This publication stands out not merely as another critique among many questioning the sufficiency of Darwin's Theory of Evolution to account for the vast biodiversity, but it unravels the astonishingly complex and varied processes necessary for the development of complex life forms and the structure of multicellular organisms. It directly confronts and highlights the shortcomings of traditional evolutionary explanations in accounting for the organization of life, offering a holistic perspective on the genuine mechanisms involved. The book undertakes a detailed and critical analysis of the core aspects of Darwinian theory, with a particular focus on molecular-level insights. It refutes Darwinian evolutionary concepts, such as universal common descent and the Tree of Life, and shows the inadequacy of natural selection, genetic drift, and gene flow as sufficient explanations as the origin of complex biological form and architecture, and multitude of life forms. This work illuminates the true mechanisms behind the complexity of life with a level of detail and approach that is unprecedented in the scientific literature, and why these mechanisms cannot be explained by unguided mechanisms, but require an intelligent mind, which instantiated them from their inception fully developed, and apt to thrive, adapt, replicate, and multiply.
There are 52 key points, ranging from alternative DNA structures to the influence of viruses, highlighting the vast diversity in genetic, biochemical, and physiological mechanisms across life forms. This complexity and variability refute universal common descent and is evidence of polyphyly, which posits that life diversified, adapted, and evolved to a limited degree after the creation of many independent species. The book unravels how the architecture of multicellular organisms arises from a complex interplay of at least 47 key developmental processes, 33 variations of genetic codes, and over 223 epigenetic, manufacturing, and regulatory codes and biological languages. These elements are coordinated through hundreds of signaling networks, embodying functional communication, integration, and systemic complexity. This holistic ensemble hints at a level of organization that indicates the requirement of an intelligent cause for the instantiation of individual species at the inception of life. At close to 1400 pages, this book stands as a transformative work, offering groundbreaking insights into the interwoven complexity of organismal structures. It provides a thorough examination of the subject matter and clarity which will unravel and unmask, how naturalistic unguided causes are inadequate explanations, and can be put to rest. This book provides evidence, that a powerful, conscious, intelligent designer is the best case-adequate explanation, that created life and biodiversity.
https://reasonandscience.catsboard.com/t3264-refuting-darwin-confirming-design
Life's Blueprint: The Essential Machinery for Life to Start
https://reasonandscience.catsboard.com/t3383-life-s-blueprint-the-essential-machinery-for-life-to-start
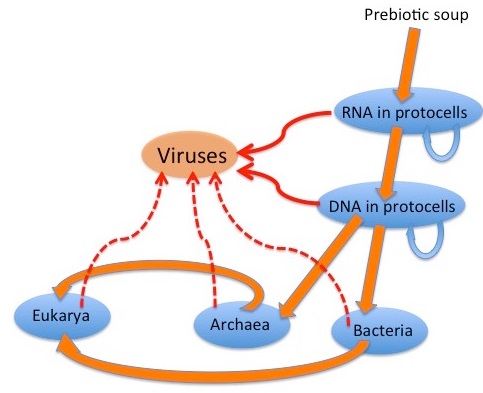
A list of evidence that points to polyphyly, rather than monophyly, and universal common descent
https://reasonandscience.catsboard.com/t2239-evolution-common-descent-the-tree-of-life-a-failed-hypothesis#10954
Bacteria
https://reasonandscience.catsboard.com/t3379-bacteria-the-first-domain-of-life
Archaea
https://reasonandscience.catsboard.com/t3378-archaea-the-second-domain-of-life
Key Developmental Processes Shaping Organismal Form and Function
https://reasonandscience.catsboard.com/t2316p25-evolution-where-do-complex-organisms-come-from#10629
The Brain of Homo Sapiens and Chimps: Distinctive Characteristics are reasons to be skeptic about shared descent
https://reasonandscience.catsboard.com/t2272-chimps-our-brothers#11024
Processes involved in embryogenesis
https://reasonandscience.catsboard.com/t3381-processes-involved-in-embryogenesis
The Orchestration of Neurogenesis: A Study in Irreducibility and Interdependence
https://reasonandscience.catsboard.com/t3373-the-orchestration-of-neurogenesis-a-study-in-irreducibility-and-interdependence
Unraveling the Molecular Foundations of Instinctual Behavior
https://reasonandscience.catsboard.com/t1732-instinct-evolutions-major-problem-to-explain#11130
What is the Theory of Evolution?
How is the origin of biological form, biodiversity, organismal complexity, and architecture explained by current biology?
