https://reasonandscience.catsboard.com/t2134-the-amazing-design-of-bacteriophage-viruses-and-its-dna-packaging-motor
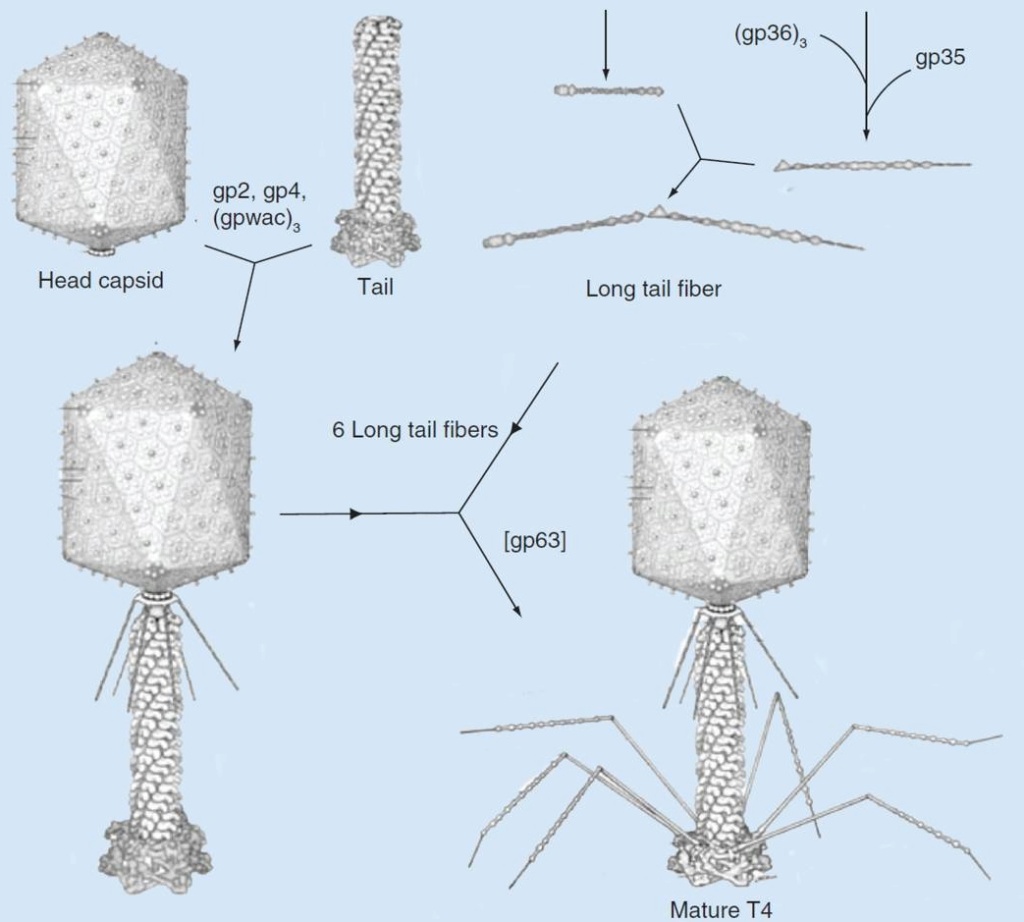
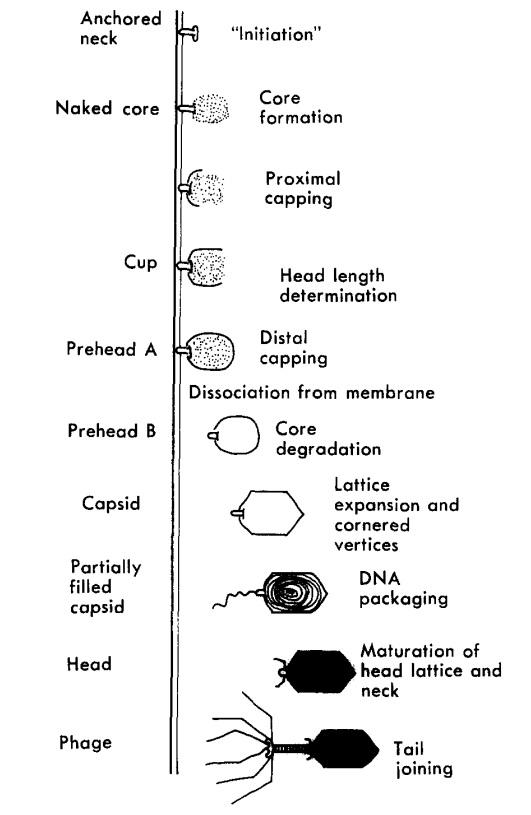
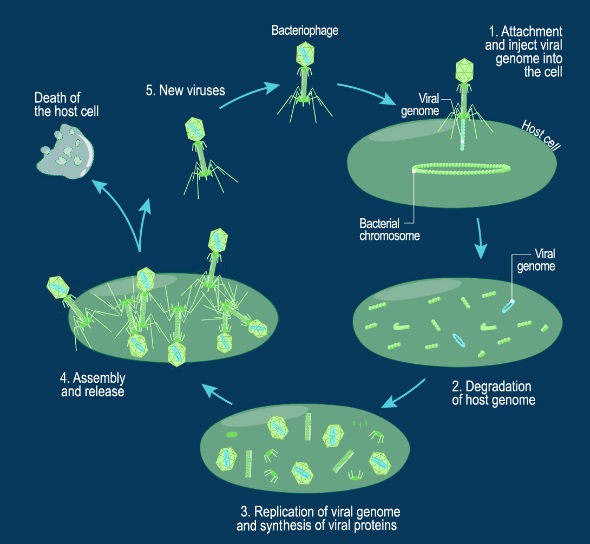
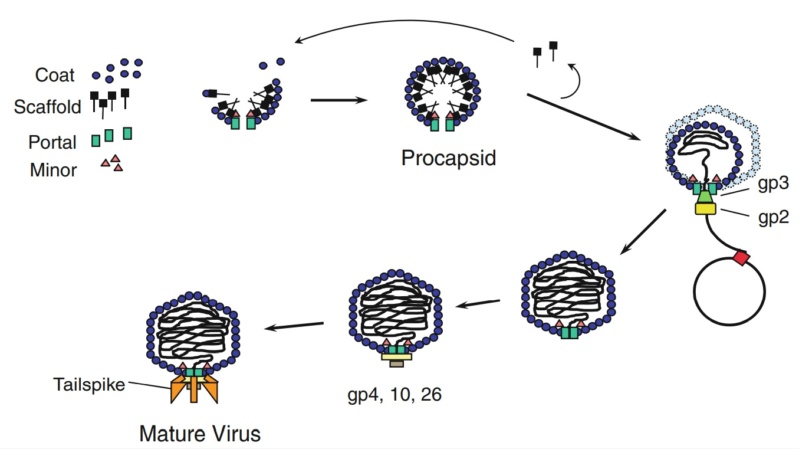
The assembly Pathway of bacteriophages
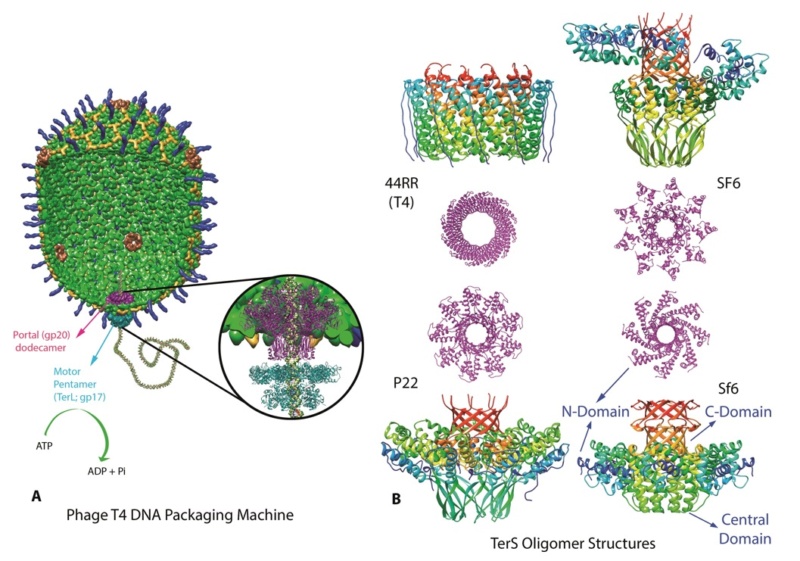
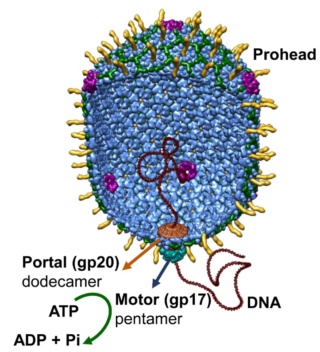
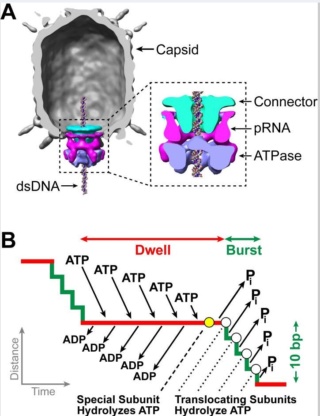
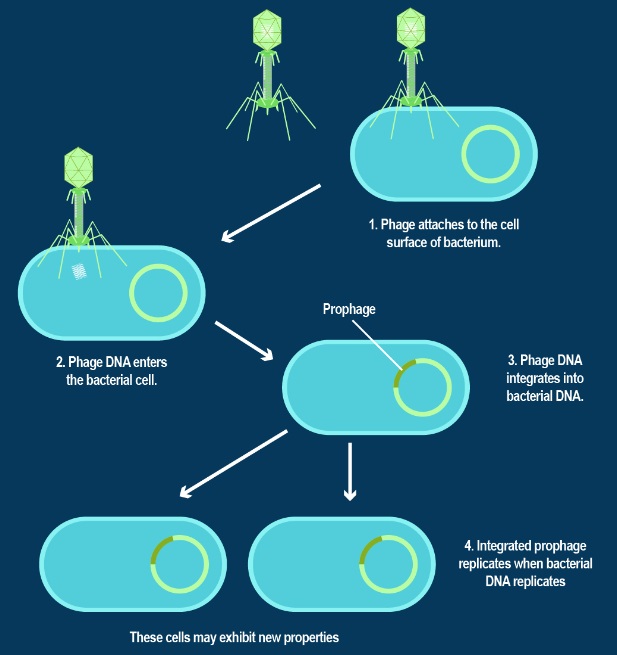
