9. Lipid Synthesis
Membranes always come from membranes
Every new cell originates from a pre-existing cell through a process of cell division. This idea is part of the Cell Theory, one of the fundamental principles of biology. When a cell divides, its plasma membrane pinches in and eventually splits to form two daughter cells, each with its own enclosing membrane. The membrane of the daughter cells arises directly from the membrane of the parent cell. As cells grow, they need to increase the surface area of their membranes. This is achieved by adding new lipid molecules (phospholipids, cholesterol, etc.) and proteins to the existing membrane. The new lipids and proteins are synthesized within the cell and then transported to the membrane, where they are incorporated. The creation of lipid asymmetry and lipid transport mechanisms is a complex topic, and much of what we understand comes from piecing together bioinformatics data, comparative biology, and structural biology. P-type ATPases, including those that function as flippases, are ancient and diverse proteins found across all domains of life: Bacteria, Archaea, and Eukarya. Given their widespread distribution and essential roles in maintaining membrane asymmetry, it's conceivable that a primitive form of flippase was present in LUCA. The phospholipid translocating flippases, especially those of the P4-ATPase family (like ATP8A1 and ATP8B1 you mentioned), are particularly interesting because they have been identified in both eukaryotes and some bacterial lineages. ATP-binding cassette (ABC) transporters, like the floppases you mentioned, are also ancient and ubiquitous, found across all three domains of life. Their primary roles often involve the transport of various substrates across cellular membranes. Given their broad distribution and diversity, it's plausible that a primitive form of ABC transporter, perhaps with floppase-like activity, existed in LUCA.
A key aspect of membrane biology is the asymmetric distribution of lipids between the inner and outer leaflets of the lipid bilayer. This asymmetry is not a static feature but is actively maintained by various proteins that facilitate the movement of lipids across the membrane. In this paper, we will explore two major classes of lipid transporters: flippases and floppases, as well as touch upon ion transport proteins. These molecular machines work in concert to establish and maintain the unique lipid compositions of membrane leaflets, which is essential for numerous cellular processes and likely played a critical role in the emergence of life itself.
Roy Yaniv (2023): In a recent paper (Kahana, A, Lancet, D, 2021), the researchers point out that it is the modest nanoscopic micelles that had numerous advantages as early protocells, despite the fact that they did not have an inner water volume (Figure 1). Within these tiny protocellular structures, networks of molecules can collaboratively function, akin to a team, because all molecules are crowded in a miniscule volume, initiating a critical step towards the emergence of life. Scientists are now exploring how simple lipid molecules, copiously present in ancient oceans, could have autonomously come together. Importantly, these lipid micelles are far from random assemblies; they possess an innate capacity for self-organisation. However, this organisation is not in terms of spatial position or order of amino acids as in a protein. Instead, the organisation is expressed in terms of composition. In a simplified example, imagine an environment in which all types of lipids have the same concentration. Upon micelle growth driven by molecule accretion, the network dynamics are capable of biasing the inner composition, with some being in high amounts and others being small or rejected entirely. This behaviour is analogous to highly specific membrane transport mechanisms controlling the content of present-day cells. Figure 1: Nanoscopic micelles: Seeking early protocellular simplicity and efficacy (Kahana, A, Lancet, D, 2021). The truly surprising aspect is that not only do lipid micelles have capacity to self-organise, but they can also maintain a constant composition upon growth. This means that these micelles have a built-in system to ensure that their lipid composition would remain stable as they get bigger. This is called ‘homeostatic growth’, another capability of reproducing living cells. When these entities split into two, the offspring are very similar to each other, just like when living cells reproduce. One of the most important findings of the research is that the catalytic networks within lipid micelles (a team of molecules working together, where certain molecules speed up the entry of some others) might have enabled self-reproduction, meaning micelles could reproduce themselves by a mechanism analogous to metabolism in living cells (Figure 2) (Lancet, D, Zidovetzki, R, Markovitch, O, 2018). 1

Nanoscopic micelles: Seeking early protocellular simplicity and efficacy (Kahana, A, Lancet, D, 2021). Link
Unresolved Challenges in Early Micelle-Based Protocellular Structures
1. Self-Organisation Without Spatial Order
The self-organisation observed in micelle-based protocells is expressed in their composition, not in a spatial or structural sense like in modern cells. While the micelles' lipid composition adjusts dynamically, it is unclear how such sophisticated compositional control could emerge unguided.
Conceptual problem: Lack of Spatial Order in Organization
- No mechanism to explain how molecular networks can function cooperatively without spatial coordination
- Difficulty explaining how compositional biases emerge in the absence of external regulation or enzymatic catalysis.
2. Homeostatic Growth in Primitive Micelles
The ability of lipid micelles to maintain a constant composition during growth, termed 'homeostatic growth,' is a trait usually associated with living cells. This phenomenon requires a robust system that can stabilize and monitor internal lipid content during size expansion, a process not clearly understood in prebiotic conditions.
Conceptual problem: Spontaneous Emergence of Homeostatic Control
- No known prebiotic mechanism to explain how primitive micelles can regulate and maintain stable compositions during growth
- Homeostatic growth typically requires feedback systems absent in early environments.
3. Catalytic Networks in Lipid Micelles
Lipid micelles appear capable of forming catalytic networks where certain molecules assist in the transport or catalysis of others, mimicking metabolic activities. This coordinated network suggests a high degree of functional complexity, difficult to explain without guided interactions.
Conceptual problem: Emergence of Catalytic Complexity
- No natural unguided pathway explains how molecules could spontaneously form highly organized catalytic networks
- Without proteins or ribozymes, there is no clear method for efficient catalytic activity within micelles.
4. Spontaneous Formation of Amphipathic Lipids
Lipid micelles depend on amphipathic molecules (lipids with hydrophilic heads and hydrophobic tails) for their structural integrity. The synthesis of such molecules is a multi-step process, traditionally reliant on enzymatic catalysis. In prebiotic environments, where no enzymes existed, it is unclear how these molecules could form.
Conceptual problem: Prebiotic Synthesis of Lipids
- The multi-step process of lipid formation lacks plausible prebiotic catalysts
- Environmental conditions necessary for spontaneous lipid formation remain speculative, with no direct evidence of sustained favorable conditions.
5. Absence of Selective Permeability in Micelles
Selective permeability is a key feature of living cells, enabling them to control the flow of substances in and out. However, early micelle structures would have lacked proteins such as transporters or channels, raising the question of how these micelles could support basic proto-cellular functions without these crucial mechanisms.
Conceptual problem: Lack of Permeability Control
- Primitive membranes likely lacked the selectivity required to differentiate between nutrient intake and waste removal
- No known primitive mechanism explains how micelles could develop selective permeability without proteins.
