https://reasonandscience.catsboard.com/t2024-the-rna-world-and-the-origins-of-life
The Death Knell for Life from an RNA world
https://www.youtube.com/watch?v=xSewB3Tzpvs
Robert P. Bywater On dating stages in prebiotic chemical evolution 15 February 2012
Despite the wide repertoire of chemical and biological properties of RNA, which make it such an appealing contender for being the first type of molecular species to usher in life onto this planet, there is no explanation for how such a complex chemical species could have arisen in the absence of sophisticated chemical machinery. The generation of complex chemicals require many millions of cycles of synthesis, partial degradation, concentration, selection and reannealing in combinatorially new ways such that sufficiently diverse species could be produced and reproduced, from which particularly suitable entities survived 34
Claim: Donald R.Prothero, The evolving Earth, page 290
Many scientists have suggested that this is an overly complicated and implausible hypothesis: build a genetic code of proteins first, then replace it with another, more complex one. Instead, they argue, it makes more sense to evolve the genetic code in nucleic acids from the very beginning, even if nucleic acids are harder to produce in chemical reactions than are proteins. Thus, we have a classic “chicken or egg” problem. Which came first: the protein replication system or the nucleic acid replication system? Fortunately, there is a way to resolve this conundrum. In the early 1980s scientists discovered certain types of RNA, known as ribozymes, that perform multiple functions. The RNA in these molecules not only acts as a genetic code but also catalyzes reactions and binds together proteins. In fact, the functional part of the ribosome in the cell, which translates the cellular RNA into proteins, is a ribozyme. Thus, ribozymes perform not only their familiar role as replicators but also the role that proteins play. Further research led to the idea that the simplest scenario for the origin of living, self-replicating systems would be an “RNA world.” The very first self-replicating form of life would be a single-stranded RNA, perhaps enclosed in a lipid bilayer membrane and perhaps using simple carbohydrates for food storage. Using both its replication powers and enzymatic powers, it would make more copies of itself and perform the role of the proteins as well until later, more complex reactions involving many different proteins could evolve. Every year, more discoveries are made that add details to our understanding of the origin of life and the RNA world. For example, according to Lehmann and colleagues (2009), coding sequences of amino acids are easily built on small RNA templates in normal prebiotic conditions. Experiments by Kun and others (2005) show that the first ribozymes in the RNA world were much longer and more stable. According to Costanza and colleagues (2009), experiments have shown that nucleotides easily merge in water to form RNA over 100 nucleotides long. Pino and colleagues (2008) demonstrated that RNA molecules link up into long chains easily under normal earth conditions. And finally, a range of experiments by Long and colleagues (2003) and by Patthy (2003) showed that new genes have been produced repeatedly by evolution. The RNA world hypothesis is now accepted as the most likely scenario for the origin of the first selfreplicating system that can be truly called “life,” although there are still additional conundrums that are being worked on: How did the RNA world get replaced by the DNA world of today? And what preceded the RNA world? Could it have been (some suggest) a PNA world (peptide-nucleic acid) system that had amino acids in the nucleic acid chains instead of the sugar ribose? Or something else? Like any good scientific problem, the
solution of one mystery leads to new and more interesting problems to solve.
Response: Leslie Orgel at the University of Oxford, UK, was among the first to propose RNA as a catalyst of the chemical reactions to make itself. A new theory was born, later dubbed the ‘RNA world hypothesis’.
The RNA world hypothesis, to be true, however, has to overcome major hurdles:
1. Life uses only right-handed RNA and DNA. The homochirality problem is unsolved. This is an “intractable problem” for chemical evolution
2. RNA has been called a “prebiotic chemist's nightmare” because of its combination of large size, carbohydrate building blocks, bonds that are thermodynamically unstable in water, and overall intrinsic instability. Many bonds in RNA are thermodynamically unstable with respect to hydrolysis in water, creating a “water problem”. Finally, some bonds in RNA appear to be “impossible” to form under any conditions considered plausible for early Earth. In chemistry, when free energy is applied to organic matter without Darwinian evolution, the matter devolves to become more and more “asphaltic”, as the atoms in the mixture are rearranged to give ever more molecular species. In the resulting “asphaltization”, what was life comes to display fewer and fewer characteristics of life.
3. Systems of interconnected software and hardware like in the cell are irreducibly complex and interdependent. There is no reason for information processing machinery to exist without the software and vice versa.
4. A certain minimum level of complexity is required to make self-replication possible at all; high-fidelity replication requires additional functionalities that need even more information to be encoded
5. RNA catalysts would have had to copy multiple sets of RNA blueprints nearly as accurately as do modern-day enzymes
6. In order a molecule to be a self-replicator, it has to be a homopolymer, of which the backbone must have the same repetitive units; they must be identical. In the prebiotic world, the generation of a homopolymer was however impossible.
7. Not one self-replicating RNA has emerged to date from quadrillions (10^24) of artificially synthesized, random RNA sequences.
8. Over time, organic molecules break apart as fast as they form
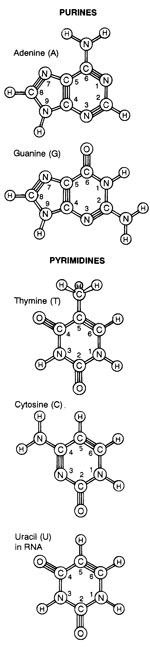
9. How could and would random events attach a phosphate group to the right position of a ribose molecule to provide the necessary chemical activity? And how would non-guided random events be able to attach the nucleic bases to the ribose? The coupling of ribose with a nucleotide is the first step to form RNA, and even those engrossed in prebiotic research have difficulty envisioning that process, especially for purines and pyrimidines.”