The amazing bacteriophage DNA packaging motor
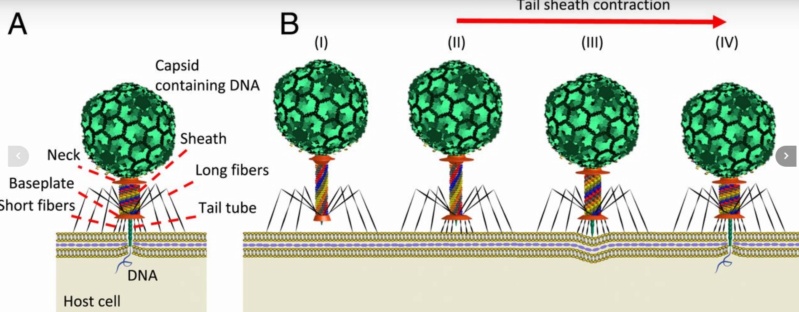
The bacteriophage DNA injection machine, and cell-puncturing device
INFECTION PROCESS OF BACTERIOPHAGE T4 BASED ON THE STRUCTURE
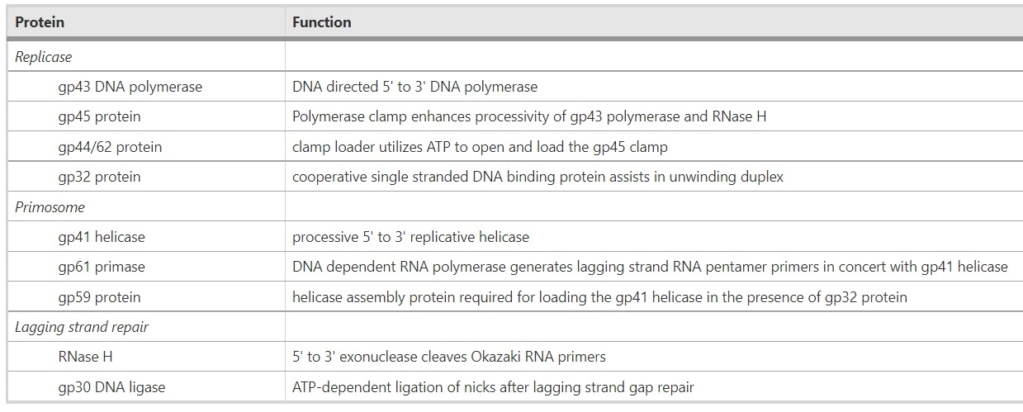
Proteins of the bacteriophage T4
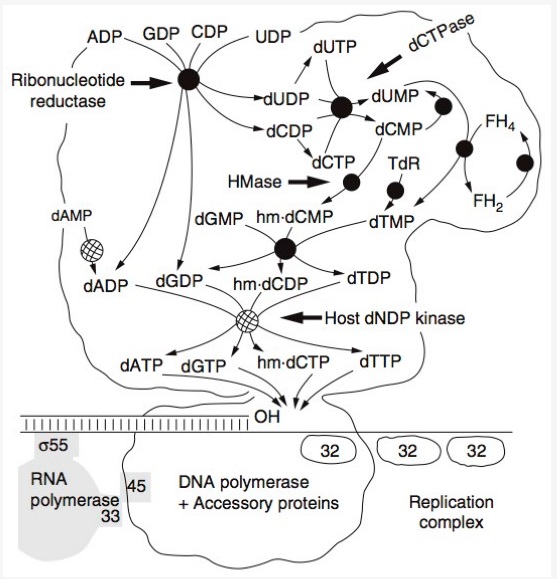
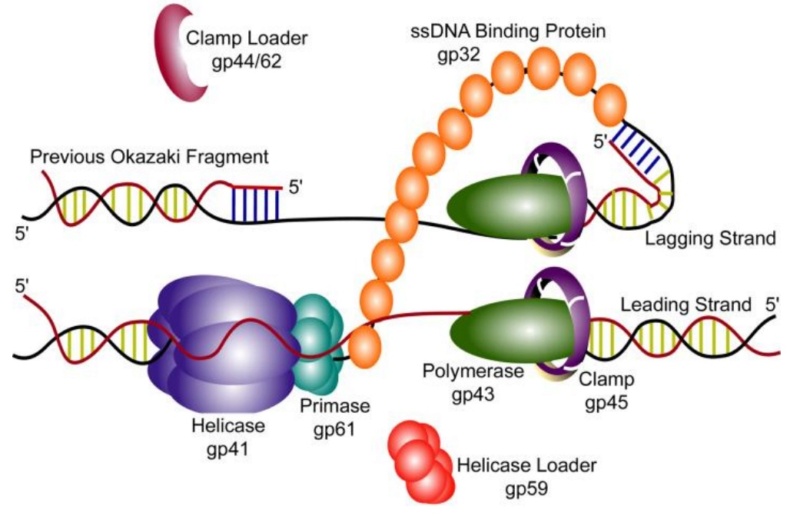
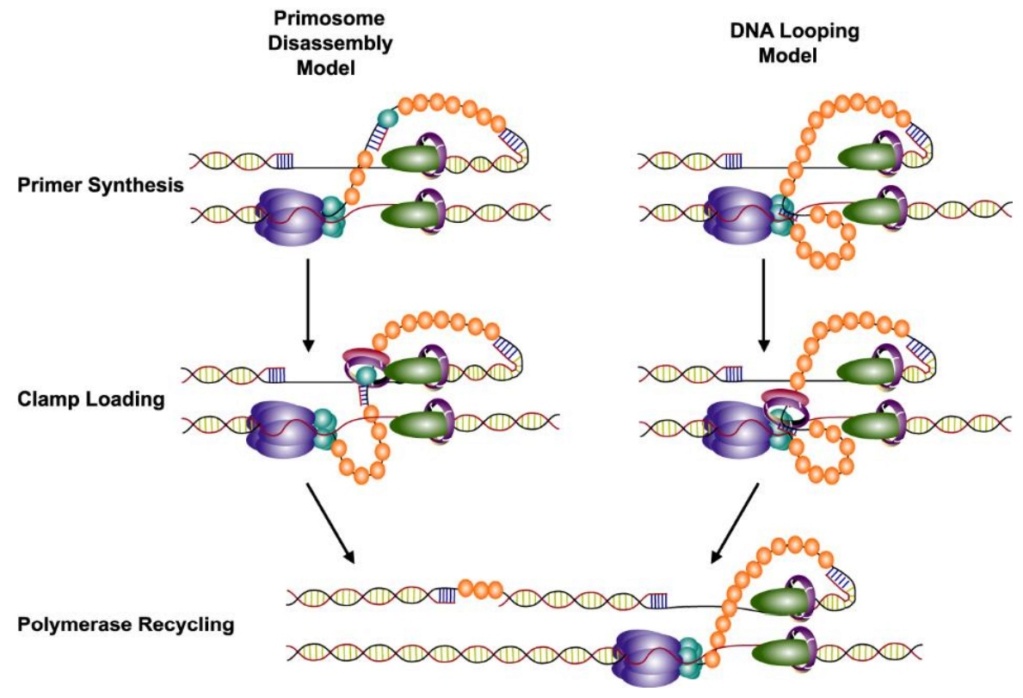
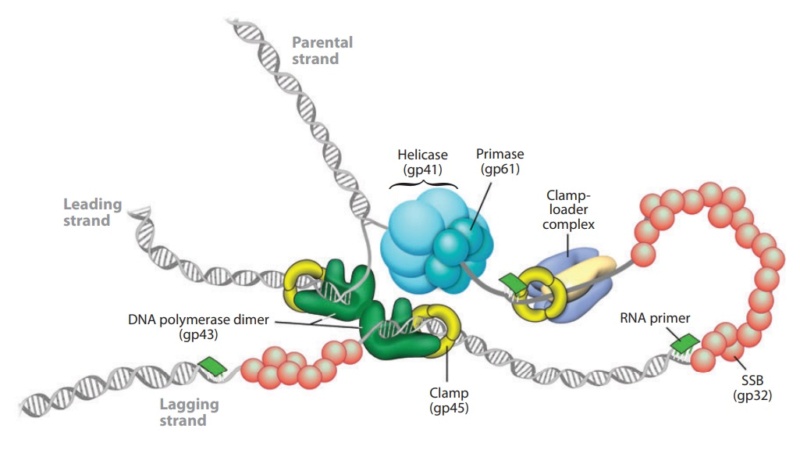
DNA replication by the bacteriophage T4 replisome
Michael Rossmann, Purdue - T4 Bacteriophage Assembly
https://vimeo.com/10700469
Michael Rossman, Purdue - T4 Bacteriophage Infection
https://vimeo.com/10701736
Fly Through Bacteriophage T4
https://vimeo.com/167273755
Introduction
The T4 phage, acts like a spring-loaded syringe and looks like something out of an industrial parts catalog. It can stick to a bacterium, punch a hole, and inject viral DNA (yes, even bacteria suffer infections). Like a conqueror seizing factories to build more tanks, this DNA then directs the cell’s machines to build more viral DNA and syringes. Like all organisms, these viruses exist because they are fairly stable and are good at getting copies of themselves made. Whether in cells or not, nanomachines obey the universal laws of nature. Ordinary chemical bonds hold their atoms together, and ordinary chemical reactions (guided by other nanomachines) assemble them. Protein molecules can even join to form machines without special help, driven only by thermal agitation and chemical forces. By mixing viral proteins (and the DNA they serve) in a test tube, molecular biologists have assembled working T4 viruses. This ability is surprising: imagine putting automotive parts in a large box, shaking it, and finding an assembled car when you look inside! Yet the T4 virus is but one of many self-assembling structures .