6. Energy Requirements for Micelle Stability and Growth
In modern cells, processes such as membrane growth and lipid synthesis are energy-intensive and depend on molecules like ATP. The lack of prebiotic energy equivalents challenges the possibility of maintaining micelle stability and supporting growth mechanisms.
Conceptual problem: Energy Source for Lipid Dynamics
- Lack of ATP or similar high-energy molecules in early Earth environments complicates explanations for the energy-intensive processes involved in micelle growth
- Without external energy sources, the stability and persistence of lipid micelles are difficult to justify.
7. Environmental Instability and Lipid Degradation
Lipid micelles are vulnerable to environmental degradation, particularly from UV radiation and oxidation, which would have been prevalent in early Earth conditions. The absence of protective mechanisms in these primitive structures further exacerbates the problem of maintaining lipid integrity long enough for them to participate in protocellular processes.
Conceptual problem: Stability of Lipids in Harsh Environments
- Early Earth’s conditions, such as radiation and fluctuating temperatures, would likely degrade lipids before they could contribute to protocell formation
- No protective systems existed in early micelles to shield lipids from environmental degradation.
8. Self-Reproduction in Micelles without Prebiotic Machinery
The Kahana and Lancet (2021) paper suggests that lipid micelles may have had the ability to self-reproduce, which would require the coordination of complex molecular networks similar to metabolic systems in living cells. However, the mechanisms driving this self-reproduction in the absence of biological machinery remain unknown.
Conceptual problem: Reproduction Without Metabolic Networks
- Reproduction of micelles in a manner analogous to cellular metabolism lacks a clear, unguided pathway
- Without enzymes or ribozymes, it is unclear how molecular interactions could replicate the complexity of metabolic processes necessary for self-reproduction.
9. Prebiotic Bias Towards Specific Lipid Compositions
The concept of lipid micelles developing compositional biases through accretion mechanisms akin to modern membrane transport systems poses a significant challenge. Prebiotic environments likely had a uniform distribution of lipid types, making it difficult to explain how specific lipids could have been favored in the absence of a selective mechanism.
Conceptual problem: Emergence of Lipid Compositional Bias
- The bias in lipid composition suggests a level of selectivity typically seen in cellular transport systems, which would not have been available prebiotically
- No clear mechanism exists to explain how micelles could have developed compositional diversity spontaneously.
10. Interdependence of Lipid Networks and Other Biochemical Systems
For micelles to function as protocells, they would need to interact with genetic material or other biomolecules, such as peptides or sugars, to establish the cooperative networks necessary for life. The simultaneous emergence of these interdependent systems presents a formidable challenge without invoking guided or designed processes.
Conceptual problem: Co-Emergence of Lipids and Biochemical Networks
- Lipid micelles alone cannot explain the full complexity required for life without the concurrent emergence of other biomolecules
- No natural process has been identified that could account for the coordinated emergence of lipid and other biomolecular systems.
11. Prebiotic Membrane Chirality Selection
Modern membranes exhibit chirality, which is essential for their function. However, prebiotic synthesis of lipids would likely produce racemic mixtures, meaning an equal proportion of right- and left-handed molecules, which would compromise membrane function.
Conceptual problem: Lack of Mechanism for Chirality Selection
- Prebiotic environments would not naturally select for one chiral form over another, yet functional membranes require specific chirality
- No known mechanism explains how primitive micelles could have developed the necessary chiral purity for functional membranes.
12. Integration with Other Molecular Systems
Even if lipid micelles could form under early Earth conditions, their integration with other systems, such as genetic material and proteins, is required for the full development of proto-cellular life. The simultaneous emergence of these diverse systems presents an unresolved problem, as no known natural mechanism can explain their coemergence.
Conceptual problem: Lack of Mechanism for Integrated Systems
- The integration of lipid micelles with other molecular systems would require simultaneous, coordinated development, which remains unexplained
- Without genetic material or primitive proteins, it is unclear how lipid micelles alone could have achieved the complexity necessary for life.
9.1. Fatty acid synthesis
The synthesis of fatty acids and phospholipids is a fundamental process that underpins the very existence of cellular life as we know it. This complex biochemical pathway not only provides essential components for cell membranes but also plays essential roles in energy storage, signaling, and maintaining cellular homeostasis. The importance of these molecules cannot be overstated, as they form the structural backbone of all living cells and enable the compartmentalization necessary for complex biological functions. At the heart of this process lies acetyl-CoA, a versatile molecule derived from glucose metabolism or other carbon sources. Acetyl-CoA serves as the primary building block for fatty acid synthesis, highlighting the interconnectedness of cellular metabolic pathways. The ability to generate and utilize acetyl-CoA would have been essential for any early form of life, as it bridges central carbon metabolism with lipid biosynthesis. The synthesis of fatty acids is a highly coordinated and energy-intensive process, requiring a suite of specialized enzymes working in concert. The fatty acid synthase complex, a marvel of molecular engineering, efficiently catalyzes a series of reactions that elongate the growing fatty acid chain two carbons at a time. This process involves multiple steps, including condensation, reduction, dehydration, and another reduction, each catalyzed by a specific enzyme or enzyme domain. The initiation of fatty acid synthesis begins with the carboxylation of acetyl-CoA to form malonyl-CoA, catalyzed by acetyl-CoA carboxylase. This step is often considered the committed step in fatty acid biosynthesis and is subject to tight regulation. The subsequent transfer of the malonyl group to the acyl carrier protein sets the stage for the cyclical process of chain elongation.
As the fatty acid chain grows, it undergoes a series of modifications that determine its final structure and properties. The introduction of double bonds by desaturases, for instance, produces unsaturated fatty acids, which are critical for maintaining membrane fluidity and function across a range of temperatures. The synthesis of phospholipids builds upon the fatty acid synthesis pathway, incorporating these hydrophobic tails into more complex molecules that form the bilayer structure of cell membranes. This process involves the addition of polar head groups to diacylglycerol, creating amphipathic molecules capable of self-assembling into the lipid bilayers that define cellular boundaries. The intricate nature of fatty acid and phospholipid synthesis, with its multiple steps and regulatory mechanisms, raises profound questions about the origin and evolution of these pathways. The complexity and interdependence of the enzymes involved challenge simplistic explanations for their emergence. Each enzyme in the pathway must function with remarkable specificity and efficiency, and the entire process must be tightly coordinated to produce fatty acids of the correct length and degree of saturation. Moreover, the fatty acid synthase complex itself, with its multiple functional domains working in a coordinated fashion, represents a level of molecular sophistication that defies easy explanation through gradual, stepwise acquisition of function. The precise arrangement of these domains is crucial for the efficiency of the overall process, suggesting a need for an all-or-nothing emergence of this complex. The biosynthesis of fatty acids and phospholipids exemplifies the principle of irreducible complexity in biological systems. Each component of the pathway is necessary for the production of functional lipids, and the removal of any single enzyme would render the entire process inoperative. This interdependence extends beyond the immediate pathway to encompass the broader metabolic network of the cell, including the generation of precursors and cofactors essential for lipid synthesis. The essential nature of these pathways for all cellular life, combined with their complexity and interdependence, invites deeper consideration of the mechanisms underlying the origin and diversification of biological systems.
