X-ray Of Life: Mapping the First Cells and the Challenges of Origins
The Problems: Elucidating the Gap Between Non-Life and Life

Summary of Major Steps and Categorizations:
I. Prebiotic Chemistry: Formation of basic organic molecules from inorganic substances.
II. Self-Replicating Molecules: Emergence of RNA with catalytic and genetic roles.
III. RNA-Peptide Interactions: Transition to systems where proteins begin to play catalytic roles.
IV. Protocell Formation: Development of membrane-bound structures encapsulating biological molecules.
V. Genetic Information Processing: Emergence of DNA, RNA transcription, and protein translation mechanisms.
VI. Metabolic Pathways: Establishment of biochemical reactions for energy production and synthesis of biomolecules.
VII. Early Cellular Life: Formation of cells with defined structures and basic life processes.
VIII. Development of Genetic and Epigenetic Codes, Regulatory and Signaling Mechanisms
IX. Specialized Functions: Adaptations leading to increased complexity and survival capabilities.
X. Integration into Complex Cellular Life
This classification outlines the progression from simple chemicals to complex life, highlighting the major steps and how each item fits into the development of life on Earth. Each category represents a significant leap in complexity, contributing to our understanding of abiogenesis and the origin of biological systems.
1. Prebiotic world
2. RNA world
3. The RNA-Peptide World
4. Proto-Cellular World
5. Early Cellular World
6. Complex Cellular Systems
7. Amino Acid Biosynthesis
8. Carbohydrate Synthesis
9. Lipid Synthesis
10. Cofactors
11. The Complex Web of Central ( Oxaloacetate) Metabolism
12. DNA Replication/Repair
13. Transcription
14. Translation/Ribosome Formation
15. Cellular Transport Systems
16. Cell Division and Structure
17. Epigenetic, manufacturing, signaling, and regulatory codes in the first life forms
18. Signaling and Regulation in Early Life
19. RNA Processing in Early Life: A Complex System of Interdependent Components
20. Cellular Defense and Stress Response
21. Proteolysis in Early Life Forms
22. Thermoprotection in the First Life Forms
23. Metal Clusters and Metalloenzymes
24. Polyamine Synthesis
25. Motility in Early Life Forms: A Case for Primitive Flagella
26. General Secretion Pathway Components
27. Formation of enzymatic proteins
28. Cellular Quality Control Mechanisms
Book Abstract
"The First Cell: Elucidating the Gap Between Non-Life and Life" is a comprehensive exploration of the origin of life, focusing on the transition from prebiotic chemicals to the first living, self-replicating cell. This book examines the molecular machinery, biosynthetic pathways, and information systems required for life to emerge, questioning the plausibility and feasibility of unguided, naturalistic processes in this extraordinary event. From the prebiotic world of simple organic compounds to the emergence of sophisticated cellular functions, each chapter goes into elucidating the specific systems and components necessary for life. The book scrutinizes the quantum leap from non-living matter to living organisms, addressing the challenges of forming critical biomolecules, establishing replication mechanisms, and developing metabolic processes. By presenting the latest scientific findings, this work aims to provide a balanced, in-depth analysis of the origin of life question. It concludes by examining the implications of the presented evidence, inviting readers to critically evaluate the adequacy of purely naturalistic explanations for life's origins.
Introduction
The origin of life stands as one of the most challenging questions in science. How did the first living cell arise from non-living matter? Our journey begins in the prebiotic world, where we examine the synthesis of organic compounds and the formation of autocatalytic reaction sets. We then explore the proposed RNA World hypothesis, considering the challenges of achieving homochirality and the potential roles of RNA in early life. As we progress, we go further into the formation of more complex systems, including the synthesis of proteins and the encapsulation of these components within vesicles. We examine the requirements for the first enzyme-mediated cells and the development of sophisticated cellular functions. Throughout this exploration, we pay close attention to the information content required in the genome to specify these complex systems. We consider the interplay between nucleic acids, proteins, and metabolic processes, and how these might have co-emerged to produce the first living cell.
At each stage, we highlight the open questions and challenges faced by researchers in this field. We examine the probabilistic hurdles, the issue of generating and maintaining biological information, and the problem of achieving integrated functionality in a prebiotic setting. This book does not presuppose any particular framework for the origin of life. Instead, it aims to present the empirical data and theoretical models currently available, encouraging readers to critically evaluate the evidence. By the end of this journey, readers will have a comprehensive understanding of the immense complexity involved in the origin of life. We will consider whether the cumulative evidence supports the idea that life could have emerged through unguided, naturalistic processes on the early Earth, or whether the data points to alternative explanations. This exploration invites us to marvel at the sophistication of even the simplest living cell and challenges us to grapple with one of the most fundamental questions in science: how did life begin?
