have cascading effects on others. For these complex regulatory systems to function coherently, all components must be in place simultaneously and working together. The likelihood of all these intricate mechanisms evolving independently through random mutations and natural selection is extremely low, given the precise coordination required for cellular function.
Many of the over 223 epigenetic codes can cross-talk with each other due to the interconnectedness of cellular processes. Cross-talk refers to the communication and interaction between different signaling pathways and regulatory mechanisms in the cell.
The 31 Genetic Codes
1. The Acetylation Code: A post-translational modification involving the addition of an acetyl group to proteins, influencing their function.
2. The Acoustic codes: Patterns of sound waves processed by the auditory system, conveying information about the environment.
3. The Adhesion Code: Molecular interactions that determine how cells adhere to each other and to surfaces.
4. The Adenylation Code: A process where adenosine monophosphate (AMP) is added to various molecules, often part of activation processes.
5. The Allosteric Code: The intricate regulation of protein function by molecules binding to sites other than the active site.
6. The Angiotensin Receptor Code: Signaling pathways involving angiotensin receptors that play a role in blood pressure regulation.
7. The Antioxidant Code: The mechanisms and molecules that protect cells from oxidative damage.
8. The Antibiotic Resistance Code: The genetic and biochemical basis of bacterial resistance to antibiotics.
9. The Apoptosis Code: Genetic and molecular mechanisms that govern programmed cell death.
10. The Archetype Code: Patterns and symbols that hold universal significance in human culture and psychology.
11. The Arrestin Receptor Code: The role of arrestin proteins in regulating G-protein-coupled receptor signaling.
12. The Assembly Code: Molecular rules governing the proper assembly of multi-component complexes.
13. The Auxin Metabolism Code: How auxin hormones are synthesized, transported, and regulated in plants.
14. The Axon Guidance Codes: Molecular signals guiding the growth of axons in neural development.
15. The Autocrine Signaling Code: Mechanisms by which cells release signaling molecules that affect their own activity.
16. The Autophagy Code: Molecular pathways that regulate autophagy, a process for recycling cellular components.
17. The BAFF Immune Code: Signaling pathways involving B-cell activating factor (BAFF) in the immune system.
18. The Bile Acid Code: The roles and regulation of bile acids in digestion and metabolism.
19. The Binaural Code: Neural processing of auditory information from both ears to localize sound sources.
20. The Bioelectric Code: Patterns of electrical signaling that influence cellular behavior and development.
21. The Biophoton Code: Hypothetical biophotonic emissions from living organisms and their potential significance.
22. The Biosynthetic Code: Genetic and biochemical pathways responsible for producing complex molecules.
23. The Universal Brain Code: The underlying principles governing neural networks and cognitive processes.
24. The Cadherin Neuronal Code: The role of cadherin molecules in neuronal adhesion and circuit formation.
25. The Calcium Signaling Code: Molecular pathways that regulate calcium-mediated intracellular signaling.
26. The Cell Cycle Checkpoint Code: Mechanisms ensuring proper progression through the cell cycle.
27. The Cell-Cell Communication Code: Molecular signals that mediate communication between neighboring cells.
28. The Cell Access Code: The regulation of cellular uptake of nutrients, ions, and other molecules.
29. The Cell Fate Determination Code: Molecular processes that dictate a cell's developmental fate.
30. The Cell Migration Code: Molecular cues guiding cells in movement during development, wound healing, etc.
31. The Cell Polarity Code: Molecular pathways that establish and maintain cellular asymmetry.
32. The Cell Surface Recognition Code: Molecules that mediate cell interactions through recognition of surface markers.
33. The Cerebral Resistance Code: The molecular mechanisms that govern the brain's ability to resist damage or adapt to challenges.
34. The Chitin (Defense) Code:Molecular processes related to the synthesis, modification, and utilization of chitin in defense mechanisms, often found in insects and other organisms.
35. The Chaperone Code: The role of chaperone proteins in assisting proper protein folding.
36. The Chromatin Code: Histone modifications and other factors that regulate chromatin structure and gene expression.
37. The Chromosomal Imprinting Code: Epigenetic marks on specific genes inherited from one parent that affect gene expression patterns based on parental origin.
38. The Chromosome Segregation Code: Molecular mechanisms that ensure accurate distribution of chromosomes during cell division to prevent errors in genetic inheritance.
39. The Circular motif ( ribosome) Code: Consists of specific RNA sequences in circular RNA molecules, which may have regulatory roles in gene expression.