Primary, and secondary speciation
Gene duplications
The major ( hypothesized) transitions in evolution
Falsifying Universal Common Ancestry

Unraveling the real mechanisms giving rise to biological adaptation, development, complex organismal forms, anatomical novelty, and biodiversity
Acknowledgements
In traditional book writing, an author is often likened to a craftsman, meticulously choosing each word and weaving them together to create a narrative. However, in the age of artificial intelligence and advanced algorithms, the nature of this craft has evolved, leading to a shift in the role and definition of an "author". As the individual behind this book, I find the term "author" insufficient to describe my role in its creation. While every word you read has been generated by ChatGPT, an AI language model, my involvement has been more akin to a conductor of a symphony. Just as a conductor doesn't play each instrument but orchestrates an entire ensemble to create harmonious music, I've not manually penned each line but have strategically directed and channeled the AI's vast knowledge and capabilities to construct this narrative. This approach to writing involves curating and arranging prompts, refining AI-generated content, and ensuring the final product aligns with a coherent vision. It's a dance between human intention and machine efficiency, where I decide the theme, tone, and direction, and the AI fills in with its vast database of information. The result is a unique blend of human creativity and machine precision. So, as you delve into the pages of this book, I invite you to view it not just as a work authored in the traditional sense, but as a collaborative symphony between human intuition and artificial intelligence. Together, in this new era of content creation, we explore the frontiers of knowledge and storytelling, redefining what it means to be an author in the digital age.
Prologue
Over 160 years have passed since Darwin's "On the Origins of Species". His theory of evolution by natural selection is a cornerstone of modern biology. It claimed to provide a unifying explanation for the diversity of life on Earth. The theory is based on the idea that organisms vary, and these variations can be inherited by the next generation. Natural selection acts on these variations, favoring those that are advantageous for survival and reproduction in a given environment. Over time, this process can supposedly lead to the evolution of new species.
Since its introduction, the theory of evolution has been claimed to be supported by a vast amount of evidence from a wide range of scientific disciplines, including genetics, paleontology, comparative anatomy, and biochemistry. The modern synthesis of the 20th century integrated Darwin's theory with the science of genetics. Darwin's theory also has had practical implications for science and society.
In the annals of scientific inquiry, few theories have sparked as much continuous debate. Darwins suggestion that the diverse forms of life emerged through a process of natural selection has been met with significant scrutiny, particularly from those who observe the natural world and see evidence of deliberate, designed complexity. Critics point to the intricate structures and unfathomable complexity of living organisms, arguing that these features could not have arisen from random mutations and natural selection alone. The eye, with its precise arrangement of lenses, muscles, and photoreceptor cells, stands as a testament to a level of complexity that seems to defy the gradualistic explanations offered by natural selection. This perspective suggests that such marvels of nature are not the product of an undirected process but rather the work of an intelligent designer.
Moreover, the fossil record, often cited in support of Darwinian evolution, presents its own set of challenges to the theory. The expected gradual transitions between species are conspicuously absent in many cases, leading to question the validity of the theory as an explanation for the origin of the vast diversity of life. Instead, the sudden appearance of fully formed species in the fossil record aligns more closely with the notion of special creation, where each species was created as it is, in a form complete and functional from the beginning. The debates extend beyond the realm of biology and into the philosophical and societal implications of Darwin's theory. Critics argue that the reduction of human life to mere products of chance and competition devalues human dignity and morality. The idea that life, in all its complexity and purpose, could be the result of random processes is at odds with the sense of intention and design that we observe in the natural world. The controversy surrounding the teaching of Darwin's theory in educational institutions highlights the deep divide between the materialistic view of life's origins and the perspective that sees evidence of design and purpose in the natural world. This ongoing debate underscores the fundamental differences in how we interpret the natural world and our place within it.
In the realm of scientific inquiry, one of the most compelling and debated subjects remains the question of life's origin and the staggering complexity of biodiversity. The dominant narrative has frequently presented the theory of evolution as the primary, unguided mechanism propelling the emergence and diversity of life. Yet, within the vast expanse of scientific discourse, alternative perspectives and interpretations of the data warrant thoughtful consideration. This book aims to provide readers with a comprehensive overview of the evidence that suggests the possibility of design as a more case-adequate explanation for the origin and biodiversity of life, in contrast to solely unguided evolutionary mechanisms. Readers are invited on a journey of exploration, fostering a deeper understanding and encouraging critical thinking on one of science's most profound questions.