10. L. E. Orgel: The myth of a self-replicating RNA molecule that arose de novo from a soup of random polynucleotides. Not only is such a notion unrealistic in light of our current understanding of prebiotic chemistry, but it should strain the credulity of even an optimist's view of RNA's catalytic potential.
11. Macromolecules do not spontaneously combine to form macromolecules
125. The transition from RNA to DNA is an unsolved problem.
13. To go from a self-replicating RNA molecule to a self-replicating cell is like to go from a house building block to a fully built house.
14. Arguably one of the most outstanding problems in understanding the progress of early life is the transition from the RNA world to the modern protein-based world. 31
15. It is thought that the boron minerals needed to form RNA from pre-biotic soups were not available on early Earth in sufficient quantity, and the molybdenum minerals were not available in the correct chemical form. 33
16. Given the apparent limitation of double-stranded RNA (dsRNA) genomes to about 30 kb, together with the complexity of DNA synthesis, it appears dif¢cult for a dsRNA genome to encode all the information required before the transition from an RNA to a DNA genome. Ribonucleotide reductase itself, which synthesizes deoxyribonucleotides from ribonucleotides, requires complex protein radical chemistry, and RNA world genomes may have reached their limits of coding capacity well before such complex enzymes had evolved. 33
Tan, Change; Stadler, Rob. The Stairway To Life:
The extra-hydroxyl group in the pentose ring of RNA makes it more susceptible to hydrolysis, especially in an alkaline environment. A weak solution of sodium hydroxide will destroy RNA but will not damage DNA. Finally, replacement of thymine in DNA with uracil in RNA makes RNA more sensitive to radioactive mutation. If DNA in living organisms requires active repair mechanisms, the more delicate prebiotic RNA, it stands to reason, must also require active repair mechanisms.
Proponents of the RNA world hypothesis commonly argue that it has been proven that RNA's could self-replicate. Let's suppose that were true, that is as if self-replication could produce a hard drive. To go from a hard drive ( which by itself requires complex information to be assembled, in case of biology, DNA, not RNA since it's too unstable, ) that does not explain the origin of the information to make all life essential parts in the cell.
It is as to go just from a hard drive storage device to a self-replicating factory with the ability of self-replication of the entire factory once ready, to respond to changing environmental demands and regulate its metabolic pathways, regulate and coordinate all cellular processes, such as molecule and building block biosynthesis according to the cells demands, depending on growth, and other factors.
The ability of uptake of nutrients, to be structured, internally compartmentalized and organized, being able to check replication errors and minimize them, and react to stimuli, and changing environments. That's is, the ability to adapt to the environment is a must right from the beginning. If just ONE single protein or enzyme - of many - is missing, no life. If topoisomerase II or helicase are missing - no replication - no perpetuation of life.
Why would a prebiotic soup or hydrothermal vents produce these proteins - if by their own there is no use for them?
Robert Shapiro: Origins, a Skeptic's Guide to the Creation of Life on Earth (1986) p.186.
In other words,' I said, `if you want to create life, on top of the challenge of somehow generating the cellular components out of non-living chemicals, you would have an even bigger problem in trying to it the ingredients
together in the right way.' `Exactly! ... So even if you could accomplish the thousands of steps between the amino acids in the Miller tar-which probably didn't exist in the real world anyway-and the components you need for a
living cell-all the enzymes, the DNA, and so forth-you's still immeasurably far from life. ... the problem of assembling the right parts in the right way at the right time and at the right place, while keeping out the wrong
material, is simply insurmountable.
Paul Davies The Algorithmic Origins of Life
Despite the conceptual elegance of the RNA world, the hypothesis faces problems, primarily due to the immense challenge of synthesizing RNA nucleotides under plausible prebiotic conditions and the susceptibility of RNA oligomers to degradation via hydrolysis 21 Due to the organizational structure of systems capable of processing algorithmic (instructional) information, it is not at all clear that a monomolecular system – where a single polymer plays the role of catalyst and informational carrier – is even logically consistent with the organization of information flow in living systems, because there is no possibility of separating information storage from information processing (that being such a distinctive feature of modern life). As such, digital-first systems (as currently posed) represent a rather trivial form of information processing that fails to capture the logical structure of life as we know it.
Replicator first, and metabolism first scenarios
https://reasonandscience.catsboard.com/t1428-replicator-first-and-metabolism-first-scenarios
No evidence that RNA molecules ever had the broad range of catalytic activities
https://reasonandscience.catsboard.com/t2243-no-evidence-that-rna-molecules-ever-had-the-broad-range-of-catalytic-activities
The hardware and software of the cell, evidence of design
https://reasonandscience.catsboard.com/t2221-the-hardware-and-software-of-the-cell-evidence-of-design
The origin of replication and translation and the RNA World
https://reasonandscience.catsboard.com/t2234-the-origin-of-replication-and-translation-and-the-rna-world
Tom Robbins: The time argument is worthless. As over time, organic molecules break apart as fast as they form - thus the monkey's on a typewriter argument does not work, as the INFORMATION represented on the paper when they strike a key, disappears off the paper as they type. Given enough time, CERTAIN things will probably happen, but only things that are not impossible (and of course there was a finite amount of time from the creation of the earth). Nature can't create specified, dedicated, self-replicating, self-repairing, self-editing information - THE ONLY source that we know of that can do this, is MIND..