Comment: Imagine what it would take for protein engineers to produce nanomachines that would need no nano arms and nano hands to assemble complex nanomachines, but parts of these machines that would be able to assemble on their own just by shaking them, like motors, bearings, and moving parts coming together randomly, and then self-assemble into a fully operational nano-machine. The engineers would need to know the single individual forces and how they would interact with other forces from the other parts. The problem becomes even more apparent when we consider that one of the forces that influence proteins are for example Van der Waals forces which operate based on quantum mechanical principles. R. W. Newberry (2019): The dominant contributors to protein folding include the hydrophobic effect and conventional hydrogen bonding, along with Coulombic interactions and van der Waals interactions. What a feat would THAT be!
Frederick William Twort FRS was an English bacteriologist and was the original discoverer in 1915 of bacteriophages. 40% of all bacteria in the oceans are killed by bacteriophages, every single day.
M. YANAGIDA (1984): The virus particle contains a double-stranded DNA molecule of 170 X 10^3 base pairs and more than 3,000 protein subunits of some 30 polypeptide species. As a virus, phage T4 has two fundamental attributes in common with cells or higher forms of life: a definite architecture and the ability to replicate that architecture according to the genetic instructions encoded in molecules of DNA. Experimental results may give new insight into design principles underlying the large and complex bacteriophage T4 head. 27
F.Arisaka (2005): Bacteriophage is an elaborate molecular machine that carries its genomic DNA and efficiently injects it into bacteria. It has a complicated assembly mechanism, where proteins as scaffolding proteins and cleavage of polypeptide bonds in some cases are involved. T4 phage belongs to a family, Caudovirales, which designates a group of phages that has a tail. More than 95% of phages have tails, and possession of the tail is unique to bacteriophages. Bacteria as single cell organisms have a much more strongly constructed membrane structure than eukaryotic cells. For example, E. coli, a gram-negative bacterium, has triple-layered cell membranes; namely, outer membrane, peptidoglycan layer, and inner membrane. Phages have such a complex structure as a tail to invade the tough barrier of the host cells. 24

Vergote (2018): The virus bacteriophage T4 resembles the Lunar Lander that was used in the 70’s by the Apollo space program. It has a landing system, duplicates of one protein in the head and a tail used to pass that DNA to infect bacteria. If you are looking for the best design, nature is the perfect place to start. 20
Is it a coincidence? The Bacteriophage T4 consists of a capsid shell, a head where it stores and protects its genome and a syringe-like structure used to insert the DNA into a host. The tail terminates with a multiprotein baseplate that changes its conformation from a “high-energy” dome-shaped to a “low-energy” star-shaped structure during infection. It also has an ultrafast DNA packaging motor to translocate or pack, long stretches of the virus's genetic material into its capsid shell.
Bacteriophages are molecular machines – created for one reason – to kill bacteria – to control bacterial species population.