Eugene V. Koonin: The Logic of Chance (2012): Despite many interesting results to its credit, when judged by the straightforward criterion of reaching (or even approaching) the ultimate goal, the origin of life field is a failure—we still do not have even a plausible coherent model, let alone a validated scenario, for the emergence of life on Earth. Certainly, this is due not to a lack of experimental and theoretical effort, but to the extraordinary intrinsic difficulty and complexity of the problem. A succession of exceedingly unlikely steps is essential for the origin of life, from the synthesis and accumulation of nucleotides to the origin of translation; through the multiplication of probabilities, these make the final outcome seem almost like a miracle.1
This quest involves chemistry, biology, and physics, pushing the boundaries of our understanding and challenging our preconceptions about the nature of life itself. Throughout this period, we have witnessed a consistent commitment to finding naturalistic explanations for life's origin. Researchers have moved from broad concepts to more specific chemical scenarios, acknowledging the challenges while remaining dedicated to scientific explanations. This progression reflects the scientific method in action: starting with hypotheses, conducting experiments, refining ideas, and gradually building a more comprehensive understanding. However, as our knowledge of life's complexity has grown, so too has the challenge of explaining its emergence. Each new discovery seems to widen the gap between our understanding of chemistry and our grasp of biology's complex systems. The transition from non-life to life, once thought to be a small step, now appears to be a quantum leap of staggering proportions.
Here, we explore the current state of origin-of-life research, examining the paradoxical situation we find ourselves in: armed with more knowledge than ever before, yet seemingly further from a complete explanation of life's beginnings. We will go into the challenges that have emerged, the new questions that have arisen, and the potential pathways forward in this critical area of scientific inquiry. As we embark on this journey, we will confront the possibility that the origin of life may require us to rethink our fundamental assumptions about the nature of matter, energy, and information. We will explore cutting-edge theories and experiments that seek to bridge the chasm between non-life and life, and consider the implications of this research for our understanding of life, the universe, and our place within it.
Throughout time, we see a consistent commitment to finding naturalistic explanations for life's origin. Researchers have moved from broad concepts (Oparin, Haldane) to more specific chemical scenarios (Miller), acknowledging the challenges (Crick) while remaining committed to scientific explanations (Dawkins, Szostak, Lane).
Alexander Oparin (1930s): The first stage in the origin of life was the formation of simple organic compounds from the atmospheric gases.
J.B.S. Haldane (1940s): The origin of life was essentially a chemical process.
Stanley Miller (1950s): The idea that the organic compounds that serve as the basis of life were formed when the primitive Earth had a reducing atmosphere is commonly accepted today.
Adler (1959) in the book " How life began" 1 wrote: The development of life appears as something that just happened, without any design or purpose. It started from the accidental mixing and combining of chemicals in the primitive sea. But the direction of development it took was not all accidental. It was influenced by the natural preferences the chemical elements have for each other. It was built on the basis of carbon's ability to form long-chain compounds. In the later stages of chemical evolution, it was also directed by the effects of natural selection. The rule of survival of the fittest guided evolution toward the development of more complicated and more efficient organisms, and finally toward the emergence of intelligent beings.
Francis Crick (1960s): The origin of life appears at the moment to be almost a miracle, so many are the conditions which would have had to have been satisfied to get it going.
Richard Dawkins (1980s): The illusion of design is a trap that has ensnared many people. The beauty of Darwin's theory is that it explains the illusion of design without requiring a designer.
In 1996, Lynn Margulis was interviewed by John Horgan, and it was published in Horgan's book: The End of Science 2 The smallest bacterium, she ( Margulis ) noted, “is so much more like people than Stanley Miller’s mixtures of chemicals, because it already has these system properties. So to go from a bacterium to people is less of a step than to go from a mixture of amino acids to that bacterium.”
Today, we find ourselves even further from solving this puzzle than we were 70 years ago, when abiogenesis research began in earnest with the discovery of DNA's structure by Watson and Crick and the Miller-Urey experiment synthesizing amino acids under simulated early Earth conditions. Despite our increasing understanding of life's complex machinery, the fundamental question of how inanimate matter transformed into the first living cell remains more elusive than ever. This book elucidates the current state of origin-of-life research, exploring the widening chasm between our knowledge of life's complexity and our ability to explain its emergence through natural processes. It examines the challenges, paradoxes, and potential new directions in the quest to unravel one of science's most enduring enigmas: the nature of life's quantum leap from chemistry to biology.
Jack Szostak (2000s): The origin of life is a problem in chemistry that we hope to solve in the next few decades.
Nick Lane (2010s): Life is not a miracle, but a natural consequence of the laws of physics and chemistry.
This progression reflects the scientific method in action: starting with hypotheses, conducting experiments, refining ideas, and gradually building a more comprehensive understanding. While the origin of life remains an open question, these researchers share a common belief that the answer lies within the realm of natural processes, governed by chemistry and physics. This is paradoxa, because, rather than bringing us closer to understanding how the transition from non-life to life could have occurred by natural means, these discoveries have paradoxically widened the gaps in our knowledge.
What is Life?
