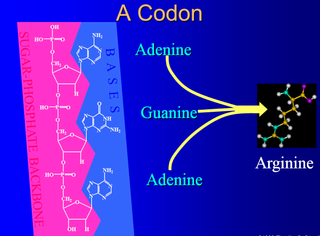
https://reasonandscience.catsboard.com/t2057-origin-of-translation-of-the-4-nucleic-acid-bases-and-the-20-amino-acids-and-the-universal-assignment-of-codons-to-amino-acids
My comment: What must be explained, is the arrangement of the codons in the standard codon table which is highly non-random, and serves to translate into the amino acid sequence to make proteins, and the origin of the assignment of the 64 triplet codons to the 20 amino acids. That is the origin of its translation. The origin of an alphabet through the triplet codons is one thing, but on top, it has to be translated to another " alphabet " constituted through the 20 amino acid sequence. That is, to explain the origin of the capability to translate the English language into Chinese. On top of that, the machinery itself to promote the process itself has also to be explained, that is the hardware. When humans translate English to Chinese, for example, we recognize the English word, and the translator knows the equivalent Chinese symbol and writes it down.

My comment: In the cell, Aminoacyl tRNA synthetase recognizes the triplet anticodon of the tRNA, and attach the equivalent amino acid to the tRNA. How could random chemical reactions have produced this recognition? Let's suppose rather than intelligence, the chance was the mechanism. The imaginary cell would have to select randomly any of the amino acids, restrict by an unknown mechanism to the 20 used for life, since there are more out there, select by an unknown mechanism only left-handed ones, and make a test drive and produce a polymer chain and see what happens. Some theories try to explain the mechanism, but they all remain unsatisfactory. Obviously. Furthermore, Aminoacyl tRNA synthetase is complex enzymes. For what reason would they have come to be, if the final function could only be employed after the whole translation process was set in place, with a fully functional ribosome being able to do its job? Remembering the catch22 situation, since they are by themselves made through the very own process in question?
And even it lets say, the whole thing was fully working and in place, what good would it be for without all the other parts required, that is, the DNA double helix, its compactation through histones and chromatins and chromosomes, its highly complex mechanism of information extraction and transcription into mRNA? Had the whole process, that is INITIATION OF TRANSCRIPTION, CAPPING, ELONGATION, SPLICING, CLEAVAGE, POLYADENYLATION, AND TERMINATION, EXPORT FROM THE NUCLEUS TO THE CYTOSOL, INITIATION OF PROTEIN SYNTHESIS (TRANSLATION), COMPLETION OF PROTEIN SYNTHESIS AND PROTEIN FOLDING, and its respective machinery not have to be all in place? Does that not constitute an interdependent and irreducibly complex system?
http://web.archive.org/web/20131209121838/http://gencodesignal.org/faq/
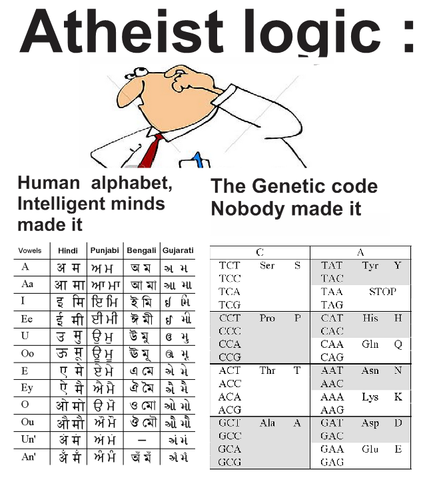
The confusion comes from the ambiguity in using the term “genetic code”. Here is a quote from Francis Crick, who seems to be the one who coined this term:
Unfortunately the phrase “genetic code” is now used in two quite distinct ways. Laymen often use it to mean the entire genetic message in an organism. Molecular biologists usually mean the little dictionary that shows how to relate the four-letter language of the nucleic acids to the twenty-letter language of the proteins, just as the Morse code relates the language of dots and dashes to the twenty-six letters of the alphabet… The proper technical term for such a translation is, strictly speaking, not a code but a cipher. In the same way the Morse code should really be called the Morse cipher. I did not know this at the time, which was fortunate because “genetic code” sounds a lot more intriguing than “genetic cipher” (from “What Mad Pursuit”, 1988)
Transfer RNA, Delivery Vehicle for Amino Acids 11
While the mRNA is being processed by the ribosome in order to assemble amino acids into a protein, how will these amino acids actually be brought into the proper order? There does not seem to be any innate attraction or affinity between an amino acid and the RNA letters which code for it. In the early research after the Watson-Crick breakthrough, it became apparent that there must be intermediates to bring the amino acids to the ribosome in proper order. Two such vital go-betweens were finally located. One serves as a transport molecule. It is called transfer-RNA, which is a different form of RNA from that which has been described. Transfer-RNA, written tRNA, is a comparatively short chain of RNA containing some seventy-five or eighty ribonucleotides.