The debate over whether biodiversification and the complexity of life can be fully explained by unguided evolutionary mechanisms or whether the involvement of an intelligent agent is necessary is a fundamental and longstanding philosophical and scientific discussion. This dichotomy reflects a broader conflict between naturalism and theism, two contrasting worldviews that shape our understanding of the origin and development of life. Naturalism is the philosophical perspective that asserts that all phenomena, including the diversity of life, can be explained by natural processes operating according to physical laws. In the context of biology, naturalism holds that evolution through mechanisms such as natural selection, genetic variation, and environmental pressures can account for the complexity and diversity of living organisms. Proponents of naturalism argue that no supernatural or divine intervention is required to explain the natural world. Theism, on the other hand, posits that the existence and characteristics of the natural world are best explained by the presence of an intelligent and purposeful creator or divine being. In this view, the complexity of life, the intricate design of organisms, and the emergence of biodiversity are seen as indicative of intentional design rather than solely the outcome of unguided natural processes. The dispute between naturalism and theism centers on the interpretation of evidence and the underlying assumptions about the nature of reality.
Making sense of the vast diversity of life is still today one of the greatest, if not the greatest intellectual challenge, together with the Origin of Life. The quest about if evolution is true is more than a scientific question. It is a battle that goes beyond science. It is a culture war between naturalism/strong atheism, and creationism/Intelligent Design. If the literal interpretation of the Genesis account in the Bible is true, then Darwin's Theory of Evolution is false, and vice-versa.
Frank Zindler, President of American Atheists, in 1996:
The most devastating thing though that biology did to Christianity was the discovery of biological evolution. Now that we know that Adam and Eve never were real people the central myth of Christianity is destroyed. If there never was an Adam and Eve there never was an original sin. If there never was an original sin there is no need of salvation. If there is no need of salvation there is no need of a Savior. And I submit that puts Jesus, historical or otherwise, into the ranks of the unemployed. I think that evolution is absolutely the death knell of Christianity.

Conservative Protestants in the 1920s also saw themselves in the midst of a great culture war, with the Bible (depicted here as the Rock of Gibraltar) coming under fierce attack by “battle-ships of unbelief.”
Leaving the Bible aside, the dispute is not about religion versus science, but between case-adequate inferences based on scientific evidence and unwarranted conclusions. The big question is: Is the origin of biodiversity, Darwin's hypothesis of universal common ancestry, and the tree of life, supported by the evidence unraveled by scientific facts, as the scientific establishment and consensus among professionals in the field advocate for, calling Darwin's Theory, and the recently modified versions of it, an undisputable scientific fact, or does the data lead to another direction? We can also ask a deeper question, and dissect the issue to the core question: Which of the two has more creative power: Design, or non-design? Intelligence, or non-intelligence? Agency, or non-agency? Conscious creation, or undirected natural, non-intelligent processes?

Claim: Herbert Spencer: Those who cavalierly reject the Theory of Evolution as not being adequately supported by facts, seem to forget that their own theory is supported by no facts at all. Like the majority of men who are born to a given belief, they demand the most rigorous proof of any adverse belief, but assume that their own needs none.
Richard Dawkins: "It is absolutely safe to say that, if you meet somebody who claims not to believe in evolution, that person is ignorant, stupid or insane (or wicked, but I'd rather not consider that)."1
John Joe McFadden (2008): Quite simply, Darwin and Wallace destroyed the strongest evidence left in the 19th century for the existence of a deity. Biologists have since used Darwin's theory to make sense of the natural world. Contrary to the arguments of creationists, evolution is no longer just a theory. It is as much a fact as gravity or erosion. 2
Reply: Opinions like Richard Dawkins have contributed to stigmatizing the proposition of intelligent design as pseudo-science, or as unscientific altogether. But is it justified? Many books have been published on the subject, and articles are frequently being written, defending both views and positions. Those that advocate in favor, often resort to the fact that a majority of biologists are on their side, and argue, because there is a widespread consensus, it must be true.
A 2019 survey of American biologists found that 98% of them agreed that "evolution by natural selection is the best explanation for the diversity of life on Earth." This survey was conducted by the Pew Research Center, a nonpartisan fact tank that conducts public opinion research. Similar surveys have been conducted in other countries, and the results have been consistent. For example, a 2018 survey of British biologists found that 97% of them agreed that "evolution by natural selection is the best explanation for the diversity of life on Earth." These surveys suggest that the vast majority of biologists worldwide accept Darwin's theory of evolution as the best explanation for biodiversity. While there may be a small minority of biologists who do not accept this theory, they are a very small minority.