40. The Coactivator/corepressor/epigenetic Code: Describes how coactivator and corepressor proteins, along with epigenetic modifications, influence gene expression regulation.
41. The Code of human language: The intricate system of sounds, words, and grammar rules that humans use to communicate complex thoughts and ideas.
42. The Cohesin-Dockerin Code: Refers to the interaction between cohesin and dockerin domains in some bacteria, which plays a role in cellulosome assembly and cell attachment.
43. The Cytokine Codes: Signaling molecules produced by immune cells that influence cellular communication and responses during immune and inflammatory processes.
44. The Compartment Code: Molecular processes that determine the organization and segregation of cellular components within specific subcellular compartments.
45. The Cholesterol Recognition/Mirror Code: The interaction of cholesterol with proteins and its impact on cellular processes.
46. The Cilia Code: Molecular mechanisms underlying the structure and function of cilia.
47. The Circardian Rhythm Codes: Genetic and molecular pathways that regulate circadian rhythms.
48. The Cytoskeleton Code: The organization and dynamics of the cytoskeleton and its role in cell function.
49. The Connexin Code: encompasses the intricate molecular mechanisms involving connexin proteins, vital components of gap junctions.
50. The DNA Repair / Damage Codes: Molecular pathways that repair DNA damage and maintain genomic integrity.
51. The DNA-Binding Code: Molecular interactions between proteins and DNA sequences.
52. The DNA methylation Code: Epigenetic modifications involving the addition of methyl groups to DNA.
53. The DNA Zip Code / Peripheral Targeting Code: DNA sequences that dictate subnuclear localization.
54. The Discriminator Codes: involve molecular mechanisms that distinguish between different cellular components, signals, or states.
55. The Differentiation Code: Signals and factors that drive cells to specialize into specific cell types.
56. The Domain substrate specificity Code of Nonribosomal peptide synthetases (NRPS): Mechanisms underlying the synthesis of complex peptides in bacteria.
57. The Endocytosis Code: Molecular mechanisms governing the process of cellular endocytosis.
58. The Endocrine Signalling Codes: Signaling pathways involving hormones and their effects on target cells.
59. The (Epigenetic) Body Plan Code: Epigenetic mechanisms shaping the development of body structures.
60. The Epigenetic Cancer Code: Epigenetic changes associated with cancer development and progression.
61. The Epidermal Growth Factor (EGF) Code: Signaling pathways involving epidermal growth factor and its receptors.
62. The Epitranscriptomic Code: Post-transcriptional modifications to RNA molecules that affect their function.
63. The Error correcting Code: Mechanisms that ensure proper DNA replication and repair errors.
64. The Epigenetic Imprinting Code: Epigenetic modifications that lead to parent-of-origin-specific gene expression.
65. The Export & Exit Codes: Molecular mechanisms that direct proteins and RNA out of the cell.
66. The Extracellular Matrix (ECM) Code: Composition and organization of the ECM and its impact on cell behavior.
67. The Forkhead Transcription Factor Code: Functions and regulation of the forkhead box (FOX) family of transcription factors.