Given the complexity of the simplest ribozymes mediating transcription and translation and the ongoing failure to obtain activated ribonucleotides from ribose and nucleobases, the RNA-world hypothesis faces substantial challenge 23
Paul C. W. Davies The algorithmic origins of life 2013 Feb 6
RNA has been called a “prebiotic chemist's nightmare” because of its combination of large size, carbohydrate building blocks, bonds that are thermodynamically unstable in water, and overall intrinsic instability. Many bonds in RNA are thermodynamically unstable with respect to hydrolysis in water, creating a “water problem”. Finally, some bonds in RNA appear to be “impossible” to form under any conditions considered plausible for early Earth. In chemistry, when free energy is applied to organic matter without Darwinian evolution, the matter devolves to become more and more “asphaltic”, as the atoms in the mixture are rearranged to give ever more molecular species. In the resulting “asphaltization”, what was life comes to display fewer and fewer characteristics of life.
Biologists routinely observe the opposite. In the biosphere, when free energy is provided to organic matter that does have access to Darwinian evolution, that matter does not become asphaltic. Instead, “life finds a way” to exploit available raw materials, including atoms and energy, to create more of itself and, over time, better of itself. This observation is made across the Earth, from its poles to the equator, from high in the atmosphere to the deepest oceans, and in humidities that cover all but the very driest. The contrast between these commonplace observations in chemistry versus commonplace observations in biology embodies the paradox that lies at the center of the bio-origins puzzle. Regardless of the organic materials or the kinds of energy present early on Earth, chemists expect that a natural devolution took them away from biology toward asphalt. To escape this asphaltic fate, this devolution must have transited a chemical system that was, somehow, able to sustain Darwinian evolution. Otherwise, the carbon on Earth would have ended up looking like the carbon in the Murchison meteorite (or the La Brea tar pits without the fossils).
We have not addressed the “chirality problem”. Here, the challenge arises as we attempt to select from among a large number of possible approaches for chiral enrichment, ranging from the interaction of organic species with chiral minerals (e.g., quartz) to the resolution at the time of oligomerization. Current experiments suggest that RNA molecules that catalyze the degradation of RNA are more likely to emerge from a library of random RNA molecules than RNA molecules that catalyze the template-directed synthesis of RNA, especially given cofactors (e.g., Mg2þ). This could, of course, be a serious (and possibly fatal) flaw to the RNA-first hypothesis for bioorigins.
We need to explain the origin of both the hardware and software aspects of life, or the job is only half finished. Explaining the chemical substrate of life and claiming it as a solution to life’s origin is like pointing to silicon and copper as an explanation for the goings-on inside a computer. It is this transition where one should expect to see a chemical system literally take on “a life of its own”, characterized by informational dynamics which become decoupled from the dictates of local chemistry alone (while of course remaining fully consistent with those dictates). Thus the famed chicken-or-egg problem (a sole hardware issue) is not the true sticking point. Rather, the puzzle lies with something fundamentally different, a problem of a causal organization having to do with the separation of informational and mechanical aspects into parallel causal narratives. The real challenge of life’s origin is thus to explain how instructional information control systems emerge naturally and spontaneously from mere molecular dynamics.
But now we hit a snag. The second step on the road to life, or at least the road to proteins, is for amino acids to link together to form molecules known as peptides. A protein is a long peptide chain, or a polypeptide. Whereas the spontaneous formation of amino acids from an inorganic chemical mixture is an allowed downhill process, coupling amino acids together to form peptides is an uphill process. It therefore heads in the wrong direction, thermodynamically speaking. Each peptide bond that is forged requires a water molecule to be plucked from the chain. In a watery medium like a primordial soup, this is thermodynamically unfavorable. Consequently, it will not happen spontaneously: work has to be done to force the newly extracted water molecule into the water saturated medium. Obviously, peptide formation is not impossible because it happens inside living organisms. But there the uphill reaction is driven along by the use of customized molecules that are pre-energized to supply the necessary work. In a simple chemical soup, no such specialized molecules would be on hand to give the reactions the boost they need. So a watery soup is a recipe for molecular disassembly, not self-assembly.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3565706/
Systems of interconnected software and hardware like in the cell are irreducibly complex and interdependent. There is no reason for information processing machinery to exist without the software and vice versa.
Proof by self-replicating RNA
1. Till now, after more than 50 years of biochemical experiments, there were no self-replicating RNA molecules generated in any different laboratory conditions that resemble the prebiotic period of creation.