The RNA strand doubles back on itself, and base-pairs with its own chain in some places. The overall shape of the tRNA molecule in some ways resembles a key or a cloverleaf. If tRNA is to do its job properly, the shape must be very precise, and this seems to depend in part upon the right temperature and the correct concentration of certain ions (e.g., magnesium and sodium) in the cell fluid. Transfer-RNA is perfectly fitted for its mission. First of all, each tRNA type attaches to only one variety of the twenty amino acids. Secondly, the particular tRNA delivers that amino acid in the proper sequence for the forming protein. This is possible because the tRNA molecule has at one end a special RNA triplet of code letters which match the mRNA codon which specifies that particular amino acid. When these complementary codons come together by base-pairing, the amino acid being transported by that tRNA is thus in position to be linked to the growing protein chain in the correct order. All this takes place at the ribosome, which is like a mobile assembly machine as it moves along the mRNA strand (or as the mRNA tape passes through the ribosomes).
https://www.youtube.com/watch?v=D5vH4Q_tAkY
https://vimeo.com/114101147
The cell converts the information carried in an mRNA molecule into a protein molecule. This feat of translation was a focus of attention of biologists in the late 1950s, when it was posed as the “coding problem”: how is the information in a linear sequence of nucleotides in RNA translated into the linear sequence of a chemically quite different set of units—the amino acids in proteins?
The first scientist after Watson and Crick to find a solution of the coding problem, that is the relationship between the DNA structure and protein synthesis was Russian physicist George Gamow. Gamow published in the October 1953 issue of Nature a solution called the “diamond code”, an overlapping triplet code based on a combinatorial scheme in which 4 nucleotides arranged 3-at-a-time would specify 20 amino acids. Somewhat like a language, this highly restrictive code was primarily hypothetical, based on then-current knowledge of the behavior of nucleic acids and proteins. 3
The concept of coding applied to genetic specificity was somewhat misleading, as translation between the four nucleic acid bases and the 20 amino acids would obey the rules of a cipher instead of a code. As Crick acknowledged years later, in linguistic analysis, ciphers generally operate on units of regular length (as in the triplet DNA scheme), whereas codes operate on units of variable length (e.g., words, phrases). But the code metaphor worked well, even though it was literally inaccurate, and in Crick’s words, “‘Genetic code’ sounds a lot more intriguing than ‘genetic cipher’.”
An mRNA Sequence Is decoded in sets of three nucleotides
Once an mRNA has been produced by transcription and processing, the information present in its nucleotide sequence is used to synthesize a protein. Transcription is simple to understand as a means of information transfer: since DNA and RNA are chemically and structurally similar, the DNA can act as a direct template for the synthesis of RNA by complementary base-pairing. As the term transcription signifies, it is as if a message written out by hand is being converted, say, into a typewritten text. The language itself and the form of the message do not change, and the symbols used are closely related.
In contrast, the conversion of the information in RNA into protein represents a translation of the information into another language that uses quite different symbols. Moreover, since there are only 4 different nucleotides in mRNA and 20 different types of amino acids in a protein, this translation cannot be accounted for by a direct one-to-one correspondence between a nucleotide in RNA and an amino acid in protein. The nucleotide sequence of a gene, through the intermediary of mRNA, is translated into the amino acid sequence of a protein. This code was deciphered in the early 1960s.
Question: how did the translation of the triplet anticodon to amino acids, and its assignment, arise? There is no physical affinity between the anticodon and the amino acids. What must be explained, is the arrangement of the codon " words " in the standard codon table which is highly non-random, redundant and optimal, and serves to translate the information into the amino acid sequence to make proteins, and the origin of the assignment of the 64 triplet codons to the 20 amino acids. That is the origin of its translation. The origin of an alphabet through the triplet codons is one thing, but on top, it has to be translated to another " alphabet " constituted through the 20 amino acids. That is to explain the origin of the capability to translate the English language into Chinese. We have to constitute the English and Chinese language and symbols first, in order to know its equivalence. That is a mental process.
tRNA's are adapter molecules. A cipher or translation system permits the translation of the information from DNA’s four-character base sequences into the twenty-character “language” of proteins. the cell needs a means of translating and expressing the information stored on DNA. Groups of three nucleotides (called codons) on the mRNA specify the addition of one of the twenty protein-forming amino acids during the process of protein synthesis. Other scientists discovered that the cell uses a set of adapter molecules to help convert the information on mRNA into proteins.
how did biological specificity and functional information arise? The proteins would have to possess the correct sequences of amino acids in order to be able to unwind and copy genetic information; the ribosomal proteins and RNAs would need to be sequenced precisely in order to fold into subunits that fit together to form a functional ribosome; the transfer RNAs would have to mediate specific associations in order to convert the random sequences of bases on the polynucleotides into specific amino-acid sequences; and the sequences of amino acids thus produced would have to be arranged precisely in order to fold into stable three-dimensional structures.

The sequence of nucleotides in the mRNA molecule is read in consecutive groups of three. RNA is a linear polymer of four different nucleotides, so there are 4 x 4 x 4 = 64 possible combinations of three nucleotides: the triplets AAA, AUA, AUG, and so on. However, only 20 different amino acids are commonly found in proteins. Either some nucleotide triplets are never used, or the code is redundant and some amino acids are specified by more than one triplet. The second possibility is, in fact, the correct one, as shown by the completely deciphered genetic code shown below:


Each group of three consecutive nucleotides in RNA is called a codon, and each codon specifies either one amino acid or a stop to the translation process.
In principle, an RNA sequence can be translated in any one of three different reading frames, depending on where the decoding process begins (Figure below). However, only one of the three possible reading frames in an mRNA encodes the required protein. We see later how a special punctuation signal at the beginning of each RNA message sets the correct reading frame at the start of protein synthesis.
AUG is the Universal Start Codon. Nearly every organism (and every gene) that has been studied uses the three ribonucleotide sequence AUG to indicate the "START" of protein synthesis (Start Point of Translation).