Eric S Miller (2003): T4 bacteriophages constitute a beautifully integrated system of biological machines and networks 13
Eric S Miller (2010): Phage T4 is one of the most extensively investigated viruses and has been the central focus of several monographs and reviews over the last 25 years. The T4 biological system is amenable to investigation by genetic, phylogenetic, biochemical, biophysical, structural, computational, and other tools. 12
A. Roberts (2015): Phage help maintain microbial diversity and balance within Earth’s biosphere. Phage are thought to turn over 20–50 percent of the biomass in Earth’s oceans daily! In the absence of these microbial predators it is hard to imagine how our planet would ever sustain life beyond mere microbes. The planet would be covered with microbial competition specialists, sequestering all of Earth’s resources necessary for advanced life. If not for bacterial predation via phage, bacteria would certainly dominate life to the exclusion of advanced organisms. 2
Vincent R. Racaniello (2015): There are more than 10^30 bacteriophage particles in the world’s oceans, enough to extend out into space for 200 million light-years if arranged head to tail. The average human body contains approximately 10^13 cells, but these are outnumbered 10-fold by bacteria and as much as 100-fold by virus particles. 14
Nicola Twilley (2015): There are an estimated 10^31—ten million trillion trillion—phages on Earth, more than every other organism, including bacteria, put together. According to researchers in Vancouver, these tiny viruses cause a collective trillion trillion successful infections per second, in the process destroying up to forty percent of all bacterial cells in the ocean every single day. Following their deaths at the hands of phages, those carbon-containing microorganisms sink down into the marine sediment, effectively removing greenhouse gases from circulation. Anything that bacteria do, from breaking down the carcasses of dead animals to converting atmospheric nitrogen into plant food, is at the mercy of the phages that infect, kill, or otherwise transform them. Phages are the puppet masters; they insure that essential biochemical processes run smoothly. 11
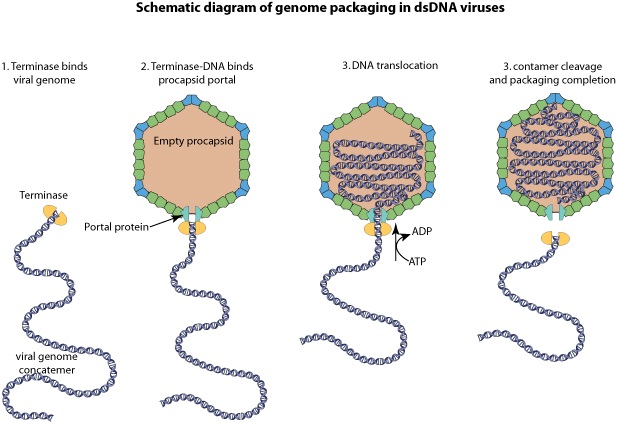
J.Sarfati (2008): Viruses are particles so tiny that they can’t be seen by an ordinary light microscope, but only under an electron microscope. Viruses come in many different sizes, shapes and designs, and they operate in diverse ways. They are composed of DNA (or RNA in the case of RNA viruses, including retroviruses) and protein. They are not living organisms because they cannot carry out the necessary internal metabolism to sustain life, nor can they reproduce themselves. They are biologically inert until they enter into host cells. Then they start to propagate using host cellular resources. The infected cell produces multiple copies of the virus, then often bursts to release the new viruses so the cycle can repeat. One of the most common types is the bacteriophage (or simply ‘phage’) which infects bacteria. It consists of an infectious tailpiece made of protein, and a head capsule (capsid) made of protein and containing DNA packaged at such high pressure that when released, the pressure forces the DNA into the infected host cell. How