2. RNA has no self-replicating power.
3. Without self-replicating RNA there is neither natural selection nor evolution.
4. Therefore, there must have been another original cause of existence and that cause is God.
The classic evolutionary problem of 'which came first, protein or DNA' has not been solved by the 'self-reproducing' RNA theory as many textbooks imply. The theory is not credible as it was based on laboratory simulations which were highly artificial, and was carried out with a 'great deal of help from the scientists'.19
For 40 years, efforts to understand the prebiotic synthesis of the ribonucleotide building blocks of RNA have been based on the assumption that they must have assembled from their three molecular components: a nucleobase (which can be adenine, guanine, cytosine or uracil), a ribose sugar and phosphate. Of the many difficulties encountered by those in the field, the most frustrating has been the failure to find any way of properly joining the pyrimidine nucleobases — cytosine and uracil — to ribose3 . The idea that a molecule as complex as RNA could have assembled spontaneously has therefore been viewed with increasing scepticism. 20
[/b]
The most widely accepted hypothesis among biologists, the RNA world hypothesis, still has strong supporters [1] but difficulties with the hypothesis are recognized, especially the problem of synthesizing RNA in the absence of enzymes 15
One of the principal problems concerning the hypothesis of the RNA world is that it appears quite unlikely that a prebiotic environment could have existed containing the mixture of activated nucleotides favoring the formation and replication of ribozymes, as well as their evolution through natural selection. Even if there were several candidate reactions for an efficient prebiotic synthesis of nucleic bases, access to monomeric nucleotides by chemical pathways, in fact, comes up against several obstacles. If one goes no further than mimicking the biochemical pathway, the first difficulty that occurs is that of synthesizing ribose, which is formed in just negligible quantities within the complex mixture obtained by polymerization of formaldehyde, and, what is more, has a limited lifetime. The bond between a nucleic base and ribose that produces a nucleoside is then a very difficult reaction. There still remains the matter of obtaining a nucleotide by phosphorylation, which leads to mixtures because three positions remain available on the ribose, and then there is its activation 24

The daunting problem how to make the pentose ring of RNA and DNA
In the case of RNA, not only must phosphodiester links be repeatedly forged (if we assume joining reactions involving P–O bond formation), but they must ultimately connect the 5ʹ‑oxygen of one nucleotide to the 3ʹ‑oxygen, and not the 2ʹ‑oxygen, of the next nucleotide. 2ʹ,5ʹ‑Linkages can be tolerated functionally at low levels in certain RNAs31, but they are not inheritable in a sequence-specific manner, and for most intents and purposes, extant biology
uses 3ʹ,5ʹ‑linkages. Although we have demonstrated that 3ʹ,5ʹ‑linkages can be preferentially formed by prebiotically selective 2ʹ‑O‑acetylation and ligation of those oligonucleotides with 3ʹ‑phosphate termini in mixtures of oligonucleotides with 2ʹ‑ and 3′-phosphate termini, the synthetic selectivities and preferences are not enough to explain how RNA with all 3ʹ,5ʹ‑linkages might first have been produced. 29
James Tour wrote :
Problematic Chemical Postulates of the RNA World Scenario
Postulate 1:
Beta-D-ribonucleotides are compounds made up of a purine (adenine or guanine) or a pyrimidine (uracil or cytosine) linked to the 1'-position of ribose in the beta-configuration.
There is, in addition, a phosphate group attached to the 5'-position of the ribose. For the four different ribonucleotides in this prebiotic scenario, there would be hundreds of other possible isomers.
But each of these four ribonucleotides is built up of three components: a purine or pyrimidine, a sugar (ribose), and phosphate. It is highly unlikely that any of the necessary subunits would have accumulated in any more than trace amounts on the primitive Earth. Consider ribose. The proposed prebiotic pathway leading to this sugar, the formose reaction, is especially problematic.
prebiotically plausible sequences of steps to the precursors of this ribose derivative and from it to the standard nucleotides are not obvious. 17
While sugars have a variety of important roles in biochemistry, they are important components of nucleic acid backbones in the form of ribose and deoxyribose. 28 All canonical sugars share the empirical formula (CH2O)n, formally making them oligomers of formaldehyde (HCHO). Ribose, the sugar used in RNA, is but one of the isomeric pentamers where n = 5, as each sugar may contain a number of stereo centers. Early in the development of organic chemistry as an empirical science, it was found that basic solutions of formaldehyde could give rise to a complex mixture of compounds, which included various sugars (Butlerow 1861). The mechanism of this synthesis has since been explored extensively (Breslow 1959) (Figure 5.13).

Early in its consideration as a prebiotic process for the production of ribose and other sugars, it was pointed out that the extreme diversity of products the reaction gives rise to, as well as the ultimate instability of sugars under the conditions of synthesis, may render this an implausible source of prebiotic carbohydrates . Recently, there has been a resurgence of interest in this pathway as several new mechanisms have been discovered that produce a less diverse mixture, give rise to a higher yield of ribose, and importantly make the ultimate sugar derivatives considerably more stable than they are in the free form. For example, conducting the reaction in the presence of borate selectively gives rise to a good yield of ribose–borate derivatives
Formose reaction
The formose reaction is of great importance to the question of the origin of life as it explains part of the path from simple formaldehyde to complex sugars like ribose and from there to RNA. In one experiment simulating early Earth conditions, pentoses formed from mixtures of formaldehyde, glyceraldehyde, and borate minerals such as colemanite. Both formaldehyde and glycolaldehyde have been observed spectroscopically in outer space, making the formose reaction of particular interest to the field of astrobiology.
Aldol reaction
The aldol reaction is a means of forming carbon–carbon bonds in organic chemistry.Discovered independently by Charles-Adolphe Wurtz and Alexander Borodin in 1872, the reaction combines two carbonyl compounds (the original experiments used aldehydes) to form a new β-hydroxy carbonyl compound. These products are known as aldols, from the aldehyde + alcohol, a structural motif seen in many of the products. Aldol structural units are found in many important molecules, whether naturally occurring or synthetic. For example, the aldol reaction has been used in the large-scale production of the commodity chemical pentaerythritol[11] and the synthesis of the heart disease drug Lipitor (atorvastatin, calcium salt).
https://en.wikipedia.org/wiki/Aldol_reaction
and aldose-ketose isomerizations.
In carbohydrate chemistry, the Lobry de Bruyn–van Ekenstein transformation also known as the Lobry de Bruyn–Alberda–van Ekenstein transformation is the base or acid catalyzed transformation of an aldose into the ketose isomer or vice versa, with a tautomeric enediol as reaction intermediate. Ketoses may be transformed into 3-ketoses, etcetera. The enediol is also an intermediate for the epimerization of an aldose or ketose.
https://en.wikipedia.org/wiki/Lobry_de_Bruyn%E2%80%93van_Ekenstein_transformation
The improbability of prebiotic nucleic acid synthesis.