The same interrogation point goes here: Why and how should natural processes have " chosen " to insert a punctuation signal, a Universal Start Codon in order for the Ribosome to " know " where to start translation? This is essential in order for the machinery to start translating at the correct place.
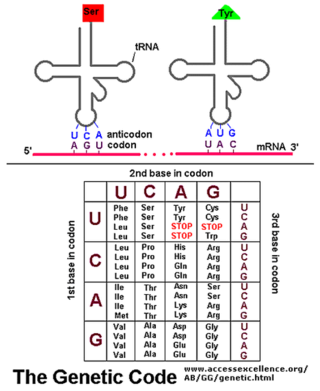
Note that three codons are referred to as STOP codons: UAA, UAG, and UGA. These are used to terminate translation; they indicate the end of the gene's coding region.

tRNA Molecules match Amino Acids to codons in mRNA
The codons in an mRNA molecule do not directly recognize the amino acids they specify: the group of three nucleotides does not, for example, bind directly to the amino acid. Rather, the translation of mRNA into protein depends on adaptor molecules that can recognize and bind both to the codon and, at another site on their surface, to the amino acid. These adaptors consist of a set of small RNA molecules known as transfer RNAs (tRNAs), each about 80 nucleotides in length.
RNA molecules can fold into precise three-dimensional structures, and the tRNA molecules provide a striking example. Four short segments of the folded tRNA are double-helical, producing a molecule that looks like a cloverleaf when drawn schematically. See below:
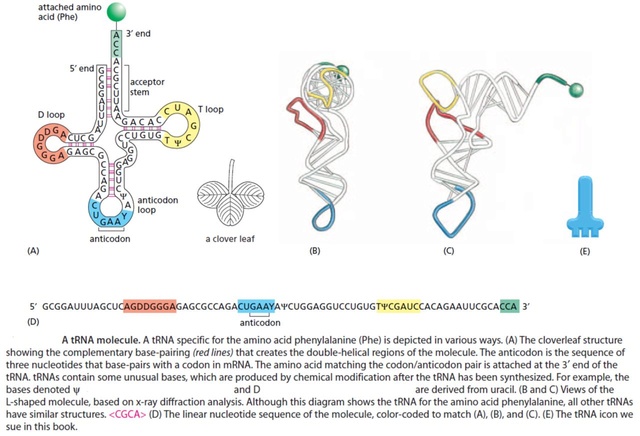
For example, a 5"-GCUC-3" sequence in one part of a polynucleotide chain can form a relatively strong association with a 5"-GAGC-3" sequence in another region of the same molecule. The cloverleaf undergoes further folding to form a compact L-shaped structure that is held together by additional hydrogen bonds between different regions of the molecule. Two regions of unpaired nucleotides situated at either end of the L-shaped molecule are crucial to the function of tRNA in protein synthesis. One of these regions forms the anticodon, a set of three consecutive nucleotides that pairs with the complementary codon in an mRNA molecule. The other is a short single- stranded region at the 3" end of the molecule; this is the site where the amino acid that matches the codon is attached to the tRNA. The genetic code is redundant; that is, several different codons can specify a single amino acid . This redundancy implies either that there is more than one tRNA for many of the amino acids or that some tRNA molecules can base-pair with more than one codon. In fact, both situations occur. Some amino acids have more than one tRNA and some tRNAs are constructed so that they require accurate base-pairing only at the first two positions of the codon and can tolerate a mismatch (or wobble) at the third position . See below

Wobble base-pairing between codons and anticodons. If the nucleotide listed in the first column is present at the third, or wobble, position of the codon, it can base-pair with any of the nucleotides listed in the second column. Thus, for example, when inosine (I) is present in the wobble position of the tRNA anticodon, the tRNA can recognize any one of three different codons in bacteria and either of two codons in eucaryotes. The inosine in tRNAs is formed from the deamination of guanine, a chemical modification that takes place after the tRNA has been synthesized. The nonstandard base pairs, including those made with inosine, are generally weaker than conventional base pairs. Note that codon–anticodon base pairing is more stringent at positions 1 and 2 of the codon: here only conventional base pairs are permitted. The differences in wobble base-pairing interactions between bacteria and eucaryotes presumably result from subtle structural differences between bacterial and eucaryotic ribosomes, the molecular machines that perform protein synthesis.
(Adapted from C. Guthrie and J. Abelson, in The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression, pp. 487–528. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press, 1982.)
This wobble base-pairing explains why so many of the alternative codons for an amino acid differ only in their third nucleotide . In bacteria, wobble base-pairings make it possible to fit the 20 amino acids to their 61 codons with as few as 31 kinds of tRNA molecules. The exact number of different kinds of tRNAs, however, differs from one species to the next. For example, humans have nearly 500 tRNA genes but, among them, only 48 different anticodons are represented.