68. The General Neural Codes: Neural patterns representing sensory information or motor commands.
69. The Genetic Recombination Codes: Mechanisms of genetic recombination that create genetic diversity.
70. The Genomic Code: Genetic information and the relationship between nucleotide sequences and phenotypes.
71. The Genomic regulatory Code: Non-coding regions of DNA that control gene expression.
72. The G-Protein Coupled Receptor (GPCR) Code: Molecular properties and signaling pathways of GPCRs.
73. The Gli Codes: Signaling pathways involving the Gli family of transcription factors.
74. The Glioma Code: Genetic and molecular aspects of glioma development and progression.
75. The Glycomic Code: Diversity and roles of glycan structures in cellular processes.
76. The Growth Codes: Molecular cues and pathways that regulate cell growth and proliferation.
77. The Hearing Code: Neural coding of auditory information and sound perception.
78. The Hedgehog Signaling Code: Signaling pathways involving Hedgehog proteins and their receptors.
79. The Heterochromatin Code: Molecular marks and proteins that regulate heterochromatin formation.
80. The Histone Sub-Code: Specific modifications of histone proteins that influence chromatin structure.
81. The Histone Variants Code: Variations in histone protein sequences that affect chromatin dynamics.
82. The Homeokinetic Muscle Code: Mechanisms underlying muscle homeostasis and adaptation.
83. The Honey Bee Dance Code: Communication through dances that inform other bees of food sources.
84. The Host Defense Code: Mechanisms by which the host defends against pathogens and infections.
85. The Hormone Receptor Code: Molecular interactions between hormones and their target receptors.
86. The HOX Code Pattern Formation: HOX gene expression patterns that guide embryonic development.
87. The Hypothalamic Code: Signaling pathways and neuropeptides involved in hypothalamic regulation.
88. The Identity Code: Mechanisms that define the unique identity of cells, tissues, and organisms.
89. The immune response code, or language: Molecular signals and pathways that orchestrate immune responses.
90. The Immune T-cell Codes: Receptor interactions and signals involved in T-cell immune responses.
91. The Importin Codes: Processes involving importin proteins that facilitate nuclear transport.
92. The Indole Physiological Code: The role of indole molecules in bacterial physiology and behavior.
93. The Inositol Phosphate Code: Signaling pathways involving inositol phosphates and their effects.
94. The Irisin (Muscle) Code: The hormone irisin and its effects on metabolism and energy expenditure.
95. The Karyotype Code: The chromosomal arrangement and number characteristic of a species.
96. The Lamin Code : Molecular interactions involving nuclear lamins that impact nuclear structure.
97. The Latency Behaviour Codes: Behaviors associated with latency periods in psychological development.
98. The Lipid Codes: Molecular structures and signals involving lipids in cellular processes.
99. The Magnitude Neuronal Codes: Neural responses that encode the intensity or magnitude of stimuli.
100. The Meiosis Codes: Molecular processes that ensure proper chromosome segregation during meiosis.
101. The Membrane Code: Properties of cellular membranes and their interactions with molecules.
102. The Memory Code: Neural mechanisms that encode and retrieve memories.
103. The Metabolic Signaling Code: Molecular pathways that link cellular metabolism with signaling.
104. The Methylation Code: The role of DNA and protein methylation in gene expression and regulation.
105. The Microbiome Code: Genetic and functional diversity of microbial communities in and on the body.
106. The Micro-RNA Codes: Small RNA molecules that regulate gene expression at the post-transcriptional level.
107. The Mnemonic codes: Mechanisms by which memory is encoded and retrieved.
108. The Modularity Codes: Molecular units and patterns that enable the assembly of complex structures.
109. The Molecular Codes: A collective term encompassing various specific codes governing cellular processes.
110. The Morphogenetic Code: Signaling molecules that direct tissue and organ development.
111. The Myelin code: Molecular cues that regulate myelin formation and maintenance in the nervous system.
112. The Molecular Recognition Code: Molecular interactions that enable specific recognition between molecules.
113. The Navigation / Orientation / Movement Codes: Neural pathways and signals that guide navigation and movement.
114. The Neuronal Activity-Dependent Gene Expression Code
115. The Neuronal Code for Reading: Neural pathways and processes involved in reading.
116. The Neuronal Hippocampal Codes: contribute to the organization and regulation of neural activity within the hippocampus.
117. The Neural Motion Codes: Neural patterns that encode and control motor movements.
118. The Neural Perception & Recognition Codes: Neural responses that process and recognize sensory information.
119. The Neural, Social Information Code: How neural circuits process social cues and interactions.
120. The neuronal Oscillatory /Frequency Codes: Neural oscillations and frequencies that regulate brain activity.
121. The Neuronal spike-rate Code: involves patterns of neuronal firing rates that convey information within the nervous system.
122. The Neuronal Taste Code: Neural pathways and signals that encode taste perception.
123. The Neuron Light Code: signifies rapid patterns of neuronal activity that transmit information through light-like signals, facilitating neural communication.
124. The Neuropeptide Code: The role of neuropeptides in neural signaling and behavior.
125. The NF-kappa-B Code: Molecular pathways involving the NF-kappa-B family of transcription factors.
126. The Nitric Oxide (NO) Signaling Code: Signaling pathways involving nitric oxide and its effects.
127. The N-Glycan Code: Diversity and roles of N-linked glycan structures in cellular processes.
128. The Nomenclatural Code: Rules for naming biological taxa and species.
129. The Non-Ribosomal Code: Mechanisms of protein synthesis by non-ribosomal peptide synthetases.
130. The Notch Code: Signaling pathways involving Notch receptors and their role in development.
131. The Nuclear Signalling Code: Molecular pathways involving nuclear signaling events.
132. The Nutrient Transport Code: Molecular mechanisms for transporting nutrients across cell membranes.
133. The Olfactory Code: Neural coding of olfactory information and smell perception.
134. The Nucleosome Code: involves molecular arrangements that dictate DNA packaging and gene accessibility using nucleosomes.