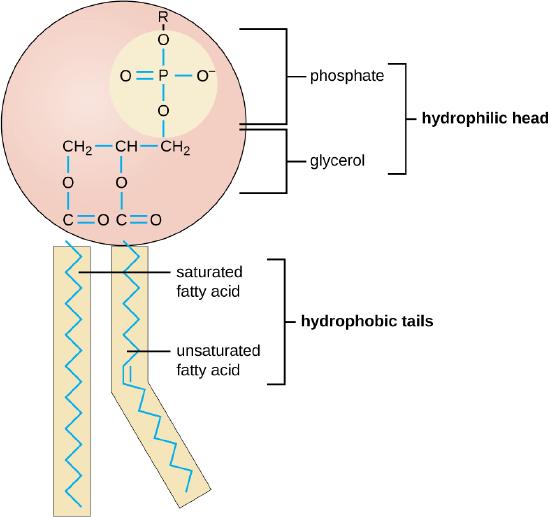
Lipids can be distinguished between mono - or diacyl glycerols (“incomplete lipids”, ILs) or phospholipids (“complete lipids”, CLs). 28
All unstructured text is available under the Creative Commons Attribution-ShareAlike License;
Acetyl-CoA, derived from glucose metabolism or other carbon sources, serves as the basic building block for fatty acid synthesis. The glycolytic pathway or a variant of it would have been essential for LUCA to produce Acetyl-CoA.
To form a complete list that encompasses the synthesis of fatty acids through the Fatty Acid Synthase Complex and complements the earlier list you provided, we can follow a logical order from initiation to elongation. Here's a comprehensive, ordered list:
9.1.1. Initiation of Fatty Acid Synthesis
Fatty acid synthesis is a fundamental metabolic process that produces fatty acids from acetyl-CoA and malonyl-CoA precursors. The initiation phase of this pathway is crucial as it sets the stage for the subsequent elongation cycle. This process is essential for membrane lipid biosynthesis, energy storage, and various cellular functions involving lipids.
Key enzymes involved in the initiation of fatty acid synthesis:
Acetyl-CoA Carboxylase (ACC) (EC 6.4.1.2): Smallest known: 2,346 amino acids (Homo sapiens)
Catalyzes the ATP-dependent carboxylation of acetyl-CoA to form malonyl-CoA. This is the first committed and rate-limiting step in fatty acid synthesis. ACC plays a crucial role in regulating the balance between fatty acid synthesis and oxidation. The enzyme exists in two isoforms in mammals: ACC1 (primarily involved in fatty acid synthesis) and ACC2 (involved in regulating fatty acid oxidation). ACC is a key target for regulation of lipid metabolism and is subject to both allosteric and covalent modifications.
Malonyl-CoA-Acyl Carrier Protein Transacylase (MCAT) (EC 2.3.1.39): Smallest known: 290 amino acids (Escherichia coli)
Catalyzes the transfer of the malonyl group from malonyl-CoA to the acyl carrier protein (ACP), forming malonyl-ACP. This reaction is crucial for providing the two-carbon units needed for fatty acid chain elongation. MCAT is part of the fatty acid synthase complex in bacteria and plants, while in animals, it's a domain of the multifunctional fatty acid synthase enzyme. The malonyl-ACP produced by this enzyme serves as the primary extender unit in the fatty acid synthesis cycle.
Fatty Acid Synthase (FAS) (EC 2.3.1.85): Smallest known: 2,511 amino acids (Homo sapiens)
While not explicitly mentioned in your initial list, Fatty Acid Synthase is crucial to include in the initiation of fatty acid synthesis. In animals, FAS is a large, multifunctional enzyme that carries out all the reactions of fatty acid synthesis, including the functions of MCAT. It contains seven catalytic domains and an acyl carrier protein domain. The initiation step involves the transfer of an acetyl group from acetyl-CoA to the ACP domain, setting the stage for elongation.
The initiation of fatty acid synthesis enzyme group consists of 3 enzymes. The total number of amino acids for the smallest known versions of these enzymes is 5,147.
Proteins with metal clusters or cofactors:
Acetyl-CoA Carboxylase (ACC) (EC 6.4.1.2): Requires biotin as a covalently bound cofactor. Also needs ATP, Mg2+ or Mn2+, and HCO3- for catalysis. Some forms are activated by citrate.
Malonyl-CoA-Acyl Carrier Protein Transacylase (MCAT) (EC 2.3.1.39): Does not require metal ions or additional cofactors for catalysis. However, it interacts with the 4'-phosphopantetheine prosthetic group of the acyl carrier protein.
Fatty Acid Synthase (FAS) (EC 2.3.1.85): Contains multiple cofactors across its various domains:
- Requires NADPH as a reducing agent
- Contains a 4'-phosphopantetheine prosthetic group on its ACP domain
- The ketoacyl synthase domain requires a catalytic cysteine residue
- The dehydratase domain uses a histidine-aspartate catalytic dyad
This overview highlights the complexity and importance of the initiation phase of fatty acid synthesis. These enzymes work together to begin the process of fatty acid production, which is critical for numerous cellular functions. The regulation of these enzymes, particularly ACC, is crucial for controlling lipid metabolism in response to cellular energy status and hormonal signals. Understanding these enzymes and their regulation is important for research into metabolic disorders, obesity, and potential therapeutic interventions targeting lipid metabolism.
9.1.2. Elongation through Fatty Acid Synthase Complex
Fatty acid synthesis is a cyclical process that extends a growing fatty acid chain by two carbons in each round. In eukaryotes, this process is carried out by a large, multifunctional enzyme complex called Fatty Acid Synthase (FAS). Each domain of FAS catalyzes a specific step in the synthesis cycle. In prokaryotes, these activities are typically performed by separate enzymes.
Key enzyme domains involved in the fatty acid synthesis cycle:
Fatty Acid Synthase - Malonyl/Acetyltransferase (MAT) (EC 2.3.1.39): Smallest known: 290 amino acids (Escherichia coli, as a separate enzyme)
This domain is responsible for loading malonyl groups from malonyl-CoA onto the acyl carrier protein (ACP) domain of FAS. It also loads the initial acetyl group to start the fatty acid chain. This step is crucial for providing the two-carbon units needed for chain elongation in each cycle.