Specific enzymes couple each Amino Acid to its appropriate tRNA Molecule
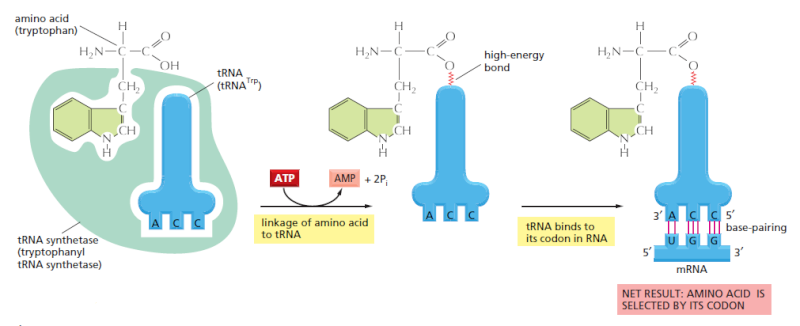
We have seen that, to read the genetic code in DNA, cells make a series of different tRNAs. We now consider how each tRNA molecule becomes linked to the one amino acid in 20 that is its appropriate partner. Recognition and attachment of the correct amino acid depends on enzymes called aminoacyl-tRNA synthetases, which covalently couple each amino acid to its appropriate set of tRNA molecules


Most cells have a different synthetase enzyme for each amino acid (that is, 20 synthetases in all); one attaches glycine to all tRNAs that recognize codons for glycine, another attaches alanine to all tRNAs that recognize codons for alanine, and so on. Many bacteria, however, have fewer than 20 synthetases, and the same synthetase enzyme is responsible for coupling more than one amino acid to the appropriate tRNAs. In these cases, a single synthetase places the identical amino acid on two different types of tRNAs, only one of which has an anticodon that matches the amino acid. A second enzyme then chemically modifies each “incorrectly” attached amino acid so that it now corresponds to the anticodon displayed by its covalently linked tRNA. The synthetase-catalyzed reaction that attaches the amino acid to the 3" end of the tRNA is one of many reactions coupled to the energy-releasing hydrolysis of ATP , and it produces a high-energy bond between the tRNA and the amino acid. The energy of this bond is used at a later stage in protein synthesis to link the amino acid covalently to the growing polypeptide chain. The aminoacyl-tRNA synthetase enzymes and the tRNAs are equally important in the decoding process

These enzymes are not gentle with tRNA molecules. The structure of glutaminyl-tRNA synthetase with its tRNA (entry 1gtr) is a good example ( see above ) The enzyme firmly grips the anticodon, spreading the three bases widely apart for better recognition. At the other end, the enzyme unpairs one base at the beginning of the chain, seen curving upward here, and kinks the long acceptor end of the chain into a tight hairpin, seen here curving downward. This places the 2' hydroxyl on the last nucleotide in the active site, where ATP and the amino acid (not present in this structure) are bound.
My comment: The tRNA and ATP fit precisely in the active site of the enzyme, and the structure is configured and designed to function in a finely tuned manner. How could such a functional device be the result of random unguided forces and chemical reactions without an end goal?
The genetic code is translated by means of two adaptors that act one after another. The first adaptor is the aminoacyl-tRNA synthetase, which couples a particular amino acid to its corresponding tRNA; the second adaptor is the tRNA molecule itself, whose anticodon forms base pairs with the appropriate codon on the mRNA. An error in either step would cause the wrong amino acid to be incorporated into a protein chain. In the sequence of events shown, the amino acid tryptophan (Trp) is selected by the codon UGG on the mRNA.
This was established by an experiment in which one amino acid (cysteine) was chemically converted into a differentamino acid (alanine) after it already had been attached to its specific tRNA. When such “hybrid” aminoacyl-tRNA molecules were used for protein synthesis in a cell-free system, the wrong amino acid was inserted at every point in the protein chain where that tRNA was used. Although, as we shall see, cells have several quality control mechanisms to avoid this type of mishap, the experiment establishes that the genetic code is translated by two sets of adaptors that act sequentially. Each matches one molecular surface to another with great specificity, and it is their combined action that associates each sequence of three nucleotides in the mRNA molecule—that is, each codon—with its particular amino acid.
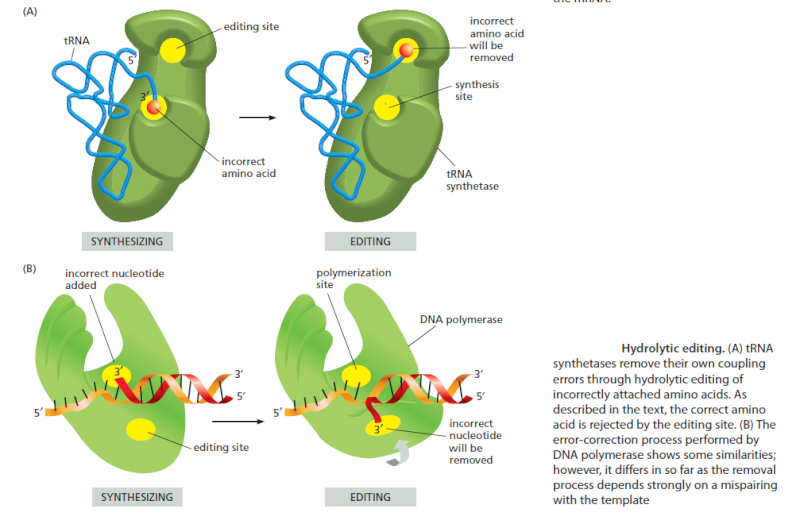
Editing by tRNA Synthetases Ensures Accuracy
Several mechanisms working together ensure that the tRNA synthetase links the correct amino acid to each tRNA. The synthetase must first select the correct amino acid, and most synthetases do so by a two-step mechanism. First, the correct amino acid has the highest affinity for the active-site pocket of its synthetase and is therefore favored over the other 19. In particular, amino acids larger than the correct one are effectively excluded from the active site. However, accurate discrimination between two similar amino acids, such as isoleucine and valine (which differ by only a methyl group), is very difficult to achieve by a one-step recognition mechanism. A second discrimination step occurs after the amino acid has been covalently linked to AMP. When tRNA binds the synthetase, it tries to force the amino acid into a second pocket in the synthetase, the precise dimensions of which exclude the correct amino acid but allow access by closely related amino acids. Once an amino acid enters this editing pocket, it is hydrolyzed from the AMP (or from the tRNA itself if the aminoacyl-tRNA bond has already formed), and is released from the enzyme. This hydrolytic editing, which is analogous to the exonucleolytic proofreading by DNA polymerases , raises the overall accuracy of tRNA charging to approximately one mistake in 40,000 couplings.