Many accounts of the origin of life assume that the spontaneous synthesis of a self-replicating nucleic acid could take place readily. Serious chemical obstacles exist, however, which make such an event extremely improbable. Prebiotic syntheses of adenine from HCN, of D,L -ribose from adenosine, and of adenosine from adenine and D-ribose have in fact been demonstrated. However these procedures use pure starting materials, afford poor yields, and are run under conditions which are not compatible with one another. Any nucleic acid components which were formed on the primitive earth would tend to hydrolyze by a number of pathways. Their polymerization would be inhibited by the presence of vast numbers of related substances which would react preferentially with them. It appears likely that nucleic acids were not formed by prebiotic routes
The nitrogenous substances react with formaldehyde, the intermediates in the pathways to sugars, and with sugars themselves to form non-biological materials10. Furthermore, as Stanley Miller and his colleagues recently reported, "ribose and other sugars have surprisingly short half-lives for decomposition at neutral pH, making it very unlikely that sugars were available as prebiotic reagents."
Or consider adenine. Reaction pathways proposed for the prebiotic synthesis of this building block start with HCN in alkaline (pH 9.2) solutions of NH4OH.12 These reactions give small yields of adenine (e.g., 0.04%) and other nitrogenous bases provided the HCN concentration is greater than 0.01 M. However, the reaction mixtures contain a great variety of nitrogenous substances that would interfere with the formose reaction. Therefore, the conditions proposed for the prebiotic synthesis of purines and pyrimidines are clearly incompatible with those proposed for the synthesis of ribose. Moreover, adenine is susceptible to deamination and ring-opening reactions (with half-lives of about 80 years and 200 years respectively at 37º C and neutral pH), making its prebiotic accumulation highly improbable. This makes it difficult to see how any appreciable quantities of nucleosides and nucleotides could have accumulated on the primitive Earth. If the key components of nucleotides (the correct purines and pyrimidines, ribose, and phosphate) were not present, the possibility of obtaining a pool of the four beta-D-ribonucleotides with correct linkages would be remote indeed.
If this postulate, the first and most crucial assumption, is not valid, however, then the entire hypothesis of an RNA World formed by natural processes becomes meaningless.
Pentose sugars, constituents of RNA and DNA, can be synthesised in the formose reaction, given the presence of formaldehyde (HCHO). The products are a melange of sugars of various carbon lengths which are optically left- and right-handed (d and l). With few exceptions sugars found in biological systems are of the d type; for instance, β-d-ribose of RNA, which is always produced in small quantities abiotically.Hydrocyanic acid (HCN) undergoes polymerisation to form diaminomaleonitrile which is on the pathway to producing adenine, hypoxanthine, guanine, xanthine and diaminopurine. These are purines: there is difficulty in producing pyrimidines (cytosine, thymine and uracil) in comparable quantities Neither preformed purines nor pyrimidines have been successfully linked to ribose by organic chemists. An attempt to make purine nucleosides resulted in a “dizzying array of related compounds”.39 This is expected if sugars and bases were randomly coupled. The prebiotic production of numerous isomers and closely related molecules hinders the likelihood of forming desirable mononucleosides. Furthermore, unless ribose and the purine bases form nucleosides rapidly they would be degraded quite quickly.
Producing ribose under realistic conditions has proven even more problematic. Prebiotic chemists have proposed that ribose could have arisen on the early earth as the by-product of a chemical reaction called the formose reaction. The formose reaction is a multistep chemical reaction that begins as molecules of formaldehyde in water react with one another. Along the way, the formose reaction produces a host of different sugars, including ribose, as intermediate by-products in the sequence of reactions. But, as Shapiro has pointed out, the formose reaction will not produce sugars in the presence of nitrogenous substances.11 These include peptides, amino acids, and amines, a category of molecules that includes the nucleotide bases. This obviously poses a couple of difficulties. First, it creates a dilemma for scenarios that envision proteins and nucleic acids arising out of a prebiotic soup rich in amino acids. Either the prebiotic environment contained amino acids, which would have prevented sugars (and thus DNA and RNA) from forming, or the prebiotic soup contained no amino acids, making protein synthesis impossible. Of course, RNA-first advocates might try to circumvent this difficulty by proposing that proteins arose well after RNA. Yet since the RNA-world hypothesis envisions RNA molecules coming into contact with amino acids early on within the first protocellular membranes (see above), choreographing the origin of RNA and amino acids to ensure that the two events occur separately becomes a considerable problem.
Nucleosides and Nucleotides
The construction of nucleosides depends on the union of a nitrogenous base, via the correct linkage, with a sugar derivative. 28 Some success in prebiotic synthesis has been achieved in this area. For example, it has been found that heating pure ribose with purines gives rise to small yields of purine ribosides, though a variety of isomers are produced. The equivalent reaction using pyrimidines does not work well, however. Recently, though, it has been found that a nonnatural pyrimidine can be linked under the same condition to give a pyrimidine nucleoside . The phosphorylation of nucleosides to give nucleotides has been accomplished in a variety of manners that are conceivably prebiotic. Dry heating various mixtures of nucleosides in the presence of ammonium salts and orthophosphate or apatite and cyanate gives decent yields of pyrimidine nucleotides. The limitations of the synthesis of pyrimidine nucleosides led Orgel and coworkers to examine other disconnects to find less obvious, unorthodox syntheses, which have been elucidated and explored further more recently. This approach is novel in that the sugar and nitrogenous base are constructed simultaneously and phosphate is incorporated prior to the completion of the nucleoside’s synthesis. This has proven effective for the pyrimidines, but to date, a complete analogous synthesis for the purine nucleotides has proven elusive.