Paul Davies suggests that life can be understood as the combination of chemistry and information, emphasizing that both components are essential to define and explain living systems. Life, in its various forms, is a multifaceted phenomenon. From the simplest single-celled organisms to the most complex ecosystems, living entities exhibit a range of defining characteristics that set them apart from non-living systems. While life is notoriously difficult to define, certain properties emerge as key markers of living systems. These properties encompass not only biological replication and metabolism but also the transfer of information, structural organization, and the interplay between permanence and change. Understanding the essential characteristics of life is vital for comprehending its underlying principles, both in present forms and in its earliest origins on Earth.
Reproduction: Reproduction is a fundamental characteristic of life, enabling organisms to propagate their genetic material across generations. However, reproduction is more than just copying genetic information; it also involves replicating the apparatus necessary for this process. Interestingly, some nonliving entities, like crystals and fires, exhibit a form of replication, while viruses, which many consider borderline living, cannot reproduce without a host. Notably, certain living organisms like mules are sterile, yet undeniably alive. For life to sustain itself beyond a single generation, the replication apparatus must be faithfully copied alongside the genes themselves, ensuring that life continues to propagate effectively.
Metabolism: At the core of life lies metabolism – the complex web of chemical reactions that organisms use to process energy and sustain their activities. Living organisms metabolize nutrients to release energy for various functions such as movement, reproduction, and growth. Dormant microorganisms, which suspend their metabolic functions for extended periods, illustrate that metabolism is a hallmark of life, but not always continuously active. Their ability to return to a living state upon favorable conditions suggests that metabolic capability, rather than constant metabolic activity, is key to defining life.
Nutrition: Closely tied to metabolism, nutrition involves the intake and processing of matter and energy necessary to fuel an organism's survival. While plants harness solar energy through photosynthesis and animals consume organic matter, nutrition alone does not define life. A continual flow of energy through non-living systems, such as Jupiter's Great Red Spot, demonstrates that life requires not just energy but the ability to harness useful, free energy for sustaining complex biochemical processes.
Complexity: Life is characterized by its extraordinary complexity, with even the smallest bacteria exhibiting intricate internal structures and processes. Living organisms are not just complicated but organized in such a way that they maintain their functions and adaptability. While non-living systems like hurricanes and galaxies can also be highly complex, it is the organized and purposeful nature of biological complexity that distinguishes life from non-life.
Organization: It is not merely complexity that defines life, but organized complexity. Living organisms exhibit a high degree of internal coordination, where all components—whether cells, tissues, or organ systems—work in harmony. This organization is not random; each part of the organism must function about the whole. For example, arteries, veins, and the heart work together to circulate blood, just as proteins and enzymes within cells coordinate to facilitate biochemical reactions. Without such organized complexity, life would cease to function coherently.
Growth and Development: Living organisms not only grow but also undergo development, which involves the differentiation and maturation of structures over time. While non-living entities, such as crystals, can grow by accumulating material, biological growth is coupled with complex developmental processes. Variation and novelty, particularly in response to environmental pressures, lead to adaptation and evolution, key factors that distinguish living organisms from inanimate objects.
Information Content: One of the hallmarks of life is the transmission of genetic information from one generation to the next. This transfer is not just a simple copying process; it involves highly specified information encoded within genes. Life, in this sense, can be seen as a form of information technology, where genetic instructions drive the formation and function of organisms. The meaningfulness of this information—how it is interpreted and used by cellular machinery—makes it fundamentally different from random patterns seen in non-living systems.
Hardware/Software Entanglement: Life's complexity arises from the interplay between nucleic acids (DNA and RNA) and proteins, which form a sophisticated system of biological hardware and software. Nucleic acids store the instructions for life, while proteins carry out the physical work. This entanglement is mediated by the genetic code, a communication system that ensures the proper translation of genetic information into functional proteins. This interdependence between biological "hardware" and "software" is unique to living systems.
Permanence and Change: A paradox at the heart of life is the balance between conservation and variation. Genes are designed to replicate and preserve the genetic message across generations. However, variation is essential for adaptation and survival, as it allows populations to evolve in response to environmental changes. This dynamic balance between permanence and change is a defining feature of life and plays a central role in the continued success of living organisms on Earth.