Sailing against an unfavorable wind is undoubtedly a challenging and energy-consuming endeavor. However, the pursuit of truth remains the guiding force, pushing us to brave these turbulent waters. In today's world, many individuals may lose faith in a creator due to a lack of proper education to critically assess scientific evidence. Instead, they are swayed by those who advocate for evolution, claiming to possess evidence on their side. In stark contrast, I have dedicated years to probing this subject deeply, allowing the evidence to be my compass without yielding to the temptation of becoming just another anti-evolution book collecting dust on shelves. My purpose is to present a well-researched perspective that questions the prevailing narrative of evolution. Although some may view me as a solitary zealot, blindly adhering to religious beliefs and disregarding contemporary scientific advancements and the consensus among professional biologists, I am not alone. My findings align with those of esteemed investigators in the field. These scientists, committed to philosophical naturalism, may not draw a simplistic conclusion like ".... and therefore, God!!" as that proposition lies beyond the realm of scientific inquiry. However, they candidly acknowledge the limitations and issues within the traditional evolutionary view. Honesty and integrity underscore my approach throughout this journey. My discoveries, mirroring those of respected researchers, rest on solid factual grounds. While some authors might resort to colorful arguments,
In November 2016, there was a three-day conference in London, a scientific discussion meeting organized by the Royal Society: New trends in evolutionary biology: biological, philosophical, and social science perspectives. On the website, they wrote: Developments in evolutionary biology and adjacent fields have produced calls for revision of the standard theory of evolution 3
Intelligent Design proponents point to a chasm that divides how evolution and its evidence are presented to the public, and how scientists themselves discuss it behind closed doors and in technical publications. This chasm has been well hidden from laypeople, yet it was clear to anyone who attended the Royal Society conference in London,as did a number of ID-friendly scientists. The opening presentation by one of those world-class biologists, Austrian evolutionary theorist Gerd Müller. He opened the meeting by discussing several of the fundamental "explanatory deficits" of “the modern synthesis,” that is, textbook neo-Darwinian theory. According to Müller, the as-yet unsolved problems include those of explaining the following:
Phenotypic complexity (the origin of eyes, ears, and body plans, i.e., the anatomical and structural features of living creatures); Phenotypic novelty, i.e., the origin of new forms throughout the history of life (for example, the mammalian radiation some 66 million years ago, in which the major orders of mammals, such as cetaceans, bats, carnivores, enter the fossil record, or even more dramatically, the Cambrian explosion, with most animal body plans appearing more or less without antecedents); and finally: Non-gradual forms or modes of transition, where you see abrupt discontinuities in the fossil record between different types. As Müller has explained in a 2003 work (“On the Origin of Organismal Form,” with Stuart Newman), although “the neo-Darwinian paradigm still represents the central explanatory framework of evolution, as represented by recent textbooks” it “has no theory of the generative.” In other words, the neo-Darwinian mechanism of mutation and natural selection lacks the creative power to generate the novel anatomical traits and forms of life that have arisen during the history of life. Yet, as Müller noted, the neo-Darwinian theory continues to be presented to the public via textbooks as the canonical understanding of how new living forms arose. The conference did an excellent job of defining the problems that evolutionary theory has failed to solve, but it offered little, if anything, by way of new solutions to those longstanding fundamental problems. 4
In the early chapters of this book, I will delve into the limitations of natural selection and genetic drift in explaining the intricate forms of complex organisms. These traditional theories fall short in their predictive power, leaving unanswered questions about the true mechanisms underlying phenotypic complexity and architecture. However, the heart of this book lies in shedding light on the groundbreaking discoveries made by science in recent years. In these recent findings, we unearth layers of biological sophistication that go far beyond genetics. Our perspective shifts from the reductionist approach to a systems view, considering every actor, from the molecular level to ecology. It is an approach that acknowledges the contributions of every level of organization, from the tiny cells to entire organisms and ecosystems, finally culminating in a conclusion that aligns with the evidence and facts presented.
My background as a machine designer informs my approach to investigating and interpreting the marvels of biological systems. Like the intricate workings of human-made artifacts and devices, biological systems exhibit remarkable parallels. From computers and robotics to energy turbines and factories, we discover that cells are veritable chemical factories teeming with machines. This realization moves beyond analogy and delves into a literal understanding.
Does Darwin's Theory of Evolution replace God?