135. The Nucleotide Sequence Codes: encompass genetic information encoded in DNA sequences, shaping traits and functions.
136. The Nutrient Sensing Code: involves molecular processes that detect and respond to nutrient levels in cells, guiding metabolic and physiological responses.
137. The Omega Leaf Code: Hypothetical code indicating plant leaf arrangement based on Fibonacci numbers.
138. The Operon Code: Genetic regulation of bacterial operons and coordinated gene expression.
139. The Orthographic Reading Code: Neural processes that enable reading and recognizing written words.
140. The Pattern Formation Code: Molecular mechanisms that create ordered patterns during development.
141. The Phagocytosis Codes: Cellular processes and molecular cues governing phagocytosis.
142. The Pheromone Codes: Molecular signals that communicate information between individuals of the same species.
143. The Phonological Codes: Neural representation and processing of speech sounds.
144. The Phosphatase Code: Regulation of cellular processes by protein phosphatases.
145. The Physiological Coregulator Code: Molecular factors that modulate physiological responses.
146. The Phosphorylation Code: Regulation of protein function through phosphorylation by kinases.
147. The Phosphorylation-Dependent Protein Interaction Code: Proteins that bind to phosphorylated targets, regulating interactions.
148. The Phospholipid Code: Role of specific phospholipids in cellular membranes and signaling.
149. The Phosphoserine Code: Functions of proteins and pathways involving phosphoserine residues.
150. The Photoreceptor Sensory Code: Neural coding of visual information and light perception.
151. The Photosynthesis Code: Molecular mechanisms of photosynthesis and energy conversion.
152. The Plant Cell Wall Code: Composition and roles of plant cell walls in growth and defense.
153. The Plant Communication Codes encompass molecular processes and signaling mechanisms by which plants exchange information and respond to various environmental cues
154. The Post-translational modification Code for transcription factors: Modifications that affect the activity and function of transcription factors.
155. Protein Kinase Codes: Families of protein kinases and their roles in cellular signaling.
156. The Poly(Adenylation) Code: Role of polyadenylation in mRNA stability and translation.
157. The Polycomb & Trithorax Codes: Complexes involved in epigenetic regulation and gene expression.
158. The Polysaccharide Codes: Diversity and roles of polysaccharides in cellular processes.
159. The Post-translational Modification Codes refer to a diverse array of molecular processes that modify proteins after they are synthesized.
160. The Presynaptic Vesicle Code: Molecular processes involving neurotransmitter-containing vesicles.
161. The Protein Allosteric Code: Mechanisms by which proteins switch between different conformations.
162. The Protein Binding Code: Molecular interactions that allow proteins to bind to specific partners.
163. The Protein Folding Code: Molecular principles that dictate protein folding into functional conformations.
164. The Protein Interaction Code: Specific molecular interactions that govern protein-protein interactions.
165. The Protein Phosphorylation Code: Regulation of protein function by reversible phosphorylation.
166. The Protein Secretory Code: Processes and signals that guide protein secretion from cells.
167. The Protein Translocation Code: involves mechanisms governing the movement of proteins within cells to their designated locations.
168. The Proteomic Code: Governs processes that regulate protein degradation and renewal within cells.
169. The Regulatory Organogenesis Codes: refer to molecular mechanisms that oversee the development of tissues and organs.