The Journey from Non-Life to Life: A Scientific Perspective from Half a Century Ago
Starting about 100 years ago, scientists and researchers envisioned the transition from non-living matter to life as follows: Scientists studying life's beginnings started in the attempt of explaining how natural processes could have transformed simple carbon and nitrogen compounds in ancient oceans into living entities. Pioneers like A. I. Oparin (1924) and J. B. S. Haldane (1929) were the first to seriously tackle this enigma. Their work inspired numerous scientists to conduct laboratory experiments and formulate theories based on their findings. Notable contributors included J. D. Bernal, M. Calvin, S. W. Fox, S. L. Miller, L. E. Orgel, J. Oró, C. A. Ponnamperuma, and H. C. Urey. Researchers proposed that the chemical evolution leading to life occurred in three distinct phases. The first phase involved the creation of simple building blocks monomers. The second phase saw the development of complex molecules or polymers. The final phase encompassed the organization of these polymers into self-regulating systems that we recognize as living cells. Scientists theorized that the formation of amino acids and nitrogen bases required energy. They identified several potential energy sources present on Earth over three billion years ago, including lightning, radiation from radioactive elements and cosmic rays, solar ultraviolet rays, meteorite impacts, and heat. In 1953, S. L. Miller conducted a groundbreaking experiment. He created a mixture simulating Earth's early atmosphere and subjected it to electrical discharges, mimicking lightning. This process successfully produced amino acids, demonstrating that primordial energy sources could have generated these crucial building blocks. Subsequent experiments showed that all theorized energy sources from the early Earth could potentially form amino acids in both atmospheric and oceanic conditions. Additionally, chemical analysis of meteorites suggested that amino acids might have formed in interplanetary dust and arrived on Earth via meteorite impacts. Researchers proposed various mechanisms for concentrating monomers to facilitate their combination into biologically significant polymers. Oparin suggested coacervate formation, while Bernal proposed adsorption onto clay, which then catalyzed monomer joining. Fox hypothesized concentration through evaporation in tidal pools, followed by heat-induced joining. Calvin and Oró suggested that certain cyanogen-derived compounds could enable direct polymer formation in seawater without prior concentration. Laboratory experiments confirmed the viability of all these proposed methods, indicating multiple potential pathways for natural processes to combine amino acids into protein-like chains and to form nucleic acids from nitrogen bases, sugars, and phosphates. Researchers also noted that spontaneous amino acid combinations in laboratories showed preferences for certain pairings, mirroring patterns observed in cellular proteins. This similarity suggested that protein molecule amino acid sequences evolved from natural affinities between amino acids. Scientists theorized that the accidental joining of metal molecules (like iron) to amino acids might have initiated enzyme evolution. They also proposed that the combination of phosphates with nitrogen bases and sugars could have led to ATP formation. Additionally, experiments showed that phospholipids and proteins could spontaneously form sheets resembling cell membranes under specific conditions.
This final phase remained the most mysterious, with limited experimental evidence to support theories. Scientists were unsure whether it occurred after or concurrently with polymer formation. Despite the lack of concrete evidence, these theories were valuable in guiding future experiments and refining hypotheses. Oparin's coacervate theory provided the most detailed scenario for cell evolution. He envisioned the primordial sea as a vast "organic soup" where increasingly complex molecules formed through natural processes. Amino acids, capable of diverse combinations, produced a variety of protein molecules large enough to form colloidal particles, gradually transforming the sea into a colloidal solution. These colloidal particles sometimes formed clusters known as coacervates, separated from the surrounding sea by a thin water molecule skin. This development was seen as a crucial step towards the formation of protoplasm. The coacervate's skin acted as both a barrier and a passageway, allowing a steady flow of molecules in and out of the cluster. Over time, some coacervates developed a balance between breakdown and buildup processes, leading to stability and growth. This balance was seen as a precursor to cellular metabolism. The need for a continuous supply of molecules for the building-up process was likened to the "feeding" behavior of living organisms. Scientists proposed that a form of natural selection occurred among these coacervates, with the more stable ones persisting and evolving. They suggested that some coacervates developed catalytic processes that enhanced their efficiency, leading to the eventual evolution of enzyme systems. As organic compounds in the sea were increasingly absorbed by coacervates, competition for resources intensified. This "struggle for existence" was thought to have accelerated the process of natural selection, favoring coacervates with the most stable metabolism, efficient enzyme systems, and effective reproduction mechanisms. Through countless small changes over millions of years, researchers believed that coacervates eventually developed into the first primitive living organisms. These early life forms were imagined to be even simpler than bacteria or blue-green algae, obtaining energy through anaerobic fermentation processes. Despite their simplicity, scientists speculated that these organisms must have possessed some mechanism for energy storage and transfer, possibly involving ATP or a similar compound. This narrative represents the scientific community's best understanding of life's origins up to approximately half a century ago. It laid the groundwork for further research and continues to influence our quest to unravel the mysteries of life's beginnings.
The scientific understanding of life's origins has evolved significantly over the past half-century. In the 1980s, the discovery of ribozymes (RNA molecules with catalytic properties) by Thomas Cech and Sidney Altman led to the formulation of the RNA World hypothesis. This theory proposed that RNA could have served as both a genetic material and a catalyst in early life forms, potentially bridging the gap between prebiotic chemistry and cellular life. Researchers like Stanley Miller continued to refine prebiotic synthesis experiments, exploring a wider range of potential early Earth conditions. The discovery of extraterrestrial organic compounds in meteorites and comets strengthened the idea that some of life's building blocks might have an cosmic origin. The field of systems chemistry emerged, focusing on how complex networks of chemical reactions could lead to self-sustaining, self-replicating systems. Jack Szostak and others made significant progress in creating socalled protocells - simple membrane-enclosed structures capable of growth and division. Hydrothermal vents, particularly alkaline vents, gained attention as potential cradles of life. Researchers like Nick Lane proposed that the chemical and energy gradients in these environments could have driven the formation of early metabolic processes.