Fatty Acid Synthase - 3-ketoacyl-ACP synthase (KS) (EC 2.3.1.41): Smallest known: 412 amino acids (Escherichia coli, as a separate enzyme)
Catalyzes the condensation reaction between the growing acyl-ACP and malonyl-ACP, extending the fatty acid chain by two carbons. This is the first step in each cycle of fatty acid elongation and results in the release of CO2 from the malonyl group.
Fatty Acid Synthase - 3-ketoacyl-ACP reductase (KR) (EC 1.1.1.100): Smallest known: 244 amino acids (Escherichia coli, as a separate enzyme)
Reduces the 3-keto group formed by the KS reaction to a 3-hydroxy group, using NADPH as the reducing agent. This is the first of two reduction steps in the fatty acid synthesis cycle.
Fatty Acid Synthase - 3-hydroxyacyl-ACP dehydratase (DH) (EC 4.2.1.59): Smallest known: 171 amino acids (Escherichia coli, as a separate enzyme)
Catalyzes the dehydration of the 3-hydroxyacyl-ACP to form a trans-2-enoyl-ACP. This reaction eliminates a water molecule, creating a double bond in the fatty acid chain.
Fatty Acid Synthase - Enoyl-ACP reductase (ER) (EC 1.3.1.9): Smallest known: 262 amino acids (Escherichia coli, as a separate enzyme)
Reduces the double bond created by the DH reaction, using NADPH as the reducing agent. This final step in the cycle produces a saturated acyl-ACP, which is then ready for another round of elongation.
The fatty acid synthesis cycle enzyme group consists of 5 enzyme domains. The total number of amino acids for the smallest known versions of these enzymes (as separate entities in E. coli) is 1,379.
Proteins with metal clusters or cofactors:
Fatty Acid Synthase - Malonyl/Acetyltransferase (MAT) (EC 2.3.1.39): Does not require metal ions or additional cofactors for catalysis. However, it interacts with the 4'-phosphopantetheine prosthetic group of the ACP.
Fatty Acid Synthase - 3-ketoacyl-ACP synthase (KS) (EC 2.3.1.41): Requires a catalytic cysteine residue for its condensation reaction. No metal ions or additional cofactors are needed.
Fatty Acid Synthase - 3-ketoacyl-ACP reductase (KR) (EC 1.1.1.100): Requires NADPH as a cofactor for the reduction reaction.
Fatty Acid Synthase - 3-hydroxyacyl-ACP dehydratase (DH) (EC 4.2.1.59): Does not require metal ions or additional cofactors. It uses a histidine-aspartate catalytic dyad for its dehydration reaction.
Fatty Acid Synthase - Enoyl-ACP reductase (ER) (EC 1.3.1.9): Requires NADPH as a cofactor for the reduction reaction. Some bacterial forms may use NADH instead.
This overview highlights the complexity and efficiency of the fatty acid synthesis cycle. In eukaryotes, these enzyme activities are combined into a single, large, multifunctional FAS enzyme, which enhances the efficiency of the process by keeping intermediates bound to the enzyme complex. The cycle repeats until the fatty acid reaches the desired length, typically 16 or 18 carbons in most organisms. Understanding this process is crucial for research into lipid metabolism, metabolic disorders, and the development of antibiotics targeting bacterial fatty acid synthesis.
9.1.3. Termination and Modification
The termination and modification of fatty acids are crucial steps that determine the final products of fatty acid synthesis. These processes involve the release of the completed fatty acid from the synthesis machinery and subsequent modifications to produce various types of fatty acids needed for cellular functions.
Key enzymes involved in the termination and modification of fatty acid synthesis:
Fatty Acid Synthase (FAS) (EC 2.3.1.86): Smallest known: 2,511 amino acids (Homo sapiens)
FAS is a large, multifunctional enzyme complex that catalyzes all steps of fatty acid synthesis, including termination. In mammals, it's responsible for synthesizing palmitate (16:0) as the primary product. The thioesterase domain of FAS, which is not always included in the EC number 2.3.1.86, is crucial for termination:
Thioesterase domain: This domain hydrolyzes the thioester bond between the completed fatty acid (usually palmitate) and the acyl carrier protein (ACP), releasing the free fatty acid. This step terminates the fatty acid synthesis cycle. FAS integrates multiple catalytic activities, including acetyl transferase, malonyl transferase, ketoacyl synthase, ketoacyl reductase, dehydratase, enoyl reductase, and thioesterase. Its complex structure allows for efficient synthesis of long-chain saturated fatty acids.
Stearoyl-CoA Desaturase (SCD) (EC 1.14.19.1): Smallest known: 355 amino acids (Mycobacterium tuberculosis)
Catalyzes the introduction of the first double bond at the Δ9 position of saturated fatty acyl-CoAs. This enzyme is crucial for the production of monounsaturated fatty acids, primarily oleic acid (18:1) from stearic acid (18:0). Key features include:
1. Substrate specificity: Primarily acts on palmitoyl-CoA and stearoyl-CoA.
2. Reaction mechanism: Introduces a cis-double bond between carbons 9 and 10, counting from the carboxyl end.
3. Importance: Balances the ratio of saturated to unsaturated fatty acids, which is critical for membrane fluidity and various cellular processes.
Fatty Acyl-CoA Elongase (ELOVL) (EC 2.3.1.199): Smallest known: 267 amino acids (Homo sapiens, ELOVL3)
While not mentioned in your initial list, Fatty Acyl-CoA Elongases are crucial for the production of very long-chain fatty acids (VLCFAs). They extend the fatty acid chain beyond the 16-18 carbon atoms produced by FAS. There are seven ELOVL enzymes (ELOVL1-7) in mammals, each with different substrate specificities and tissue distributions.
The termination and modification of fatty acid synthesis enzyme group consists of 3 enzymes. The total number of amino acids for the smallest known versions of these enzymes is 3,133.
Proteins with metal clusters or cofactors:
Fatty Acid Synthase (FAS) (EC 2.3.1.86):
- Requires NADPH as a reducing agent
- Contains a 4'-phosphopantetheine prosthetic group on its ACP domain
- The ketoacyl synthase domain requires a catalytic cysteine residue
- The dehydratase domain uses a histidine-aspartate catalytic dyad
Stearoyl-CoA Desaturase (SCD) (EC 1.14.19.1):
- Contains a di-iron center in its active site
- Requires molecular oxygen and electrons from cytochrome b5 for catalysis
- Uses NADH or NADPH as the ultimate electron donor
Fatty Acyl-CoA Elongase (ELOVL) (EC 2.3.1.199):
- Does not require metal ions or additional cofactors for catalysis
- Works in conjunction with other enzymes of the elongation complex, which use NADPH and malonyl-CoA
This overview highlights the complexity of fatty acid termination and modification processes. These enzymes work together to produce a diverse array of fatty acids essential for various cellular functions:
1. FAS terminates the synthesis of long-chain saturated fatty acids.
2. SCD introduces double bonds, creating monounsaturated fatty acids.
3. ELOVLs extend fatty acids to produce very long-chain fatty acids.
Understanding these enzymes and their regulation is crucial for research into lipid metabolism, metabolic disorders, and the development of therapies targeting lipid-related diseases. The balance and diversity of fatty acids produced by these enzymes are critical for membrane structure, energy storage, and signaling molecules in cells.