Often atheists claim that Darwin's Theory of Evolution replaces God. Richard Dawkins famously noted that: “Darwin made it possible to be an intellectually fulfilled atheist.” While Darwin supposedly encountered an alternative explanation for the origin of biodiversity, that does not include an explanation for:
It's striking how the universe and living organisms display a remarkable level of complexity and intricacy, resembling the work of a designer. When I examine the intricate structures of cells, the interdependent relationships between organisms and their environments, and the elegant laws governing the universe, it's challenging to dismiss the possibility of a deliberate creator. While evolutionary theory offers insights into how species change over time, it doesn't fully address the question of the origin of life or the underlying design that seems to permeate the natural world. The concept of the universe as a winding clock suggests a beginning point, a moment of creation. The Big Bang theory provides an explanation for the origin of the universe, but it doesn't explain the ultimate cause behind this cosmic event. The fact that the universe had a definite starting point raises profound questions about what might have initiated this process. Evolution may account for the diversity of life within the universe, but it doesn't delve into the origins of the universe itself. The finely-tuned constants and laws of physics that enable the existence of life are remarkable. The precision required for a universe that can support life is truly astounding. Evolutionary theory, while illuminating how species change and adapt over time, doesn't explain why the universe seems meticulously fine-tuned to allow for the emergence of life. The existence of these finely tuned parameters and physical laws raises the question of whether they are the result of chance or intention. When I contemplate the inner workings of a cell, I'm struck by its astonishing complexity. Cells are akin to miniature cities, complete with factories, machinery, and information processing systems. The intricate molecular processes and structures within cells appear to point toward a purposeful design. While evolution provides insights into how species diversify, it doesn't account for the origin and complexity of cellular systems. The presence of complex genetic codes and information within living organisms is a profound mystery. DNA's role as a blueprint for life, along with the intricate processes of gene expression, challenges our understanding of how such sophisticated interdependent information systems could arise solely through unguided processes. Evolutionary mechanisms can account for changes within populations, but the origin of the genetic information, the genetic code and language, and the coding systems themselves remain an unsolved question. Not because science has not investigated it, but because unguided mechanisms are inadequate explanations. Considering all these aspects, it's not a matter of simply arguing from ignorance or inserting a "God of the Gaps." Instead, it's a rational inference based on the observations and evidence at hand. From what I can discern, the presence of intricate designs, fine-tuned parameters, complex information systems, and the orchestrated interplay of diverse components points toward the involvement of an intelligent agent. Just as my own experience tells me that intelligence can produce sophisticated structures, systems, and information, I find it a logical and reasonable inference to conclude that an intelligent creator is the best explanation for the origins and complexities we observe in the universe and life.
Comparing the Evolution from a calculator to a computer, to the evolution from simple, to complex organisms
Let's embark on an imaginative journey into the world of manufacturing and explore the plausibility of a factory evolving from producing calculators to manufacturing computers. In this scenario, we encounter a factory where occasional manufacturing errors introduce variations in the calculators being produced. By a stroke of serendipity, one of these variations unexpectedly enhances the calculator's functionality, capturing the hearts of users and prompting the factory to permanently incorporate the change. However, the transformation from a calculator factory into a computer factory presents a formidable set of challenges. A calculator, with its simple design, performs basic arithmetic operations and typically features a limited number of buttons for numerical input. On the other hand, a computer encompasses complex processing capabilities, storage, input/output devices, an operating system, and a plethora of software applications. Suppose a manufacturing error results in a calculator with slightly more memory or a larger display. While these changes might enhance the calculator's functionality, they fall short of enabling it to become a computer. Additional components such as a keyboard, storage units, a monitor, and interfaces for peripherals would be required. Alas, these components cannot be easily modified or derived from the existing calculator parts. Even if, by a twist of fate, a neighboring factory inadvertently supplies a computer's motherboard to the calculator factory, numerous modifications would still be necessary to integrate it with the existing calculator components. The buttons on the calculator would need to be reconfigured as keys, the display would have to undergo an upgrade to become a full-fledged monitor, and an array of new interfaces and connections would need to be developed from scratch.
The transition from a calculator to a computer entails not only significant changes in manufacturing processes but also a fundamental shift in production flow. Computer manufacturing requires advanced techniques such as printed circuit board assembly, soldering, and chip integration, which differ substantially from the processes employed in calculator production. The factory would find itself on an adventurous path, requiring the acquisition of new machinery, retraining of its workforce, and the establishment of new quality control measures tailored to computer production. Furthermore, the transition would necessitate the introduction of entirely different raw materials and supply chains. Computer components, including integrated circuits, processors, memory modules, and hard drives, would need to be sourced and seamlessly integrated into the production process. This would entail forging relationships with new suppliers, implementing specialized import mechanisms, and incorporating additional testing and validation procedures to ensure the quality and functionality of the computer's components. Additionally, the factory would need to adapt its production lines and infrastructure to accommodate the assembly of computers. The manufacturing process would grow in complexity, involving the installation of various components, the integration of software systems, and the meticulous testing and quality assurance of the final product. The transition from a calculator factory to a computer factory transcends simple modifications and adaptations within the existing production process. It requires the integration of specialized components, the development of intricate interactions and systems, the acquisition of new machinery, the implementation of advanced manufacturing techniques, the sourcing of different raw materials, and the establishment of new supply chains and quality control measures. Applying biological evolution through the gradual accumulation of unguided errors is inasmuch an invalid concept within the natural realm, as applying it to the extraordinary transition from a calculator to a computer. It presents an array of challenges that extend beyond the scope of simple modifications and adaptations within an existing production process.