Editing significantly decreases the frequency of errors and is important for translational quality control, and many details of the various editing mechanisms and their effect on different cellular systems are now starting to emerge. 8
High Fidelity
Aminoacyl-tRNA synthetases must perform their tasks with high accuracy. Every mistake they make will result in a misplaced amino acid when new proteins are constructed. These enzymes make about one mistake in 10,000. For most amino acids, this level of accuracy is not too difficult to achieve. Most of the amino acids are quite different from one another, and, as mentioned before, many parts of the different tRNA are used for accurate recognition. But in a few cases, it is difficult to choose just the right amino acids and these enzymes must resort to special techniques.
Isoleucine is a particularly difficult example. It is recognized by an isoleucine-shaped hole in the enzyme, which is too small to fit larger amino acids like methionine and phenylalanine, and too hydrophobic to bind anything with polar sidechains. But, the slightly smaller amino acid valine, different by only a single methyl group, also fits nicely into this pocket, binding instead of isoleucine in about 1 in 150 times. This is far too many errors, so corrective steps must be taken. Isoleucyl-tRNA synthetase (PDB entry 1ffy) solves this problem with a second active site, which performs an editing reaction. Isoleucine does not fit into this site, but errant valine does. The mistake is then cleaved away, leaving the tRNA ready for a properly-placed leucine amino acid. This proofreading step improves the overall error rate to about 1 in 3,000. 9
My comment: This is an amazing error proofreading technique, which adds to other repair mechanisms in the cell. Once again the question arises: How could these precise molecular machines have arisen by natural means, without intelligence involved? This seems to be one more amazing example of highly sophisticated nanomolecular machinery designed to fulfill its task with a high degree of fidelity and error minimization, which can arise only by the foresight of an incredibly intelligent creator.
I. Why wasn’t one ancestor enough when they both do the same job?
II. How did the two types of ancestral synthetases avoid competition that might have eliminated the inferior Class? 12
A new peer-reviewed paper in the journal Frontiers in Genetics, "Redundancy of the genetic code enables translational pausing," finds that so-called "redundant" codons may actually serve important functions in the genome. Redundant (also called "degenerate") codons are those triplets of nucleotides that encode the same amino acid. For example, in the genetic code, the codons GGU, GGC, GGA, and GGG all encode the amino acid glycine. While it has been shown that such redundancy is actually optimized to minimize the impact of mutations resulting in amino acid changes, it is generally assumed that synonymous codons are functionally equivalent. They just encode the same amino acid, and that's it.
The ribosome is capable of reading both sets of commands -- as they put it, "the ribosome can be thought of as an autonomous functional processor of data that it sees at its input." To put it another way, the genetic code is "multidimensional," a code within a code. This multidimensional nature exceeds the complexity of computer codes generated by humans, which lack the kind of redundancy of the genetic code. As the abstract states:
The codon redundancy ("degeneracy") found in protein-coding regions of mRNA also prescribes Translational Pausing (TP). When coupled with the appropriate interpreters, multiple meanings and functions are programmed into the same sequence of configurable switch-settings. This additional layer of Ontological Prescriptive Information (PIo) purposely slows or speeds up the translation decoding process within the ribosome. Variable translation rates help prescribe functional folding of the nascent protein. Redundancy of the codon to amino acid mapping, therefore, is anything but superfluous or degenerate. Redundancy programming allows for simultaneous dual prescriptions of Translational Pausing TP and amino acid assignments without cross-talk. This allows both functions to be coincident and realizable. We will demonstrate that the TP schema is a bona fide rule-based code, conforming to logical code-like properties. Second, we will demonstrate that this TP code is programmed into the supposedly degenerate redundancy of the codon table. We will show that algorithmic processes play a dominant role in the realization of this multi-dimensional code.
The paper even suggests, "Cause-and-effect physical determinism...cannot account for the programming of sequence-dependent biofunction."
The origin of the genetic code is acknowledged to be a major hurdle in the origin of life, and I shall mention just one or two of the main problems. Calling it a ‘code’ can be misleading because of associating it with humanly invented codes which at their core usually involve some sort of pre-conceived algorithm; whereas the genetic code is implemented entirely mechanistically – through the action of biological macromolecules. This emphasises that, to have arisen naturally – e.g. through random mutation and natural selection – no forethought is allowed: all of the components would need to have arisen in an opportunistic manner.
Crucial role of the tRNA activating enzymes 7
To try to explain the source of the code various researchers have sought some sort of chemical affinity between amino acids and their corresponding codons. But this approach is misguided:
First of all, the code is mediated by tRNAs which carry the anti-codon (in the mRNA) rather than the codon itself (in the DNA). So, if the code were based on affinities between amino acids and anti-codons, it implies that the process of translation via transcription cannot have arisen as a second stage or improvement on a simpler direct system - the complex two-step process would need to have arisen right from the start.