170. The Regulatory Network Codes encompass intricate signaling networks that control cellular responses.
171. The Renal Codes pertain to molecular processes specific to kidney function and regulation.
172. The Representation Codes involve molecular mechanisms underlying information encoding and processing.
173. The Retinal Codes concern molecular events and patterns of activity within the retina, vital for vision.
174. The RNA-Interference Codes relate to the regulatory roles of RNA interference in gene expression.
175. The RNA Polymerase Modification Codes involve modifications affecting RNA polymerase function in transcription.
176. The RNA Recognition Code involves molecular interactions between RNA molecules and other cellular components.
177. The Redox Code encompasses processes influenced by cellular redox (oxidation-reduction) states.
178. The Regeneration Codes involve molecular cues and mechanisms that guide tissue and organ regeneration.
179. The Retinoic Acid Signaling Code relates to signaling pathways activated by retinoic acid and their effects.
180. The Ribonucleic Acid Modification Code (RNA Modification Code) pertains to post-transcriptional RNA modifications.
181. The Ribosomal Code concerns molecular interactions and functions of ribosomal components.
182. The Riboswitch Code involves RNA structures that modulate gene expression in response to ligand binding.
183. The Quorum Sensing Code: Bacterial communication through the release and sensing of signaling molecules.
184. The RNA Code: Genetic information carried by RNA molecules, including coding and non-coding roles.
185. The RNA Editing Code: Post-transcriptional modifications that change RNA sequences.
186. The RNA Modification Code: Various modifications that alter the structure and function of RNA.
187. The RNA Splicing Code: Processes that remove introns and join exons in mRNA molecules.
188. The RNA Transport Code: Mechanisms that guide RNA molecules to specific cellular locations.
189. The Semaphoring Codes: Signaling pathways involving semaphorin proteins and their role in axon guidance.
190. The Serotonin Code involves molecular processes related to the signaling and effects of serotonin, a neurotransmitter influencing mood and behavior.
191. The Sexual Dimorphic Codes Codes encompass molecular mechanisms underlying the development of gender-specific traits.
192. The Signal Integration Codes involve processes that combine multiple cellular signals for coordinated responses.
193. The Sperm RNA Code relates to the unique RNA molecules found in sperm cells, potentially influencing early development.
194. The Signal Integration Codes encompass processes that harmonize and interpret various cellular signals.
195. The Synaptic Adhesive Code refers to molecular interactions that guide the adhesion and connectivity of neurons at synapses.
196. The Stem Cell Code encompasses molecular cues that regulate stem cell behavior and differentiation.
197. The Sumoylation Code relates to the post-translational modification known as sumoylation, which influences protein activity and interactions.
198. The Skin Inflammation Code Code involves molecular pathways that contribute to inflammation and immune responses in the skin.
199. The Sodium/Calcium Channel Gating Code involves molecular mechanisms that regulate the opening and closing of sodium and calcium ion channels.
200. The Speech Code relates to neural and cognitive processes underlying the production and comprehension of speech.
201. The Spliceosome Code: Molecular machinery responsible for mRNA splicing.
202. The Substrate Specificity Code pertains to the molecular factors determining the selection and interaction of enzymes with specific substrates.
202. The Sugar Code encompasses the roles of sugar molecules in cell-cell interactions, signaling, and recognition.
203. The Sulfation Code involves the addition of sulfate groups to molecules, influencing their functions and interactions.
204. The Sulfur Code relates to the roles and effects of sulfur-containing molecules in various cellular processes.
205. The Synaptic Code: Molecular and cellular processes that underlie synaptic transmission.
206. The Toll-like Receptor Codes: Signaling pathways involving Toll-like receptors in the immune system.
207. The Transcription Factor Binding Code: Mechanisms by which transcription factors interact with DNA.
208. The Transcriptional Regulatory Code: Molecular mechanisms that control gene expression.
209. The Transmembrane Protein Code: Structure and function of proteins that span cellular membranes.
210. The tRNA Code: Transfer RNA molecules that decode mRNA into protein sequences.
211. The Ubiquitin Code: Post-translational modification involving ubiquitin and protein degradation.
212. The Tactile Neural Codes involve patterns of neural activity that transmit tactile sensations and touch-related information.
213. The Talin Code refers to the molecular processes related to talin protein's role in cell adhesion and signaling.
214. The Terpene Biosynthesis Code involves the genetic and biochemical pathways responsible for producing terpenes in organisms.
215. The Thermal / Temperature Neuronal Codes relate to patterns of neural activity that convey temperature-related sensory information.
216. The Translational Control Code: Regulation of gene expression at the level of translation initiation and elongation.
217. The Tight Junction Codes pertain to molecular interactions and functions of tight junctions, important for cell barrier formation.
218. The Tissue Code encompasses molecular characteristics specific to different types of tissues in multicellular organisms.
219. The Tissue Memory Code involves molecular processes that contribute to the memory or lineage history of tissues.
220. The Tubulin Code involves modifications and interactions of tubulin proteins, crucial for microtubule function.
221. The Visual Code involves neural and molecular processes that enable visual perception and processing.
222. The Wobbling Base Pairing Code relates to flexible pairing of bases in DNA/RNA, affecting translation accuracy.
223. The Zinc Finger Code: DNA-binding motifs formed by zinc finger proteins.
1) http://www.garlandscience.com/res/pdf/9780815341291_ch08.pdf
2. Barbieri: Organic codes
Last edited by Otangelo on Mon Mar 04, 2024 6:04 am; edited 91 times in total (Reason for editing : cebola de oleo)