Current Understanding
1. Multiple Hypotheses: There's no single, universally accepted theory for the origin of life. Instead, several competing hypotheses exist, including the RNA World, the Iron-Sulfur World, and the Lipid World.
2. Complexity of Early Life: We now recognize that even the simplest forms of life are incredibly complex. The gap between prebiotic chemistry and the first cell appears larger than previously thought.
3. Information Problem: A key challenge is explaining how complex, information-rich polymers like RNA or DNA could have arisen and replicated before the existence of enzymes.
4. Metabolism-First vs. Replication-First: Debates continue over whether metabolism or replication came first in the origin of life, with some researchers proposing that both might have co-evolved.
5. Role of Compartmentalization: The importance of creating bounded systems (like protocells) for concentrating reactants and enabling evolution is increasingly recognized.
6. Expanded Prebiotic Inventory: Our understanding of the chemical diversity possible under prebiotic conditions has grown, including the potential for more complex organic synthesis.
7. Importance of Energy Flows: There's increased focus on how energy fluxes and disequilibrium could have driven the emergence of life.
8. Synthetic Biology Approaches: Researchers are using synthetic biology techniques to reconstruct possible early biological systems, providing new insights into life's origins.
9. Exoplanet Studies: The discovery of numerous exoplanets has broadened our perspective on the potential conditions for life's emergence.
Despite these advancements, the origin of life remains one of science's greatest unsolved mysteries. The complexity of even the simplest living systems and the vast timescales involved continue to challenge our understanding. Current research is increasingly interdisciplinary, drawing on fields ranging from astronomy to quantum physics in the quest to understand how life began.
The Origin of Life: Various proposals over time
Numerous hypotheses attempt to unravel the processes that led to the emergence of life from non-living matter. Each theory brings its perspective, from the role of chemical reactions in primordial environments to the formation of self-replicating molecules. The journey through these ideas reveals not only the ingenuity of scientific thought but also the immense challenge of piecing together the puzzle of life's beginnings. While each hypothesis offers valuable insights, they collectively underline the fundamental issue: can naturalistic, unguided processes sufficiently account for the origin of life?
1. 1866: Haeckel's Monera Hypothesis: Proposed by Ernst Haeckel. He suggested that life originated from simple, homogeneous substances called "Monera," self-organizing into living organisms.
2. 1920s: The Oparin-Haldane Hypothesis: Proposed by Aleksandr Oparin and J.B.S. Haldane. This theory posits that life originated from organic compounds synthesized in a reducing atmosphere, with energy from lightning or ultraviolet light.
3. 1920s: Heterotroph Hypothesis: Proposed by early biologists. It suggests the first organisms were heterotrophs that consumed organic molecules, eventually leading to the development of autotrophy and oxygen release.
4. 1930s: Coacervate Hypothesis: Proposed by Oparin. He suggested that life began with the formation of coacervates—droplets of organic molecules that aggregated and began to exhibit basic metabolic activity.
5. 1950s: Fox's Microsphere Hypothesis: Proposed by Sidney Fox. He theorized that life began with the formation of microspheres, tiny droplets of amino acids, capable of growing and dividing, mimicking life-like processes.
6. 1950s: Electric Spark Hypothesis: Based on the idea that lightning could have sparked chemical reactions in the Earth's early atmosphere, producing organic compounds from simple molecules, as demonstrated by the Miller-Urey experiment.
7. 1953: Miller-Urey Experiment: Conducted by Stanley Miller and Harold Urey. This experiment demonstrated that amino acids, the building blocks of proteins, could form spontaneously under early Earth conditions, supporting Oparin-Haldane's hypothesis.
8. 1970s: Eigen's Hypercycle Hypothesis: Proposed by Manfred Eigen. This theory suggests that life began with self-replicating molecules, interacting in a hypercycle, a system of cooperative feedback loops that allowed for the evolution of complexity.
9. 1970s: Autocatalytic Networks Hypothesis: Introduced by Stuart Kauffman, suggests that life began as a set of self-replicating and self-sustaining autocatalytic chemical networks that could grow and evolve through natural selection.
10. 1977: Submarine Hot Springs Hypothesis: Proposed after the discovery of hydrothermal vents. This hypothesis suggests that the energy and chemical conditions at oceanic ridge crests may have initiated life.