In the scenario of a factory evolving from producing calculators to manufacturing computers, we can draw an analogy to real-life biological systems and explore why the challenges of such a transition apply similarly. This analogy highlights the complexities and limitations of relying solely on random errors and gradual changes to explain the emergence of complex biological structures and functions. In biological evolution, the concept of random mutations acting over long periods of time to generate complex features is often compared to the manufacturing scenario described above. Just as a calculator factory cannot easily evolve into a computer factory solely through small, accidental changes, biological systems face similar challenges when transitioning from simpler forms to more complex ones. In the biological realm, an initial mutation might introduce a minor improvement in an organism's functionality, similar to the way a manufacturing error could enhance a calculator's features. However, transitioning from simple structures to complex ones, like evolving from basic organs to sophisticated systems in organisms, requires the emergence of entirely new features. Just as a calculator's design is insufficient to accommodate the complexity of a computer, gradual modifications are unlikely to account for the emergence of intricate biological features like eyes, wings, or complex physiological processes. As the calculator factory needs to incorporate the making of new components like keyboards, storage units and monitors to become a computer factory, biological evolution would need to seamlessly integrate new anatomical structures, biochemical pathways, and regulatory mechanisms to facilitate the evolution of complex traits. Simply accumulating small, random changes is insufficient to explain the development of these integrated and coordinated systems. The transition from calculators to computers involves a fundamental shift in complexity and functionality. Similarly, the evolution of simple life forms to more advanced ones entails a profound increase in complexity, with the appearance of new genetic information, integrated with regulatory networks, signaling pathways and processing, feedback loops, codes and languages, and intricate cellular processes that work intrinsically, and extrinsically, in an interdependent fashion.
Intrinsically, because the individual parts are irreducibly complex, and extrinsically, because these parts have only function, integrated in the greater system. The random accumulation of mutations, like manufacturing errors, is not enough to bridge this significant gap. Just as the calculator factory would require new machinery, specialized manufacturing processes, and components to become a computer factory, biological evolution would require the emergence of new genes, proteins, signaling, manufacturing, and regulatory codes, languages, proteins, enzymes, metabolic pathways, elements to drive the formation of new complex traits. These components are often highly specialized and finely tuned, which poses a challenge for gradual and unguided evolutionary processes. A calculator factory's transition to a computer factory would necessitate the establishment of new supply chains, interactions, and quality control measures. Similarly, in biological systems, evolving complex traits requires the simultaneous development of multiple components that interact precisely within cellular networks. These networks are often irreducibly complex, meaning that removing any one component can render the entire system non-functional. The challenges posed by the transition from calculator production to computer manufacturing in your analogy provide insights into why the concept of gradual, unguided evolution faces significant difficulties when applied to explaining the origin of complex biological structures and functions. The need for specialized components, integrated systems, information-rich genetic, epigenetic, manufacturing, signaling, and regulatory codes, and precisely adjusted and finely orchestrated and tuned interactions aligns with the idea that intelligent design or purposeful guidance might be necessary to explain the complexity and diversity observed in the biological world.

Objection: Your argument is basically the old 'irreducible complexity' argument. That's a very weak argument, and is really just an argument from ignorance- not knowing how something evolved does not mean it didn't evolve. All we need to show that evolution is possible for such a system is to show that there are functional, related, less complicated systems possible.
Answer: Charles Darwin spoke about the "complexity of the eye" in his book "On the Origin of Species." He wrote:
"To suppose that the eye, with all its inimitable contrivances for adjusting the focus to different distances, for admitting different amounts of light, and for the correction of spherical and chromatic aberration, could have been formed by natural selection, seems, I freely confess, absurd in the highest possible degree."
Darwin goes on to explain how the eye could have evolved through incremental steps:
"Yet reason tells me, that if numerous gradations from a perfect and complex eye to one very imperfect and simple, each grade being useful to its possessor, can be shown to exist; if further, the eye does vary ever so slightly, and the variations be inherited, which is certainly the case; and if any variation or modification in the organ be ever useful to an animal under changing conditions of life, then the difficulty of believing that a perfect and complex eye could be formed by natural selection, though insuperable by our imagination, can hardly be considered real."
In essence, Darwin was presenting a potential objection to his theory. One of the core tenets of Darwinian evolution is that beneficial traits — those that confer a survival advantage in a particular environment — will be more likely to be passed on to subsequent generations. This process, known as natural selection, is a driving force behind evolutionary change. If it were conclusively shown that there existed a complex trait that could not have arisen through a series of smaller, beneficial steps, then that would pose a significant challenge to the theory of evolution as proposed by Darwin. In other words, if there was a trait for which there were no possible intermediate stages that offered any survival advantage, then natural selection could not account for the emergence of that trait.