9.1.4. Fatty Acid Elongation (if needed)
The term elongation in this context refers specifically to the extension of already synthesized fatty acid chains (usually palmitate, a 16-carbon chain) to produce long-chain fatty acids. This process also involves elongation but happens after the initial fatty acid has been synthesized. Fatty Acid Elongation is a crucial process in lipid metabolism that extends the carbon chain of fatty acids. This pathway is essential for producing long-chain fatty acids, which are vital components of cellular membranes, energy storage molecules, and signaling lipids. The elongation process typically occurs in the endoplasmic reticulum and involves a series of enzymatic reactions that add two-carbon units to the growing fatty acid chain.
Key enzyme involved:
Enoyl-ACP reductase (EC 1.3.1.9): Smallest known: 262 amino acids (Mycobacterium tuberculosis)
Catalyzes the final step in each cycle of fatty acid elongation by reducing enoyl-CoA (or enoyl-ACP) to acyl-CoA (or acyl-ACP). This enzyme is crucial for the completion of each elongation cycle and plays a key role in determining the final length of fatty acids. It's essential for maintaining the proper balance of fatty acid species in cells.
The Fatty Acid Elongation enzyme group consists of 1 enzyme domain. The total number of amino acids for the smallest known version of this enzyme is 262.
Information on metal clusters or cofactors:
Enoyl-ACP reductase (EC 1.3.1.9): Requires NADH or NADPH as a cofactor for the reduction reaction. Some variants of this enzyme, particularly in plants and bacteria, contain a [4Fe-4S] iron-sulfur cluster that is crucial for its catalytic activity. In certain organisms, like Mycobacterium tuberculosis, the enzyme uses NADH and contains no metal cofactors.
The Fatty Acid Elongation pathway, of which Enoyl-ACP reductase is a part, typically involves four main steps that are repeated cyclically:
1. Condensation: Addition of a two-carbon unit to the growing fatty acid chain.
2. Reduction: Conversion of 3-ketoacyl-CoA to 3-hydroxyacyl-CoA.
3. Dehydration: Removal of water to form enoyl-CoA.
4. Reduction: Catalyzed by Enoyl-ACP reductase, converting enoyl-CoA to acyl-CoA.
Enoyl-ACP reductase is particularly important because it catalyzes the rate-limiting step in many fatty acid elongation systems. Its activity can significantly influence the overall rate of fatty acid synthesis and the distribution of fatty acid chain lengths in the cell. The enzyme's role in fatty acid elongation makes it a target for antibacterial and antifungal drugs, as inhibiting this enzyme can disrupt the organism's ability to synthesize essential fatty acids. For example, the antibiotic isoniazid targets the enoyl-ACP reductase in Mycobacterium tuberculosis as part of its mechanism of action against tuberculosis. In addition to its role in primary metabolism, the fatty acid elongation pathway, including the action of enoyl-ACP reductase, is crucial for the production of specialized lipids such as waxes in plants and very-long-chain fatty acids in mammals. These products have diverse functions, including energy storage, water resistance in plant cuticles, and components of skin lipids in animals. Understanding the function and regulation of enoyl-ACP reductase and the fatty acid elongation pathway is crucial for various fields, including metabolic engineering for biofuel production, development of new antibiotics, and research into lipid-related disorders in humans.
Unresolved Challenges in Fatty Acid Synthesis
1. Enzyme Complexity and Specificity
The fatty acid synthesis pathway involves highly specific enzymes, each catalyzing a distinct reaction. The challenge lies in explaining the origin of such complex, specialized enzymes without invoking a guided process. For instance, acetyl-CoA carboxylase (EC 6.4.1.2) requires a sophisticated active site to catalyze the carboxylation of acetyl-CoA to form malonyl-CoA. The precision required for this catalysis raises questions about how such a specific enzyme could have arisen spontaneously.
Conceptual problem: Spontaneous Complexity
- No known mechanism for generating highly specific, complex enzymes without guidance
- Difficulty explaining the origin of precise active sites and substrate specificity
2. Multi-Domain Enzyme Complexity
The fatty acid synthase complex (EC 2.3.1.86) is a multi-domain enzyme responsible for catalyzing the synthesis of long-chain saturated fatty acids. The challenge lies in explaining how such a sophisticated multi-functional enzyme could have emerged without a guided process. Each domain must function precisely and in coordination with others for the complex to work effectively.
Conceptual problem: Coordinated Multi-functionality
- No known mechanism for the spontaneous emergence of multi-domain enzymes
- Difficulty in explaining the origin of coordinated functions within a single enzyme complex
3. Pathway Interdependence
The fatty acid synthesis pathway exhibits a high degree of interdependence among its constituent enzymes. Each step in the pathway relies on the product of the previous reaction as its substrate. This sequential dependency poses a significant challenge to explanations of gradual, step-wise origin. For example, malonyl-CoA-acyl carrier protein transacylase (EC 2.3.1.39) requires malonyl-CoA (produced by acetyl-CoA carboxylase) as its substrate.
Conceptual problem: Simultaneous Emergence
- Challenge in accounting for the concurrent appearance of interdependent components
- Lack of explanation for the coordinated development of multiple, specific molecules
4. Cofactor Requirements
Several enzymes in the fatty acid synthesis pathway require specific cofactors for their function. For instance, 3-ketoacyl-ACP reductase (EC 1.1.1.100) requires NADPH as a cofactor. The challenge lies in explaining the origin of these cofactors and their specific interactions with enzymes without invoking a guided process.
Conceptual problem: Cofactor-Enzyme Coordination
- Difficulty in explaining the simultaneous emergence of enzymes and their specific cofactors
- Lack of a mechanism for the coordinated development of enzyme active sites and cofactor binding regions
5. Regulatory Mechanisms
The fatty acid synthesis pathway is subject to complex regulatory mechanisms to ensure appropriate production levels. For example, acetyl-CoA carboxylase is regulated by both allosteric regulation and covalent modification. The challenge lies in explaining the emergence of these sophisticated regulatory mechanisms without invoking a guided process.