nucleotides do not link unless there is some type of activation of the phosphate group. The only effective activating groups for the nucleotide phosphate group (imidazolines, etc.), however, are those that are totally implausible in any prebiotic scenario. In living organisms today, adenosine-5'-triphosphate (ATP) is used for activation of nucleoside phosphate groups, but ATP would not be available for prebiotic syntheses. Joyce and Orgel note the possible use of minerals for polymerization reactions, but then express their doubts about this possibility
Joyce and Orgel then note that if there were activation of the phosphate group, the primary polymer product would have 5', 5'-pyrophosphate linkages; secondarily 2', 5'-phosphodiester linkages -- while the desired 3',5'-phosphodiester linkages would be much less abundant. However, all RNA known today has only 3',5'-phosphodiester linkages, and any other linkages would alter the three-dimensional structure and possibilities for function as a template or a catalyst.
Even waiving these obstacles, and allowing for minute amounts of oligoribonucleotides, these molecules would have been rendered ineffective at various stages in their growth by adding incorrect nucleotides, or by reacting with the myriads of other substances likely to have been present. Moreover, the RNA molecules would have been continuously degraded by spontaneous hydrolysis and other destructive processes operating on the primitive Earth.
In brief, any movement in the direction of an RNA World on a realistically-modeled early Earth would have been continuously suppressed by destructive cross-reactions.
One of the more enigmatic and difficult problems confronting the prebiotic chemistry community is identifying how the monomers of RNA, or pre-RNA, or even non-related polymeric components selectively formed and self-assembled out of the presumed random prebiotic mixtures. It is in this assembly into informational polymers (Figure 4) where significant selection processes must have occurred not only for the base composition but also for the other components of nucleic acids (or nucleic acid alternatives and precursors). Focusing on just a narrow view of RNA precursors, the linking of a nucleo-base to a ribose sugar is one such pressure. There are multiple ways in which a nucleobase can be attached to ribose via an N-glycosidic bond, but only one is found in contemporary nucleic acids (via the N9 of purines and N1 of pyrimidines). 14
Achieving regio- and stereochemical selectivity of glycosylation reactions under simulated prebiotic conditions has plagued the community ever since Orgel and others began working on this problem
Complex Chemical Systems Can Develop in an Environment That Is Far from Chemical Equilibrium 7
Simple organic molecules such as amino acids and nucleotides can associate to form polymers. One amino acid can join with another by forming a peptide bond, and two nucleotides can join together by a phosphodiester bond.
Protein synthesis (condensation of amino acids through sequential peptide bond formation) is a fundamental and ubiquitous reaction in biology. Aqueous media are the required environments in which this chemistry takes place; however, protein synthesis is unfavorable in aqueous solution. In modern biology, the condensation reactions necessary in the formation of peptide bonds are facilitated catalytically by the large subunit of the ribosome.
Peptide bond synthesis occurs in the 50S subunit ( of the ribosome ) at the peptidyl transferase center, (PTC) 4
The crucial peptide bond formation of protein synthesis is catalyzed by the ribosome in all organisms. 5
The activation of amino acids and the formation of peptides under primordial conditions is one of the great riddles of the origin of life.
The famous pioneer of evolutionary origin-of-life experiments, Stanley Miller, points out that polymers are ‘too unstable to exist in a hot prebiotic environment’ 10
Question: How could simple organic molecules such as amino acids and nucleotides associate to form polymers, one amino acid joining with another by forming a peptide bond, if peptide bonds are synthesized in the probably most complex protein complex known, the ribosome, but the ribosome was not there at this stage ?

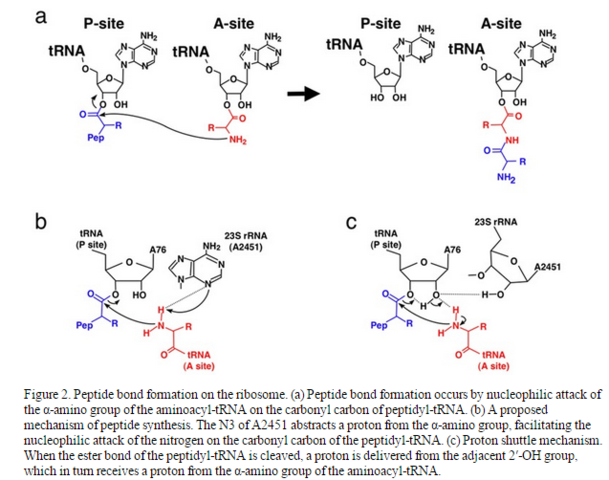
Prebiotic Peptide synthesis was likely initiated in a simple way, yet must have evolved into the contemporary complexity of the ribosome. Of course. There is no other explanation, since an Intelligent designer is excluded a priori. In order to know how the current ribosome-catalyzed reaction evolved from a primitive system, model systems based on the RNA world hypothesis with the molecules like the minihelix and tRNA were postulated. Elucidation of the evolutionary route from the simple system to the present complex ribosome is a big challenge in modern science; this gap may be filled by the concept of the proto-ribosome, which is composed of a symmetrical tRNA-like dimer. 5
The peptide synthesis hypotheses must jump over a large gap to attain ribosome-based peptide synthesis. 5
We have right at the beginning of naturalistic proposals of the origin of life the typical guess work which extends all over the key issues of the origin of life, the transition to the 3 domains of life, from unicellular to multicellular, and biodiversity on earth. That seems to be a RED LiNE extending through all scientific papers, which deal with key issues in biology. Naturalism is simply not tenable, the explanations all end at a dead end, where guesswork is common, and evolution of the gaps arguments.