Some try to counter-argue by claiming that just because we might not currently understand or have evidence for the intermediate stages of a trait's development doesn't mean they didn't exist. Demonstrating that no possible beneficial stages could have existed is a tall order. In practice, as we continue to gather more knowledge and refine our understanding of biology and genetics, many previously puzzling evolutionary developments have been illuminated.
The appeal to ignorance is the claim that if something has not been proven to be false must be true, and vice versa. (e.g., There is no compelling evidence that UFOs are not visiting the Earth; therefore, UFOs exist, and there is intelligent life elsewhere in the Universe.) Or: There is a teapot revolving in an elliptical orbit between Earth and Mars. Because nobody can prove otherwise, it's true. It also does not consider and ignore that there may have been an insufficient investigation to prove that the proposition is either true or false.
This objection becomes pointless when an exhaustive class of mutually exclusive propositions has been established, a framework of examination, and all possibilities have been carefully examined. Applying Bayesian probability, or abductive reasoning to the best explanation, combined with eliminative induction, we can come to well-informed, plausible, and rational conclusions.
In the Intelligent Design vs. Evolution debate, we have two competing hypotheses, where one can be shown with high certainty false, and the other true. Either an intelligent powerful eternal designer exists and creates the universe, and life, or not. Eliminative inductions argue for the truth of a proposition by arguing that competitors to that proposition are false. Provided the proposition, together with its competitors, forms a mutually exclusive and exhaustive class, eliminating all the competitors entails that the proposition is true. Since either there is a God, or not, either one or the other is true. Eliminative inductions, in fact, become deductions.
Life cannot come from non-life. Science has not been able to come up with even a plausible, coherent model that would show the possible pathway from random molecules to a self-replicating cell. All tests have failed. On every level. Cells have a codified description of themselves in digital form stored in genes and have the machinery to transform that blueprint through information transfer from genotype to phenotype, into an identical representation in analog 3D form, the physical 'reality' of that description. Specified complexity observed in genes dictates and directs the making of irreducible complex molecular machines, robotic molecular production lines, and chemical cell factories. All historical, observational, testable, and repeatable examples have demonstrated that information and operational functionality come from intelligent sources.
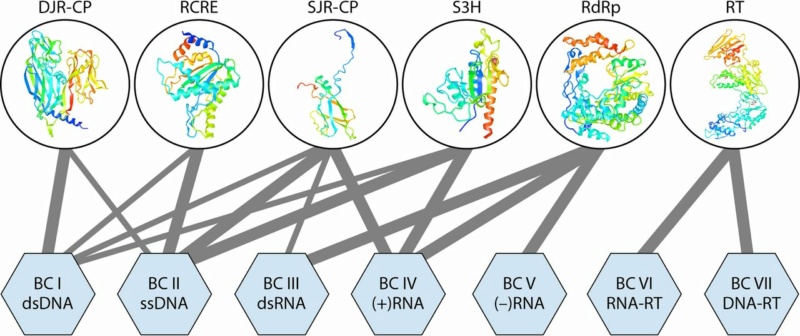
Premise 1: Life is directed and choreographed by instructional, complex, specified information. We find this in the universal genetic code, its 33 distinct variations, and over 230 interconnected manufacturing, signaling, and operational codes 4) . Furthermore, hundreds, if not thousands (in the case of prokaryotes), of complex signaling networks, are interwoven. These networks and codes communicate with and rely upon one another to shape the majestic architecture and immense biodiversity we see in multicellular organisms.
Premise 2: This prodigious reservoir of layered information is far from arbitrary. It's akin to digital semiotic languages, characterized by syntax, semantics, and pragmatics. Whether it's proteins, metabolic pathways, or biomechanical constructs, every component functions with unparalleled precision, adhering to rules optimized for distinct roles. Like an alphabet, these codes are indispensable; having only half a code is akin to possessing half an alphabet—insufficient to convey the crucial information life demands.
Premise 3: Although the molecular structures that convey life are tangible, the very essence of information is ethereal. It's a conceptual entity, operating beyond the capabilities of spontaneous and aimless physical occurrences. To suggest that random processes can spawn semiotic codes is like expecting a rainbow to craft poetry or the wind to draft an architectural blueprint. The directive and purposeful nature evident in biological coding points to intention, foresight, and objective-driven designs.
Conclusion: The breathtaking complexity and evident purpose in the design and diversity of organisms demand an explanation beyond sheer coincident mutations and natural selection. Such intricacies hint not merely at the outcomes of undirected evolution but suggest the craftsmanship of an intentional, intelligent designed setup.