Conceptual problem: Regulatory Complexity
- Difficulty in accounting for the emergence of complex regulatory mechanisms
- Lack of explanation for the coordinated development of enzymes and their regulatory systems
6. Substrate Availability
The pathway requires specific substrates, such as acetyl-CoA and malonyl-CoA, which must be available in sufficient quantities. The challenge lies in explaining how early cellular systems could have maintained a steady supply of these substrates without a fully developed metabolic network.
Conceptual problem: Substrate Accessibility
- Difficulty in accounting for the availability of specific substrates in early cellular systems
- Lack of explanation for the coordinated emergence of substrate production and utilization pathways
7. Energy Requirements
Several reactions in the pathway, such as the one catalyzed by acetyl-CoA carboxylase, require ATP. The challenge lies in explaining how early cellular systems could have met these energy requirements without a fully developed energy metabolism.
Conceptual problem: Energy Availability
- Difficulty in accounting for the availability of high-energy molecules in early cellular systems
- Lack of explanation for the coordinated emergence of energy-producing and energy-consuming pathways
8. Structural Complexity
The enzymes involved in fatty acid synthesis exhibit complex three-dimensional structures essential for their function. For instance, fatty acid synthase forms a large, multi-subunit complex. The challenge lies in explaining the emergence of such sophisticated protein structures without invoking a guided process.
Conceptual problem: Spontaneous Structural Organization
- No known mechanism for the spontaneous formation of complex protein structures
- Difficulty in explaining the origin of specific subunit organizations and quaternary structures
9. Chirality
Fatty acid synthesis involves chiral molecules and stereospecific reactions. For example, 3-hydroxyacyl-ACP dehydratase (EC 4.2.1.59) catalyzes a stereospecific dehydration reaction. The challenge lies in explaining the emergence of such stereospecificity without a guided process.
Conceptual problem: Spontaneous Stereospecificity
- No known mechanism for the spontaneous emergence of stereospecific reactions
- Difficulty in explaining the origin of enzymes capable of distinguishing between stereoisomers
10. Metabolic Integration
The fatty acid synthesis pathway is deeply integrated with other metabolic processes, including the citric acid cycle and glycolysis. The challenge lies in explaining how such intricate metabolic integration could have emerged without a guided process.
Conceptual problem: Metabolic Interconnectivity
- No known mechanism for the spontaneous emergence of integrated metabolic networks
- Difficulty in explaining the origin of pathway interconnections and metabolic flexibility
These unresolved challenges highlight the complexity of the fatty acid synthesis pathway and the significant conceptual problems faced when attempting to explain its origin through unguided processes. The high degree of specificity, interdependence, and complexity observed in these enzymes and their interactions pose substantial questions that current naturalistic explanations struggle to address adequately.
Phospholipid Synthesis
9.2. Phospholipid synthesis
The synthesis of phospholipids represents a fundamental process underpinning the essence of cellular existence. These complex molecules form the structural backbone of all biological membranes, enabling the compartmentalization that defines life at the cellular level. The ability to produce phospholipids would have been an absolute necessity for the first living organisms on Earth. At its core, phospholipid synthesis is a process of enzymatic reactions, beginning with simple precursors and forming sophisticated amphipathic ( a molecule that has both hydrophilic (water-attracting) and hydrophobic (water-repelling) parts) molecules capable of self-assembling into bilayers. This process bridges the gap between basic metabolic pathways and the complex architecture of cellular membranes, highlighting the interconnectedness of biochemical systems. The pathway begins with glycerol-3-phosphate (G3P), a pivotal molecule that serves as the backbone for phospholipid construction. The formation of G3P itself is tied to central carbon metabolism, illustrating how lipid synthesis is integrated with other essential cellular processes. From this foundation, a series of carefully orchestrated enzymatic steps attach fatty acids and diverse head groups, ultimately producing the variety of phospholipids necessary for membrane function and cellular homeostasis. The complexity of phospholipid synthesis extends beyond the mere addition of molecular components. Each step requires exquisite specificity and regulation to ensure the production of lipids with the correct composition and properties. To achieve the desired outcome, the enzymes involved must work in concert, with precise timing and spatial organization. This level of coordination raises pertinent questions about the origins of such a sophisticated system. Moreover, the diversity of phospholipids produced through these pathways is critical for the proper functioning of cellular membranes across a wide range of environments and physiological conditions. The ability to modulate membrane composition in response to environmental cues is a hallmark of cellular adaptability, further underscoring the importance of a flexible and responsive lipid synthesis machinery. The precision required at each step, from the initial formation of fatty acids to the final assembly of complex phospholipids, speaks to a level of biochemical sophistication that challenges simplistic explanations for its emergence. This introduction sets the stage for a deeper exploration of the enzymatic processes involved in phospholipid synthesis, the potential pathways, and the implications of this essential biochemistry for our understanding of cellular life's origins and fundamental nature.
Glycerol-3-phosphate (G3P) formation: G3P is a central molecule in phospholipid synthesis. The first life forms might have obtained G3P either through glycolysis or from dihydroxyacetone phosphate (DHAP), a glycolytic intermediate.
9.2.1. Attachment of two fatty acyl groups to glycerol-3-phosphate (G3P)
Attachment of Fatty Acids to G3P: Two fatty acyl groups, usually derived from acyl-CoA molecules, are esterified to the G3P at the sn-1 and sn-2 positions to produce phosphatidic acid. For the synthesis of phosphatidic acid through the attachment of two fatty acyl groups to glycerol-3-phosphate (G3P), the enzymatic steps are as follows:
Phosphatidic acid biosynthesis is a critical initial step in glycerophospholipid metabolism. This pathway is essential for the production of phospholipids, which are fundamental components of cellular membranes and play crucial roles in cellular signaling and energy storage. The process involves the sequential attachment of two fatty acyl groups to glycerol-3-phosphate (G3P), resulting in the formation of phosphatidic acid, a key intermediate in lipid biosynthesis.
Key enzymes involved:
Glycerol-3-phosphate O-acyltransferase (GPAT) (EC 2.3.1.15): Smallest known: 306 amino acids (Mycobacterium tuberculosis)
Catalyzes the initial and rate-limiting step in de novo glycerophospholipid biosynthesis. GPAT transfers an acyl group from acyl-CoA to the sn-1 position of glycerol-3-phosphate, forming lysophosphatidic acid (LPA). This enzyme is crucial for regulating the flux of fatty acids into the glycerophospholipid biosynthetic pathway and plays a significant role in triglyceride biosynthesis.
Lysophosphatidic acid acyltransferase (LPAAT) (EC 2.3.1.51): Smallest known: 257 amino acids (Chlamydia trachomatis)
Catalyzes the second acylation step in phosphatidic acid biosynthesis. LPAAT transfers an acyl group from acyl-CoA to the sn-2 position of lysophosphatidic acid, producing phosphatidic acid. This enzyme is critical for determining the fatty acid composition of membrane phospholipids and thus influences membrane fluidity and cellular function.