A restricted set of 20 amino acids constitute the universal building blocks of the proteins, while RNA and DNA molecules are constructed from just four types of nucleotides each. It is uncertain why these particular sets of monomers were selected for biosynthesis in preference to others that are chemically similar
The earliest polymers may have formed in any of several ways - for example, by the heating of dry organic compounds or by the catalytic activity of high concentrations of inorganic polyphosphates or other crude mineral catalysts. Under laboratory conditions, the products of similar reactions are polymers of variable length and random sequence in which the particular amino acid or nucleotide added at any point depends mainly on chance
The origin of life requires that in an assortment of such molecules there must have been some possessing, if only to a small extent, a crucial property: the ability to catalyze reactions that lead, directly or indirectly, to production of more molecules of the catalyst itself. Production of catalysts with this special self-promoting property would be favored, and the molecules most efficient in aiding their own production would divert raw materials from the production of other substances. In this way one can envisage the gradual development of an increasingly complex chemical system of organic monomers and polymers that function together to generate more molecules of the same types, fueled by a supply of simple raw materials in the environment. Such an autocatalytic system would have some of the properties we think of as characteristic of living matter: it would comprise a far from random selection of interacting molecules; it would tend to reproduce itself; it would compete with other systems dependent on the same feedstocks; and if deprived of its feedstocks or maintained at a wrong temperature that upsets the balance of reaction rates, it would decay toward chemical equilibrium and "die."
What we see here, is a typical fairy tale story, based on no evidence, just fantasy without a shred data to back up the story. Why should someone be credule towards such a scenario? Words like must have been, would be, one can envisage, would, would have, we think of, it would do not invoke much credibility.....
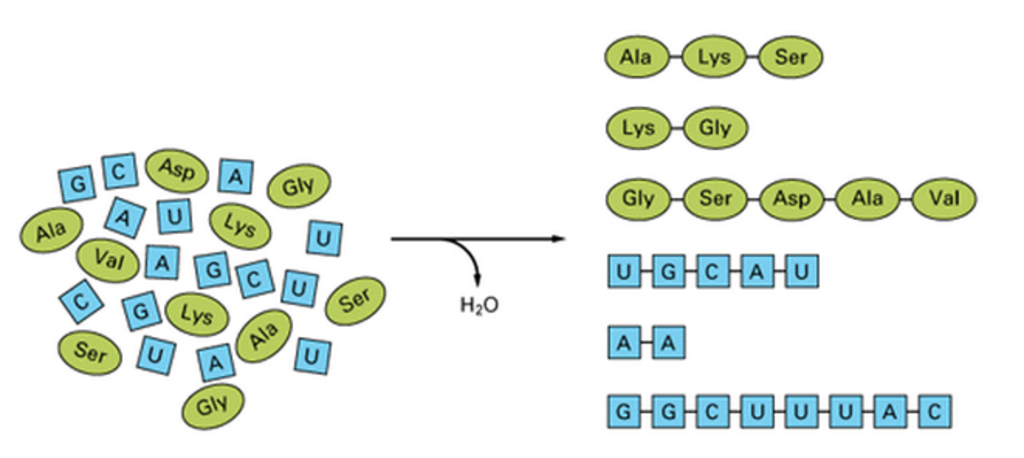
photo storage
Formation of polynucleotides and polypeptides. Nucleotides of four kinds (here represented by the single letters A, U, G, and C) can undergo spontaneous polymerization with the loss of water. The product is a mixture of polynucleotides that are random in length and sequence. Similarly, amino acids of different types, symbolized here by three-letter abbreviated names, can polymerize with one another to form polypeptides. Present-day proteins are built from a standard set of 20 types of amino acids.
Polynucleotides Can Both Store Information and Catalyze Chemical Reactions 2
Polynucleotides have properties that contrast with those of polypeptides. They have more limited capabilities as catalysts, but they can directly guide the formation of exact copies of their own sequence. This capacity depends on the complementary pairing of nucleotide subunits, which enables one polynucleotide to act as a template for the formation of another. In the simplest case, a polymer composed of one nucleotide (for example, polycytidylic acid, or poly C) can line up the subunits required to make another polynucleotide (in this example, polyguanylic acid, or poly G) along its surface, thereby promoting their polymerization into poly G

Polynucleotides as templates. Preferential binding occurs between pairs of nucleotides (G with C and U with A) by relatively weak chemical bonds (above). This pairing enables one polynucleotide to act as a template for the synthesis of another (left).
This capacity depends on the complementary base pairing of nucleotide subunits, which enables one polynucleotide to act as a template for the formation of another. As we have seen in this and the preceding chapter, such complementary templating mechanisms lie at the heart of DNA replication and transcription in modern-day cells. But the efficient synthesis of polynucleotides by such complementary templating mechanisms requires catalysts to promote the polymerization reaction: without catalysts, polymer formation is slow, error-prone, and inefficient.
Consider now a polynucleotide with a more complex sequence of subunitsspecifically, a molecule of RNA strung together from four types of nucleotides, containing the bases uracil (U), adenine (A), cytosine (C), and guanine (G), arranged in some particular sequence. Because of complementary pairing between the bases A and U and between the bases G and C, this molecule, when added to a mixture of activated nucleotides under suitable conditions, will line them up for polymerization in a sequence complementary to its own. The resulting new RNA molecule will be rather like a mold of the original, with each A in the original corresponding to a U in the copy and so on. The sequence of nucleotides in the original RNA strand contains information that is, in essence, preserved in the newly formed complementary strands: the second round of copying, with the complementary strand as a template, restores the original sequence 2

1
Replication of a polynucleotide sequence (here an RNA molecule). In step 1 the original RNA molecule acts as a template to form an RNA molecule of complementary sequence. In step 2 this complementary RNA molecule itself acts as a template, forming RNA molecules of the original sequence. Since each templating molecule can produce many copies of the complementary strand, these reactions can result in the "multiplication" of the original sequence.