You might be tempted to think, "This is the conclusion; I can stop here." However, I share this insight with you at the outset, not as a full stop, but as an invitation. My hope is to pique your interest and guide you through the journey that led me to this revelation. Instead of placing this at the end, I've chosen to begin with it, to set the stage for the narrative that unfolds.
Biological Complexity and Information: A Case for Intelligent Design
In 1973, the evolutionary biologist Theodosius Dobzhansky, famously stated that "Nothing in Biology Makes Sense Except in the Light of Evolution". This quote was written a half-century ago. Much has changed since then. Scientific inquiry has made huge leaps of progress and unraveled more than ever before, how complex life is. This has led to the conclusion by many that the intricate complexity and diversity found in biological organisms and their architecture are best explained through the lens of intelligent design rather than unguided evolutionary processes. The remarkable organismal complexity and diversity, as well as the emergence of anatomical novelties and biodiversity, are driven by complex informational codes encoded within genetic and epigenetic systems that operate in an interdependent manner together. These codes involve at least 33 variations of genetic codes and over 230 epigenetic manufacturing, signaling, and regulatory codes as the primary contributors to the formation of organismal form, architecture, and biodiversity. This informational complexity is not simply a result of physical processes but emerges from a digital semiotic language. This language encompasses syntax, semantics, and pragmatics, and it is the means through which functional outcomes are achieved. Every protein, metabolic pathway, organelle, or biomechanical structure is framed as functioning based on these variegated semiotic codes, implying an intentional and purposeful arrangement. Information is not a physical entity but a conceptual one. The generation of semiotic codes requires intentionality and foresight, which physical processes lack. Physical processes can create semiotic code is akin to suggesting that a rainbow can write poetry or a blueprint. Information has only been observed to originate from a mind with goals, intentions, and creative foresight.
Creating a cake or a complex machine both require a clear set of instructions that guide the assembly of raw materials into a functional and organized structure. These examples can help us understand the concept of informational complexity and how it relates to the origin of life and biological organisms. To make a cake, a recipe must contain precise details: the types and quantities of ingredients, the order of mixing, the temperature and baking time, and even the method of decoration. All these instructions combine to create a final product with specific characteristics such as taste, texture, and appearance. The recipe serves as a blueprint that transforms basic ingredients into a coherent and well-defined dessert. Similarly, building a machine involves a detailed blueprint that outlines the arrangement of components, their connections, and how they interact. The blueprint provides a step-by-step guide for assembling the machine in a way that ensures it functions as intended. Without this instructional information, the machine's parts would remain disparate and lack the coherence necessary for proper operation. Drawing a parallel, the machinery of life within a cell and the cell itself can be seen as akin to a complex machine and its components. The genome – the genetic blueprint of an organism – contains the instructions necessary to assemble and operate the intricate cellular machinery. Just as a recipe guides the creation of a cake and a blueprint directs the assembly of a machine, the genetic code encodes the information needed to construct proteins, regulate processes, and coordinate the activities of a living cell. The analogy holds that the origin of life and biological complexity requires an initial blueprint or recipe. The intricate interplay of molecular processes, metabolic pathways, and cellular functions depends on precise and specific information stored through the genetic code. This information directs the synthesis of proteins, the control of gene expression, and the orchestration of cell activities. Just as a recipe or blueprint originates from a mind with intelligence and foresight, the intricate molecular choreography within cells and the design of the cell itself point to an intelligent origin that conceived and guided the development of life's informational complexity. Some have objected that cells are Self-replicating, while human-made factories are not. John von Neumann's Universal Constructor is an example of a man-made self-replicating machine. The fact that life is based on self-replication is a significant hallmark of complexity that is not easily achieved through unintelligent processes. Self-replication is not only an advanced feat, but it also requires precise coordination of various processes and components. 593 proteins are involved in human DNA replication and each has essential roles in maintaining the fidelity of genetic information during replication. Comparing cellular processes to factories and manufacturing lines highlights the intricacies of biochemical pathways. The highly organized and efficient nature of these processes implies the requirement of input of a high level of intelligence and intentionality, much like the organization in a man-made factory. This book will demonstrate that everything in biology can be comprehended independently of evolution. Understanding biology is achievable through the lens of intelligent design.
1. Richard Dawkins: In short: Nonfiction
2. John Joe McFadden: Evolution of the best idea that anyone has ever had July 1, 2008
3. New trends in evolutionary biology: biological, philosophical and social science perspectives
4. P. NELSON AND D. KLINGHOFFER: Scientists Confirm: Darwinism Is Broken DECEMBER 13, 2016
Last edited by Otangelo on Thu Feb 29, 2024 6:41 am; edited 130 times in total