Second, the amino acid has no role in identifying the tRNA or the codon (This can be seen from an experiment in which the amino acid cysteine was bound to its appropriate tRNA in the normal way – using the relevant activating enzyme, and then it was chemically modified to alanine. When the altered aminoacyl-tRNA was used in an in vitro protein synthesizing system (including mRNA, ribosomes etc.), the resulting polypeptide contained alanine (instead of the usual cysteine) corresponding to wherever the codon UGU occurred in the mRNA. This clearly shows that it is the tRNA alone (with no role for the amino acid) with its appropriate anticodon that matches the codon on the mRNA.). This association is done by an activating enzyme (aminoacyl tRNA synthetase) which attaches each amino acid to its appropriate tRNA (clearly requiring this enzyme to correctly identify both components). There are 20 different activating enzymes - one for each type of amino acid.
Interestingly, the end of the tRNA to which the amino acid attaches has the same nucleotide sequence for all amino acids - which constitutes a third reason.
Interest in the genetic code tends to focus on the role of the tRNAs, but as just indicated that is only one half of implementing the code. Just as important as the codon-anticodon pairing (between mRNA and tRNA) is the ability of each activating enzyme to bring together an amino acid with its appropriate tRNA. It is evident that implementation of the code requires two sets of intermediary molecules: the tRNAs which interact with the ribosomes and recognise the appropriate codon on mRNA, and the activating enzymes which attach the right amino acid to its tRNA. This is the sort of complexity that pervades biological systems, and which poses such a formidable challenge to an evolutionary explanation for its origin. It would be improbable enough if the code were implemented by only the tRNAs which have 70 to 80 nucleotides; but the equally crucial and complementary role of the activating enzymes, which are hundreds of amino acids long, excludes any realistic possibility that this sort of arrangement could have arisen opportunistically.
Progressive development of the genetic code is not realistic
In view of the many components involved in implementing the genetic code, origin-of-life researchers have tried to see how it might have arisen in a gradual, evolutionary, manner. For example, it is usually suggested that to begin with the code applied to only a few amino acids, which then gradually increased in number. But this sort of scenario encounters all sorts of difficulties with something as fundamental as the genetic code.
1. First, it would seem that the early codons need have used only two bases (which could code for up to 16 amino acids); but a subsequent change to three bases (to accommodate 20) would seriously disrupt the code. Recognising this difficulty, most researchers assume that the code used 3-base codons from the outset; which was remarkably fortuitous or implies some measure of foresight on the part of evolution (which, of course, is not allowed).
2. Much more serious are the implications for proteins based on a severely limited set of amino acids. In particular, if the code was limited to only a few amino acids, then it must be presumed that early activating enzymes comprised only that limited set of amino acids, and yet had the necessary level of specificity for reliable implementation of the code. There is no evidence of this; and subsequent reorganization of the enzymes as they made use of newly available amino acids would require highly improbable changes in their configuration. Similar limitations would apply to the protein components of the ribosomes which have an equally essential role in translation.
3. Further, tRNAs tend to have atypical bases which are synthesized in the usual way but subsequently modified. These modifications are carried out by enzymes, so these enzymes too would need to have started life based on a limited number of amino acids; or it has to be assumed that these modifications are later refinements - even though they appear to be necessary for reliable implementation of the code.
4. Finally, what is going to motivate the addition of new amino acids to the genetic code? They would have little if any utility until incorporated into proteins - but that will not happen until they are included in the genetic code. So the new amino acids must be synthesised and somehow incorporated into useful proteins (by enzymes that lack them), and all of the necessary machinery for including them in the code (dedicated tRNAs and activating enzymes) put in place – and all done opportunistically! Totally incredible!
My comment: What must be explained, is the arrangement of the codons in the standard codon table which is highly non-random, and serves to translate into the amino acid sequence to make proteins, and the origin of the assignment of the 64 triplet codons to the 20 amino acids. That is the origin of its translation. The origin of an alphabet through the triplet codons is one thing, but on top, it has to be translated to another " alphabet " constituted through the 20 amino acid sequence. That is, to explain the origin of the capability to translate the English language into Chinese. On top of that, the machinery itself to promote the process itself has also to be explained, that is the hardware. When humans translate English to Chinese, for example, we recognize the English word, and the translator knows the equivalent Chinese symbol and writes it down.
Why is it not rational to conclude that the code itself, the software, as well as the hardware, are best explained through the invention of a highly intelligent being, rather than random chemical affinities and reactions?
And even it lets say, the whole thing was fully working and in place, what good would it be for without all the other parts required, that is, the DNA double helix, its compactation through histones and chromatins and chromosomes, its highly complex mechanism of information extraction and transcription into mRNA? Had the whole process, that is INITIATION OF TRANSCRIPTION, CAPPING, ELONGATION, SPLICING, CLEAVAGE, POLYADENYLATION, AND TERMINATION, EXPORT FROM THE NUCLEUS TO THE CYTOSOL, INITIATION OF PROTEIN SYNTHESIS (TRANSLATION), COMPLETION OF PROTEIN SYNTHESIS AND PROTEIN FOLDING, and its respective machinery not have to be all in place? Does that not constitute an interdependent and irreducibly complex system?