Such complementary templating mechanisms are elegantly simple, and they lie at the heart of information transfer processes in biological systems. The genetic information contained in every cell is encoded in the sequences of nucleotides in its polynucleotide molecules, and this information is passed on (inherited) from generation to generation by means of complementary base-pairing interactions.
Templating mechanisms, however, require additional catalysts to promote polymerization; without these the process is slow and inefficient and other, competing reactions prevent the formation of accurate replicas. Today, the catalytic functions that polymerize nucleotides are provided by highly specialized catalytic proteins that is, by enzymes. In the "prebiotic soup" primitive polypeptides might perhaps have provided some catalytic help. But molecules with the appropriate catalytic specificity would have remained rare unless the RNA itself were able somehow to reciprocate and favor their production. We shall come back to the reciprocal relationship between RNA synthesis and protein synthesis, which is crucially important in all living cells. But let us first consider what could be done with RNA itself, for RNA molecules can have a variety of catalytic properties, besides serving as templates for their own replication. In particular, an RNA molecule with an appropriate nucleotide sequence can act as a catalyst for the accurate replication of another RNA molecule - the template - whose sequence can be arbitrary.
The special versatility of RNA molecules is thought to have enabled them to play a central role in the origin of life. We have however to ignore what has stated above, namely that without catalysts the process is slow and inefficient which makes this hypothesis remotely possible. That makes the scenario very unlikely. And so far, the sequence would be completely random, no coded information.
Linking the RNA sugar-phosphate backbones together
Phosphodiester bonds
The sugar-phosphate backbone forms the structural framework of nucleic acids, including DNA and RNA. This backbone is composed of alternating sugar and phosphate groups, and defines directionality of the molecule. 3

free upload pictures

1) http://www.bioon.com/book/biology/mboc/mboc.cgi@action=figure&fig=1-4.htm
2) http://www.bioon.com/book/biology/mboc/mboc.cgi@code=010103174953651.htm
3) http://www.nature.com/scitable/definition/phosphate-backbone-273
3) http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3465415/
4) http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2926754/
5) http://journalofcosmology.com/Abiogenesis130.html
6) Alberts, molecular biology of the cell, pg.401
7) http://gowiki.tamu.edu/wiki/index.php/Category:GO:0090501_!_RNA_phosphodiester_bond_hydrolysis
7) http://www.bioon.com/book/biology/mboc/mboc.cgi@code=010102442543328.htm
8 ) http://www.arn.org/docs/odesign/od171/rnaworld171.htm
9) R. Shapiro, "The improbability of prebiotic nucleic acid synthesis," Origins of Life 14 (1984): 565-570; R. Shapiro, "Prebiotic ribose synthesis: a critical analysis," Origins of Life 18 (1988): 71-85. http://www.ncbi.nlm.nih.gov/pubmed/6462692
10) http://creation.com/origin-of-life-the-polymerization-problem
11) https://www.promega.com/~/media/files/resources/product%20guides/cloning%20enzymes/ligases.pdf?la=en
12) http://en.wikipedia.org/wiki/Ligase_ribozyme
13) http://en.wikipedia.org/wiki/Hairpin_ribozyme
14) http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4181368/
15) http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4390864/
16) http://exploringorigins.org/nucleicacids.html
17) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC18793/
18) http://creation.com/origin-of-life-critique
19) Scientific American, February, 1991 p:100-109
20) Systems chemistry on early Earth, Jack W. Szostak, nature, Vol 459|14 May 2009,
21) http://arxiv.org/pdf/1207.4803v2.pdf
22) http://arxiv.org/pdf/1207.4803v2.pdf
23) https://www.mpg.de/9333399/Origin_of_Life_basetext.pdf
24) M. Gargaud · H. Martin · P. López-García T. Montmerle · R. Pascal Young Sun, Early Earth and the Origins of Life , page 116
25) http://pubs.acs.org.sci-hub.ren/doi/abs/10.1021/ar200332w
26) Earth Evolution of a Habitable World, Second edition, page 156
27) http://inference-review.com/article/two-experiments-in-abiogenesis#endnote-1
28 ) ASTROBIOLOGY An Evolutionary Approach, page 106
29) http://sci-hub.tw/https://www.nature.com/articles/s41570-016-0012
30) https://cos.gatech.edu/hg/item/575811
31) http://www.weizmann.ac.il/sb/Pages/Yonath/Belousoff-2010SpringerBOOK.pdf
32) https://en.wikipedia.org/wiki/Formose_reaction
33. http://sci-hub.tw/https://www.sciencedirect.com/science/article/pii/S1074552100000429
34. https://sci-hub.ren/10.1007/s00114-012-0892-6
further readings: http://creation.com/native-folds-in-polypeptide-chains-1
http://www.evolutionnews.org/2015/06/on_the_origin_o_7097191.html
The Origin of RNA and “My Grandfather’s Axe”
Insuperable Problems Of The Genetic Code Initially Emerging In An RNA World
http://biorxiv.org/content/early/2017/05/22/140657
The RNA World: molecular cooperation at the origins of life
Last edited by Otangelo on Mon Jan 09, 2023 7:25 pm; edited 99 times in total