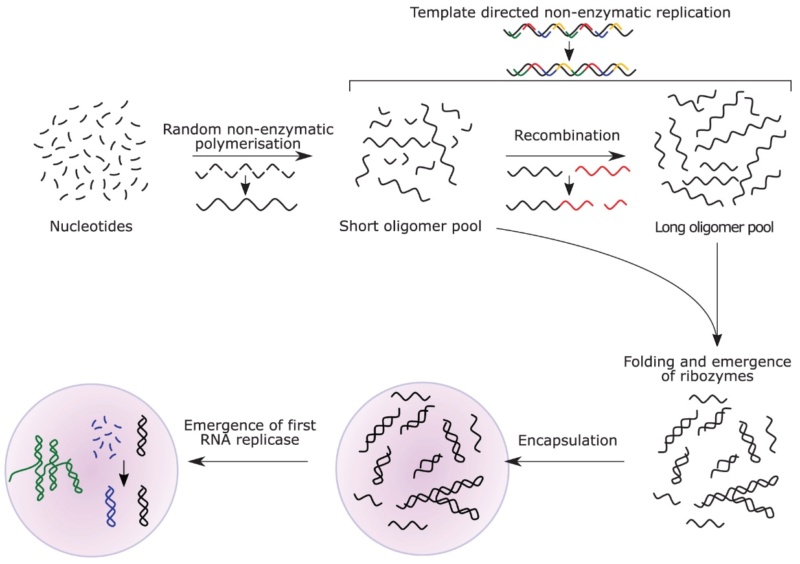
11. 1980s: RNA World Hypothesis: Proposed by several scientists, including Walter Gilbert. This hypothesis suggests that early life forms were based on RNA, which both stored genetic information and catalyzed chemical reactions.
12. 1980s: Deep Sea Vent Hypothesis: Posits that life originated around hydrothermal vents deep in the ocean, where superheated water rich in minerals provided the energy and chemical conditions necessary for early life.
13. 1980s: Iron-Sulfur Cluster Hypothesis: Suggests that iron-sulfur clusters, common in enzymes today, were among the first catalysts for life, helping drive chemical reactions critical for the origin of life.
14. 1980s: Clay Hypothesis: Proposed by Graham Cairns-Smith, suggests that life originated on the surface of clay minerals, which helped catalyze organic reactions, leading to the formation of early biochemical compounds.
15. 1988: Iron-Sulfur World Hypothesis: Proposed by Günter Wächtershäuser. This theory posits that life originated on iron and nickel sulfide surfaces near hydrothermal vents, where organic molecules were synthesized through catalysis.
16. 1988: Pyrite Formation Hypothesis: Proposed by Wächtershäuser. He claimed that the formation of pyrite (FeS2) from hydrogen sulfide and iron provided an energy source for early autotrophic life forms.
17. 1993: Bubbles Hypothesis: Proposed by researchers. It suggests that bubbles on the surface of the primordial seas could have concentrated and catalyzed organic molecules, eventually leading to the first living cells.
18. 1995: Thermoreduction Hypothesis: Proposed by evolutionary biologists. This theory posits that life originated from thermophiles in extreme heat environments, possibly linked to the Last Universal Common Ancestor (LUCA).
19. 1997: Protein Interaction World Hypothesis: Proposed by Michael Yarus. This theory suggests that life originated from a system of self-reproducing protein interactions before the emergence of nucleic acids.
20. Early 2000s: Lipid World Hypothesis: Suggests that self-replicating lipid structures formed the basis for early life, with membranes forming before genetic material like RNA or DNA.
21. Early 2000s: PAH World Hypothesis (Polycyclic Aromatic Hydrocarbons): Suggests that life may have originated from PAHs, which could have acted as scaffolding to help form more complex organic molecules like RNA.
22. 2004: Organic Aerosols Hypothesis: Proposed by K.P. Wickramasinghe. It suggests that aerosols composed of amphiphiles on the ocean's surface divided and led to chemical differentiation and the formation of protocells.
23. 2004: Hydrogel Environment Hypothesis: Proposed by Tadashi Sugawara. This theory posits that early life emerged in hydrogel environments that concentrated water, gases, and organic molecules.
24. 2005: Dual Origin Hypothesis: This hypothesis extends the dual ancestral development concept to the origin of life, explaining that two different systems may have evolved into the complex interplay of genetics and metabolism.
25. 2009: Zinc World Hypothesis: Proposed by Armen Mulkidjanian. This theory suggests that life began in hydrothermal environments rich in zinc sulfide, utilizing sunlight for organic synthesis.
26. 2010: Thermodynamic Origin of Life Hypothesis: Suggests that life emerged as a natural outcome of the Earth's thermodynamic drive to dissipate solar energy by increasing entropy through irreversible processes.
27. 2011: Asteroids and Formamide Hypothesis: Researchers showed that the combination of meteorite material and formamide could produce nucleic acids and other biomolecules under prebiotic conditions.
28. 2011: Thermodynamic Dissipation Theory: Suggests that life originated as a mechanism for increasing the Earth's entropy by absorbing and transforming sunlight into heat.
29. 2013: Self-Assembling Molecules Hypothesis: Proposed by Georgia Tech researchers. It demonstrated that RNA components could self-assemble in water, offering a pathway for RNA to form prebiotically.
30. 2015: GADV-Protein World Hypothesis: Posits that life began with peptides composed of Gly, Ala, Asp, and Val, which exhibited catalytic activity before RNA emerged.