The phospholipid biosynthesis enzyme group consists of 2 enzymes. The total number of amino acids for the smallest known versions of these enzymes is 563.
Information on metal clusters or cofactors:
Glycerol-3-phosphate O-acyltransferase (GPAT) (EC 2.3.1.15): Requires Mg²⁺ as a cofactor for optimal activity. The magnesium ion is essential for the catalytic mechanism, facilitating the transfer of the acyl group from acyl-CoA to glycerol-3-phosphate.
Lysophosphatidic acid acyltransferase (LPAAT) (EC 2.3.1.51): Does not require metal ions or cofactors for its catalytic activity. However, the enzyme's activity can be modulated by various lipids and proteins in the cellular environment.
These two enzymes work in concert to produce phosphatidic acid, a critical metabolite in lipid biosynthesis. GPAT, as the initial and rate-limiting enzyme, plays a crucial role in regulating the flux of fatty acids into glycerolipid synthesis. LPAAT, by catalyzing the second acylation step, is key in determining the molecular species of phospholipids produced. Together, they form the foundation of the glycerophospholipid biosynthetic pathway, which is essential for membrane biogenesis, lipid signaling, and energy storage in cells across various organisms.
Formation of the Phospholipid Head Group: Various head groups can be added to phosphatidic acid to form different phospholipids. The CDP-diacylglycerol pathway is one way to achieve this. For instance, in the synthesis of phosphatidylethanolamine and phosphatidylserine, the head groups ethanolamine and serine would be activated and subsequently attached.
9.2.2. Formation of phospholipid head groups
The formation of phospholipid head groups via the CDP-diacylglycerol pathway entails several enzymatic steps. Here are the primary enzymatic reactions involved:
The CDP-diacylglycerol pathway is a critical metabolic route for the biosynthesis of various phospholipids, including phosphatidylinositol, phosphatidylglycerol, and cardiolipin. This pathway is essential for membrane biogenesis and cellular signaling. The initial step in this pathway involves the conversion of phosphatidic acid to CDP-diacylglycerol, which serves as a key intermediate for subsequent phospholipid synthesis.
Key enzyme involved:
Phosphatidate cytidylyltransferase (CDS) (EC 2.7.7.41): Smallest known: 243 amino acids (Synechocystis sp.)
Catalyzes the formation of CDP-diacylglycerol from phosphatidic acid and CTP. This enzyme plays a crucial role in channeling phosphatidic acid into the CDP-diacylglycerol pathway, thus regulating the synthesis of phosphatidylinositol, phosphatidylglycerol, and cardiolipin. CDS is essential for maintaining the appropriate balance of these phospholipids in cellular membranes and is particularly important in tissues with high energy demands, such as the heart, due to its role in cardiolipin synthesis.
The CDP-diacylglycerol synthesis enzyme group consists of 1 enzyme. The total number of amino acids for the smallest known version of this enzyme is 243.
Information on metal clusters or cofactors:
Phosphatidate cytidylyltransferase (CDS) (EC 2.7.7.41): Requires divalent metal ions, typically Mg²⁺ or Mn²⁺, for catalytic activity. These metal ions play a crucial role in the enzyme's mechanism by:
1. Facilitating the binding of the CTP substrate
2. Stabilizing the transition state during the reaction
3. Promoting the release of the pyrophosphate byproduct
The metal ions coordinate with the phosphate groups of CTP and the phosphatidic acid substrate, bringing them into the correct orientation for the nucleophilic attack that forms the CDP-diacylglycerol product.
Phosphatidate cytidylyltransferase is a pivotal enzyme in phospholipid biosynthesis, acting as a metabolic branch point that directs the flow of lipid precursors into specific phospholipid classes. Its activity is tightly regulated in response to cellular lipid levels and metabolic demands. The enzyme's importance is underscored by its conservation across diverse organisms, from bacteria to humans, reflecting its fundamental role in membrane biogenesis and cellular homeostasis.
The CDP-diacylglycerol formed by this enzyme serves as a versatile precursor for the synthesis of several phospholipids:
1. Phosphatidylinositol, crucial for cell signaling and membrane trafficking
2. Phosphatidylglycerol, important for bacterial membranes and as a precursor to cardiolipin
3. Cardiolipin, essential for mitochondrial function and energy metabolism
By regulating the availability of CDP-diacylglycerol, phosphatidate cytidylyltransferase indirectly influences numerous cellular processes, including signal transduction, membrane dynamics, and energy production. This makes it a potential target for therapeutic interventions in disorders involving lipid metabolism or mitochondrial dysfunction.
9.2.3. Phosphatidylethanolamine (PE) synthesis
The biosynthesis of phosphatidylethanolamine (PE) and phosphatidylserine (PS) is crucial for maintaining cellular membrane structure and function. These phospholipids play essential roles in various cellular processes, including membrane fusion, cell signaling, and apoptosis. The CDP-diacylglycerol pathway and related enzymes are key to the synthesis of these important phospholipids.
Key enzymes involved:
Ethanolaminephosphate cytidylyltransferase (ECT) (EC 2.7.7.14): Smallest known: 367 amino acids (Saccharomyces cerevisiae)
Catalyzes the rate-limiting step in the CDP-ethanolamine pathway for PE synthesis. ECT converts phosphoethanolamine to CDP-ethanolamine, which is a crucial intermediate in PE biosynthesis. This enzyme is essential for maintaining proper PE levels in cellular membranes and is particularly important in rapidly dividing cells.
CDP-diacylglycerol—ethanolamine O-phosphatidyltransferase (EPT) (EC 2.7.8.1): Smallest known: 389 amino acids (Saccharomyces cerevisiae)
Catalyzes the final step in PE synthesis via the CDP-ethanolamine pathway. EPT transfers the phosphoethanolamine group from CDP-ethanolamine to diacylglycerol, forming PE. This enzyme is crucial for regulating the balance between PE and other phospholipids in cellular membranes.
CDP-diacylglycerol—serine O-phosphatidyltransferase (PSS) (EC 2.7.8.8 ): Smallest known: 473 amino acids (Saccharomyces cerevisiae)
Catalyzes the formation of PS by transferring a phosphatidyl group from CDP-diacylglycerol to L-serine. This enzyme is essential for PS biosynthesis and plays a crucial role in maintaining PS levels in cellular membranes, particularly in eukaryotic cells.
Phosphatidylserine decarboxylase (PSD) (EC 4.1.1.65): Smallest known: 353 amino acids (Escherichia coli)
Catalyzes the decarboxylation of PS to form PE. This enzyme provides an alternative route for PE synthesis and is particularly important in prokaryotes and in the mitochondria of eukaryotes. PSD plays a crucial role in maintaining the proper balance between PS and PE in cellular membranes.