Koonin, the logic of chance, page 237
The origin of translation: The key ideas and models
During the 40 years since the discovery of the translation mechanism and deciphering of the genetic code, numerous theoretical (inevitably, speculative, sometimes far-fetched, often highly ingenious) models of the origin and evolution of various components of the translation apparatus and different aspects of the translation process have been proposed. It is unrealistic to provide here a thorough critical review of these models. Instead, I consider a few central ideas that are germane to the thinking about the origin of translation and then discuss in somewhat greater detail the only two coherent scenarios I am aware of. The main general point about the evolution of translation is that selection for protein synthesis could not have been the underlying cause behind the origin of the translation system. To evolve this complex system via the Darwinian route, numerous steps are required, but proteins appear only at the last steps; until that point, an evolving organism “does not know” how good proteins could be.
The DNA - Enzyme System is Irreducibly Complex 10
An often undiscussed aspect of complexity is how the tRNA get assigned to the right amino acids. For the DNA language to be translated properly, each tRNA codon must be attached to the correct amino acid. If this crucial step in DNA replication is not functional, then the language of DNA breaks down. Aminoacyl - tRNA synthetases (aaRSs) ensure that the proper amino acid is attached to a tRNA with the correct codon through a chemical reaction called "aminoacylation." Accurate translation requires not only that each tRNA be assigned the correct amino acid, but also that it not be aminoacylated by any of the aaRS molecules for the other 19 amino acids. One biochemistry textbook notes that because all aaRSs catalyze similar reactions upon various similar tRNA molecules, it was thought they "evolved from an common ancestor and should therefore be structurally related." (Voet and Voet pg. 971-975) However, this was not the case as the, "aaRSs form a diverse group of [over 100] enzymes … and there is little sequence similarity among synthetases specific for different amino acids." (Voet and Voet pg. 971-975) Amazingly, these aaRSs themselves are coded for by the DNA: this forms the essence of a chicken-egg problem. The enzymes themselves build help perform the very task which constructs them!
Can the Origin of the Genetic Code Be Explained by Direct RNA Templating? August 24, 2011 Stephen C. Meyer, Paul A. Nelson
The three main naturalistic concepts on the origin and evolution of the code are the stereochemical theory, according to which codon assignments are dictated by physico-chemical affinity between amino acids and the cognate codons (anticodons).
The genetic code as we observe it today is a semantic (symbol- based) relation between (a) amino acids, the building blocks of proteins, and (b) codons, the three-nucleotide units in messenger RNA specifying the identity and order of different amino acids in protein assembly. The actual physical mediators of the code, however, are transfer RNAs (tRNAs) that, after being charged with their specific amino acids by aminoacyl tRNA synthetases (aaRSs), present the amino acids for peptide bond formation in the peptidyl-transferase (P) site of the ribosome, the molecular machine that constructs proteins.
When proteins are produced in cells based on the "genetic code" of codons, there is a precise process under which molecules called transfer RNA (tRNA) bind to specific amino acids and then transport them to cellular factories, ribosomes, where the amino acids are placed together, step by step, to form a protein. Mistakes in this process, which is mediated by enzymes called synthetases, can be disastrous, as they can lead to improperly formed proteins. Thankfully, the tRNA molecules are matched to the proper amino acids with great precision, but we still lack a fundamental understanding of how this selection takes place. 4
The secondary structure of a typical tRNA see figure below, reveals the coding (semantic) relations that Yarus et al. are trying to obtain from chemistry alone - a quest Yockey has compared to latter-day alchemy
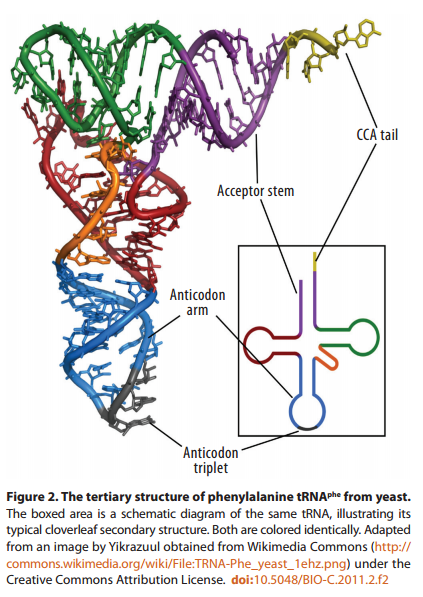
At the end of its 3' arm, the tRNA binds its cognate amino acid via the universally conserved CCA sequence. Some distance away—about 70 Å—in loop 2, at the other end of the inverted cloverleaf, the anticodon recognizes the corresponding codon in the mRNA strand. (The familiar ‘cloverleaf’ shape represents only the secondary structure of tRNA; its three-dimensional form more closely resembles an “L” shape, with the anticodon at one end and an amino acid at the other.)Thus, in the current genetic code, there is no direct chemical interaction between codons, anticodons, and amino acids. The anticodon triplet and amino acid are situated at opposite ends of the tRNA: the mRNA codon binds not to the amino acid directly, but rather to the anticodon triplet in loop 2 of the tRNA.