31. 2015: Meteorite and Solar Wind Hypothesis: Italian researchers suggest that solar wind interacting with meteorite material could have created life's building blocks before they arrived on Earth.
32. 2016: Minimotif Synthesis Hypothesis: Suggests a feed-forward catalytic system in which small peptides emerged first, followed by RNA and genetic encoding.
33. 2016: Hydrothermal Vent Models (Near-Inevitable Life): Posits that life was a near-inevitable consequence of chemical conditions at hydrothermal vents, rather than a miraculous event.
34. 2016: LUCA Near Underwater Volcanoes Hypothesis: Suggests that the Last Universal Common Ancestor (LUCA) lived near hydrothermal vents and metabolized hydrogen.
35. 2016: Primordial Soup and Shocks Hypothesis: Proposes that shocks from meteorite impacts or lightning could have contributed to the synthesis of organic molecules in the primordial soup.
36. 2017: Peptide-Nucleic Acid Replicator Hypothesis: Posits that life originated from a replicating system composed of both peptides and nucleic acids, rather than RNA alone.
37. 2017: Foldamer Hypothesis: Suggests that prebiotic polymers could grow in sequence and length through folding and self-binding, promoting self-replication.
38. 2017: Droplet Hypothesis: Suggests that droplets in a primordial soup could have exhibited replication and growth, possibly leading to early cellular life.
39. 2017: Chemically Driven RNA Hypothesis: Demonstrates how simple chemical reactions on the early Earth could have produced RNA precursors.
40. 2017: Phase Transition Hypothesis: Suggests that life emerged as a first-order phase transition, where replicators began to outcompete non-living systems, leading to rapid evolutionary diversification.
41. 2017: Modular Hierarchy Hypothesis: Suggests that molecular complementarity and modular hierarchies were essential for the chemical systems that eventually gave rise to life.
42. 2018: Viral Birth of DNA Hypothesis: Proposed by researchers studying viruses in hot, acidic environments. This hypothesis suggests that viruses may have played a key role in the transition from RNA-based life to DNA-based life by performing the transfer of genetic information from RNA to DNA.
43. 2019: Photochemical Origin of Life Hypothesis: Proposed by various researchers. This theory posits that ultraviolet light played a crucial role in driving the chemical reactions that led to the formation of organic molecules necessary for life.
44. 2019: Peptide-RNA World Hypothesis: Proposed as an extension of the RNA World Hypothesis. It suggests that early life involved both peptides and RNA co-evolving, which may have helped overcome some limitations of RNA alone as the origin of life.
45. 2020: Wet-Dry Cycle Hypothesis: Suggests that life began in environments experiencing wet-dry cycles, such as tidal pools or ponds. These cycles could have driven the formation of increasingly complex polymers like RNA and proteins by concentrating and activating building blocks.
46. 2020: Hydrothermal Cliff Hypothesis: This theory proposes that life originated near underwater cliffs or porous rock formations, where minerals and hydrothermal fluids created ideal microenvironments for the development of early metabolic systems and the formation of cell-like structures.
47. 2020: Phosphate-Driven Origin Hypothesis: This hypothesis suggests that phosphorus-containing minerals were critical in the formation of early biomolecules. Phosphorus, essential for DNA, RNA, and ATP, may have been sourced from minerals like schreibersite delivered by meteorites or found near hydrothermal vents.
48. 2021: Metabolism-First Hypothesis (Updated): Researchers expanded on the original metabolism-first hypothesis, suggesting that self-sustaining metabolic pathways could have formed in deep-sea hydrothermal vents, predating the emergence of genetic materials like RNA or DNA.
49. 2022: Chemical Evolution of Exoplanets Hypothesis: A newer hypothesis inspired by exoplanet research. It proposes that life could have originated on other planets under extreme chemical and environmental conditions, similar to those found on early Earth, and could have been transported to Earth via panspermia.
50. 2022: Electrochemical Origin Hypothesis: Suggests that electric fields, particularly in environments near volcanoes or within the Earth’s crust, might have helped concentrate key ions and organic compounds, kickstarting metabolic and replicative systems.
51. 2023: Quantum Origin of Life Hypothesis: An emerging field of research posits that quantum phenomena, such as tunneling and entanglement, could have played a fundamental role in the formation of early life by influencing molecular interactions and chemical reactions critical to the origin of life.
The search for understanding the origin of life has led to numerous hypotheses, each attempting to tackle the mystery from a unique perspective. From the early proposals of simple self-organizing entities to more recent ideas about quantum phenomena influencing life's genesis, the field has evolved with each new discovery.