The phosphatidylethanolamine and phosphatidylserine biosynthesis enzyme group consists of 4 enzymes. The total number of amino acids for the smallest known versions of these enzymes is 1,582.
Information on metal clusters or cofactors:
Ethanolaminephosphate cytidylyltransferase (ECT) (EC 2.7.7.14): Requires Mg²⁺ as a cofactor for catalytic activity. The magnesium ion helps coordinate the CTP substrate and stabilize the transition state during the reaction.
CDP-diacylglycerol—ethanolamine O-phosphatidyltransferase (EPT) (EC 2.7.8.1): Does not require metal ions or cofactors for its catalytic activity. However, its activity can be modulated by the lipid composition of the membrane environment.
CDP-diacylglycerol—serine O-phosphatidyltransferase (PSS) (EC 2.7.8.8 ): Does not require metal ions or cofactors for its catalytic activity. Like EPT, its activity can be influenced by the surrounding lipid environment.
Phosphatidylserine decarboxylase (PSD) (EC 4.1.1.65): Contains a covalently bound pyruvoyl group as a prosthetic group, which is essential for its catalytic activity. This pyruvoyl group is formed through a post-translational modification and serves as the electron sink during the decarboxylation reaction.
These enzymes work together to regulate the synthesis and interconversion of PE and PS, which are critical components of cellular membranes. The pathway provides flexibility in phospholipid synthesis, allowing cells to adjust their membrane composition in response to various physiological conditions and metabolic demands. The diverse catalytic mechanisms and regulatory properties of these enzymes highlight the complexity of phospholipid metabolism and its importance in cellular homeostasis.
9.2.4. Formation of Phospholipids
As previously discussed, two fatty acid molecules (usually in the form of acyl-CoA) are attached to a glycerol-3-phosphate (G3P) molecule through esterification reactions, resulting in the formation of phosphatidic acid (PA).
The phospholipid head group is then attached to the phosphatidic acid. In the CDP-diacylglycerol pathway, for example, the activated head group displaces the cytidyl group from CDP-diacylglycerol, leading to the formation of the final phospholipid. The nature of the head group determines the specific type of phospholipid (e.g., phosphatidylethanolamine, phosphatidylcholine, phosphatidylserine, etc.).
9.2.5. CDP-diacylglycerol pathway
Phospholipid biosynthesis is a fundamental process in all living organisms, crucial for membrane formation, cellular signaling, and energy storage. The initial steps involve the formation of phosphatidic acid from glycerol-3-phosphate and its subsequent conversion to CDP-diacylglycerol, which serves as a key intermediate for various phospholipid species.
Key enzymes involved:
Glycerol-3-phosphate O-acyltransferase (GPAT) (EC 2.3.1.15): Smallest known: 306 amino acids (Mycobacterium tuberculosis)
Catalyzes the initial and rate-limiting step in de novo glycerophospholipid biosynthesis. GPAT transfers an acyl group from acyl-CoA to the sn-1 position of glycerol-3-phosphate, forming lysophosphatidic acid (LPA). This enzyme is crucial for regulating the flux of fatty acids into the glycerophospholipid biosynthetic pathway and plays a significant role in triglyceride biosynthesis.
1-acylglycerol-3-phosphate O-acyltransferase (AGPAT) (EC 2.3.1.51): Smallest known: 257 amino acids (Chlamydia trachomatis)
Catalyzes the second acylation step in phosphatidic acid biosynthesis. AGPAT transfers an acyl group from acyl-CoA to the sn-2 position of lysophosphatidic acid, producing phosphatidic acid. This enzyme is critical for determining the fatty acid composition of membrane phospholipids and thus influences membrane fluidity and cellular function.
Phosphatidate cytidylyltransferase (CDS) (EC 2.7.7.41): Smallest known: 243 amino acids (Synechocystis sp.)
Catalyzes the formation of CDP-diacylglycerol from phosphatidic acid and CTP. This enzyme plays a crucial role in channeling phosphatidic acid into the CDP-diacylglycerol pathway, thus regulating the synthesis of phosphatidylinositol, phosphatidylglycerol, and cardiolipin. CDS is essential for maintaining the appropriate balance of these phospholipids in cellular membranes.
The glycerophospholipid biosynthesis enzyme group consists of 3 enzymes. The total number of amino acids for the smallest known versions of these enzymes is 806.
Information on metal clusters or cofactors:
Glycerol-3-phosphate O-acyltransferase (GPAT) (EC 2.3.1.15): Requires Mg²⁺ as a cofactor for optimal activity. The magnesium ion is essential for the catalytic mechanism, facilitating the transfer of the acyl group from acyl-CoA to glycerol-3-phosphate.
1-acylglycerol-3-phosphate O-acyltransferase (AGPAT) (EC 2.3.1.51): Does not require metal ions or cofactors for its catalytic activity. However, the enzyme's activity can be modulated by various lipids and proteins in the cellular environment.
Phosphatidate cytidylyltransferase (CDS) (EC 2.7.7.41): Requires divalent metal ions, typically Mg²⁺ or Mn²⁺, for catalytic activity. These metal ions play a crucial role in the enzyme's mechanism by facilitating the binding of the CTP substrate, stabilizing the transition state during the reaction, and promoting the release of the pyrophosphate byproduct.
These three enzymes work in concert to produce CDP-diacylglycerol, a critical metabolite in lipid biosynthesis. GPAT and AGPAT collaborate to form phosphatidic acid, which is then converted to CDP-diacylglycerol by CDS. This pathway is tightly regulated and plays a central role in membrane biogenesis and lipid signaling.
The sequential action of these enzymes highlights the complexity and precision of phospholipid biosynthesis:
1. GPAT initiates the pathway by attaching the first fatty acid to glycerol-3-phosphate.
2. AGPAT completes the formation of phosphatidic acid by adding the second fatty acid.
3. CDS then converts phosphatidic acid to CDP-diacylglycerol, creating a versatile precursor for various phospholipids.
This pathway is critical for maintaining the proper composition of cellular membranes and for producing lipid-based signaling molecules. The regulation of these enzymes allows cells to adjust their membrane composition in response to various physiological conditions and metabolic demands, underscoring their importance in cellular homeostasis and adaptation.
9.2.6. Enzymes involved in Phospholipid Synthesis from CDP-diacylglycerol
The synthesis of specific phospholipids from CDP-diacylglycerol is a crucial process in cellular membrane biogenesis and lipid metabolism. These enzymes catalyze the final steps in the formation of various phospholipids, each with unique functions in cellular processes.
Last edited by Otangelo on Sun Sep 22, 2024 2:50 pm; edited 15 times in total