Since all twenty amino acids, when bound to their corresponding tRNA molecules, attach to the same CCA sequence at the end of the 3’ arm, the stereochemical properties of that nucleotide sequence clearly do not determine which amino acids attach, and which do not. The CCA sequence is indifferent, so to speak, to which amino acids bind to it
Nevertheless, tRNAs are informationally (i.e., semantically) highly specific: protein assembly and biological function—but not chemistry—demand such specificity. As noted, in the current code, codon-to-amino acid semantic mappings are mediated by tRNAs, but also by the enzymatic action of the twenty separate aminoacyl-tRNA synthetases
This is a functionally interdependent system of highly specific molecules, including mRNA, a suite of tRNAs, and twenty specific aaRS enzymes, each of which is itself constructed from information stored on the very DNA strands that the system as a whole decodes.
Aminoacyl tRNA synthetase
An aminoacyl tRNA synthetase (aaRS) is an enzyme that catalyzes the esterification of a specific cognate amino acid or its precursor to one of all its compatible cognate tRNAs to form an aminoacyl-tRNA. In other words, aminoacyl tRNA synthetase attaches the appropriate amino acid onto its tRNA. This is called "charging" or "loading" the tRNA with the amino acid. Once the tRNA is charged, a ribosome can transfer the amino acid from the tRNA onto a growing peptide, according to the genetic code. Aminoacyl tRNA therefore plays an important role in DNA translation, the expression of genes to create proteins. 2
This set of twenty enzymes knows what amino acid to fasten to one end of a transfer-RNA (tRNA) molecule, based on the triplet codon it reads at the other end. It's like translating English to Chinese. A coded message is complex enough, but the ability to translate a language into another language bears the hallmarks of intelligent design. 6
Most cells use twenty aaRS enzymes, one for each amino acid. Each of these proteins recognizes a specific amino acid and the specific anticodons it binds to within the code. They then bind amino acids to the tRNA that bears the corresponding anticodon.
Thus, instead of the code reducing to a simple set of stereochemical affinities, biochemists have found a functionally interdependent system of highly specific molecules, including mRNA, a suite of tRNAs, and twenty specific aaRS enzymes, each of which is itself constructed from information stored on the very DNA strands that the system as a whole decodes.
Attempts to explain one part of the integrated complexity of the gene-expression system, namely the genetic code, by reference to simple chemical affinities lead not to simple rules of chemical attraction, but instead to an integrated system of multiple large molecular components. While this information-transmitting system exploits (i.e., uses) chemistry, it is not reducible to direct chemical affinities between codons or anticodons and their cognate amino acids.
The DRT model and the sequencing problem
One further aspect of Yarus’s work needs clarification and critique. One of the longest-standing and most vexing problems in origin-of-life research is known as the sequencing problem, the problem of explaining the origin of the specifically-arranged sequences of nucleotide bases that provide the genetic information or instructions for building proteins.
Yet, in addition to its other deficiencies it is important to point out that Yarus et al. do not solve the sequencing problem, although they do claim to address it indirectly. Instead, Yarus et al. attempt to explain the origin of the genetic code—or more precisely, one aspect of the translation system, the origin of the associations between certain RNA triplets and their cognate amino acids.
Yarus et al. want to demonstrate that particular RNA triplets show chemical affinities to particular amino acids (their cognates in the present-day code). They try to do this by showing that in some RNA strands, individual triplets and their cognateamino acids bind preferentially to each other. They then envision that such affinities initially provided a direct (stereochemical) template for amino acids during protein assembly.
Since Yarus et al. think that stereochemical affinities originally caused protein synthesis to occur by direct templating, they also seem to think that solving the problem of the origin of the code would also simultaneously solve the problem of sequencing. But this does not follow. Even if we assume that Yarus et al. have succeeded in establishing a stereochemical basis for the associations between RNA triplets and amino acids in the present-day code (which they have not done; see above), they would not have solved the problem of sequencing.
The sequencing problem requires that long RNA strands would need to contain triplets already arranged to bind their cognate amino acids in the precise order necessary to assemble functional proteins. Yarus et al. analyzed RNA strands enriched in specific code-relevant triplets, and claim to have found that these strands show a chemical affinity with their cognate amino acids. But they did not find RNA strands with a properly sequenced series of triplets, each forming an association with a code-relevant amino acid as the DRT model would require, and arranged in the kind of order required to make functional proteins. To synthesize proteins by direct templating (even assuming the existence of all necessary affinities), the RNA template must have many properly sequenced triplets, just as we find in the actual messenger RNA transcripts.
1) https://bio-complexity.org/ojs/index.php/main/article/viewArticle/43
2) https://en.wikipedia.org/wiki/Aminoacyl_tRNA_synthetase
3) http://pubs.acs.org/subscribe/archive/mdd/v04/i03/html/03timeline.html
4) http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4323281/
http://www.riken.jp/en/pr/press/2014/20140612_1_yokoyama/
http://www.evolutionnews.org/2014/07/each_time_genom088091.html
5) http://www.evolutionnews.org/2014/08/paper_finds_fun089301.html
6) http://www.evolutionnews.org/2015/06/from_chemicals096611.html
7) http://www.c4id.org.uk/index.php?option=com_content&view=article&id=211:the-problem-of-the-origin-of-life&catid=50:genetics&Itemid=43
8 )http://www.ncbi.nlm.nih.gov/pubmed/19379069
9) http://www.rcsb.org/pdb/101/motm.do?momID
10) http://www.ideacenter.org/contentmgr/showdetails.php/id/845
11) http://creationsafaris.com/epoi_c09.htm
Last edited by Otangelo on Sun Jan 03, 2021 10:56 am; edited 41 times in total