The Journey from Prebiotic Chemicals to the Progenote, to Modern Life Domains
The list shows the progression from the simplest level of organization (atoms) to the most complex (ecosystems), representing a bottom-up approach to understanding biological organization and complexity. It is a very general scheme, intended to show merely the increasing organizational complexity of several processes. Although eight steps are shown, leading from atoms to ecosystems, there is one step that far outweighs the others in enormity: the step from macromolecules to cells. All the other steps can be accounted for on theoretical grounds—if not correctly, at least elegantly. However, the macromolecule-to-cell transition is a jump of fantastic dimensions, which lies beyond the range of testable hypotheses. In this area all is conjecture. The available facts do not provide a basis for postulating that cells arose on this planet. This is not to say that some parachemical forces were at work. We wish to point out the fact that there is no scientific evidence. The physicist had learned to avoid trying to specify when time began and when the matter was created, except within the framework of frank speculation. The origin of the precursor cell appears to fall into the same category of unknowables. It is an area with fascinating conceptual changes, but at present, and perhaps forever, the facts cannot be known. To postulate that life arose elsewhere in the universe and was then brought to earth by some means would be merely begging the question; we must still answer how life arose wherever it may have done so originally.
Stages of Bottom-Up Development
1. Atoms Fundamental building blocks of matter.
2. Prebiotic chemical synthesis Formation of simple organic molecules from inorganic precursors in early Earth conditions.
3. Molecular self-assembly Creation of more complex structures like protocells through spontaneous organization of molecules.
4. RNA World Development of self-replicating RNA molecules capable of storing information and catalyzing reactions.
5. RNA-Protein World Emergence of primitive translation systems, allowing RNA to guide protein synthesis.
6. DNA-RNA-Protein World Transition to DNA as the primary genetic material, with RNA as an intermediate and proteins as functional molecules.
7. Proto-cell formation Development of lipid membranes and basic metabolic processes, creating the first cell-like structures.
8. LUCA (Last Universal Common Ancestor) Emergence of a self-replicating organism with DNA, RNA, and proteins, from which all current life descended.
9. Individuals are Distinct, self-contained living entities capable of independent existence.
10. Populations Groups of individuals of the same species living in a particular area.
11. Species, communities Interbreeding groups and diverse assemblages of different species interacting in an ecosystem.
12. Ecosystems Complex networks of living organisms interacting with their physical environment.

A (conjectured) brief sketch of the history of life. At present, life is divided into three domains: Bacteria, Archaea and Eukaryota. Following the lineages of the three domains backward in time (solid lines), we find that they coalesce into the Last Universal Common Ancestor (LUCA), approximately 3.8 Gya. The dashed red line indicates the point in time where it is thought that the Darwinian transition occurred: before that, life was evolving in a communal way (progenote); after the Darwinian transition, life evolved as described by the Modern Synthesis. 1
Last Universal Common Ancestor (LUCA) vs First Universal Common Ancestor (FUCA)
LUCA (Last Universal Common Ancestor)
Definition: Most recent common ancestor of all current life
Timeframe: ~3.5-3.8 billion years ago
Characteristics:
Simple, single-celled organism
DNA-based genetic code
Ribosomes for protein synthesis
Basic cellular machinery and metabolism
Cell membrane
FUCA (First Universal Common Ancestor)
Definition: Hypothetical first common ancestor of all life
Timeframe: Earlier than LUCA, possibly >3.8 billion years ago
Characteristics (highly speculative):
Primitive genetic material (possibly RNA)
Basic metabolic processes
Rudimentary cell-like structures
Key Differences
Complexity: LUCA more complex than FUCA
Genetic Material: LUCA likely DNA-based, FUCA possibly RNA-based
Evolutionary Stage: LUCA later stage, FUCA earlier stage
Evidence: More indirect evidence for LUCA, FUCA largely speculative
Why FUCA is Problematic in Origin of Life Studies
In Origin of Life research, focusing on FUCA presents significant challenges:
Lack of Evidence: There is virtually no direct evidence about the nature of FUCA due to its extreme antiquity.
High Speculation: Any attempt to define FUCA relies heavily on speculation rather than scientific data. The period between the origin of life and LUCA makes it difficult to pinpoint a single "first" ancestor.
Definition Issues: The boundary between non-life and life is blurry, making it challenging to define what qualifies as the "first" living entity.
Methodological Limitations: Current scientific methods are not capable of providing concrete information about life forms from such an early period.
Advantages of Focusing on LUCA in Bottom-Up Research
More Tangible Evidence: Comparative genomics provides indirect but substantial evidence about LUCA's nature.
Clearer Definition: LUCA represents a more defined point in evolutionary history.
Practical Research Target: Studying LUCA allows for more concrete hypotheses and experimental designs.
Bridge to Modern Life: LUCA serves as a crucial link between early life and the diversity we see today.
Consensus in Scientific Community: There's broader agreement on LUCA's existence and general characteristics.
While the concept of FUCA is intriguing, its highly speculative nature makes it less practical for scientific investigation. Focusing on LUCA as a threshold for investigation in bottom-up research provides a more solid foundation for understanding the origins and early evolution of life on Earth.
Last edited by Otangelo on Sat Oct 12, 2024 9:14 am; edited 26 times in total