does the virus manage to assemble this long information molecule at high pressure inside such a small package, especially when the negatively charged phosphate groups repel each other? It has a special packaging motor, more powerful than any molecular motor yet discovered, even those in muscles.The genome is about 1,000 times longer than the diameter of the virus. It is the equivalent of reeling in and packing 100 yards of fishing line into a coffee cup, but the virus is able to package its DNA in under five minutes.This motor exerts a force twice as powerful as a car engine. So the motor, a terminase enzyme complex, ‘can capture and begin packaging a target DNA molecule within a few seconds.’ Such a powerful motor must use a lot of energy, and in one second, this one goes through over 300 units of life’s energy currency ATP. The virus has a complementary motor-enzyme, ATPase, built into its packaging engine, to release the energy of the ATP. And not only is the packing motor powerful, it can change its speed as if it had gears. The researchers say that this is important, because the DNA fed to it from the cell is likely not a straightforward untangled thread. Just as it is good for a car to have brakes and gears, rather than only being able to go 60 miles per hour, the DNA-packaging motor may need to slow down, or stop and wait if it encounters an obstruction. It may permit DNA repair, transcription or recombination— the swapping of bits of DNA to enhance genetic diversity—to take place before the genetic material is packaged within the viral capsid. 4
Joseph W. Francis (2003): Microbes and viruses perform essential roles in all ecosystems of the biosphere. Microbes and viruses perform many beneficial activities in ecosystems and in symbiotic partnerships with all biological organisms. I propose that microbes were created as an organosubstrate; a link between macro-organisms and a chemically rich but inert physical environment, to provide a substrate upon which multicellular creatures can thrive and persist in intricately designed ecosystems. Viewed in this context microbes and viruses could also be thought of as a single, complex, massive, multicellular, multitaxon organism with incredible and powerful life supporting properties. Many microbes live on and within living organisms. It is estimated that the number of microbes living on the human body far exceeds the 70 trillion human cells that comprise it. The discipline of microbial ecology is increasingly revealing that microbial and viral symbionts play vitally important roles within organisms and ecosystems. In fact, axenic (germ-free life) probably does not exist in nature; all animal species with the exception of prenatal life are thought to live with microbial symbionts. A tremendous number of symbiotic relationships are being discovered. Many of these relationships involve complex lifestyles and anatomies that appear to be designed to foster the symbiotic lifestyle. A general survey of symbiotic relationships also shows that the most common functions provided by symbionts involve nutritional support, protection and reproduction/population control.
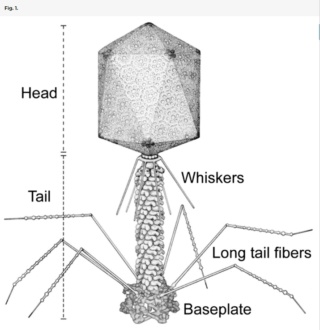
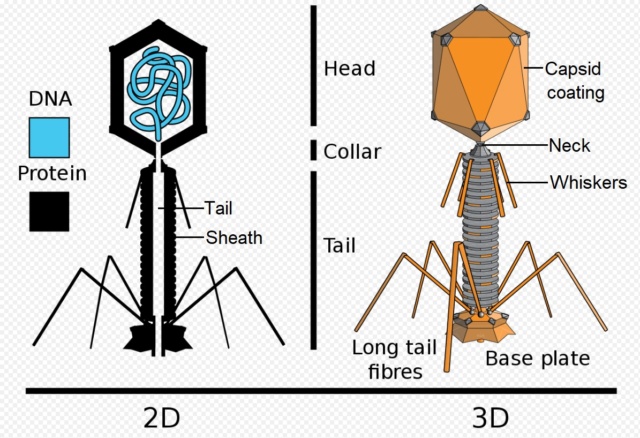
Structure of the bacteriophage T4
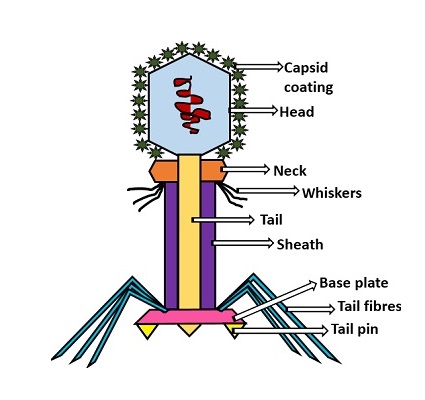
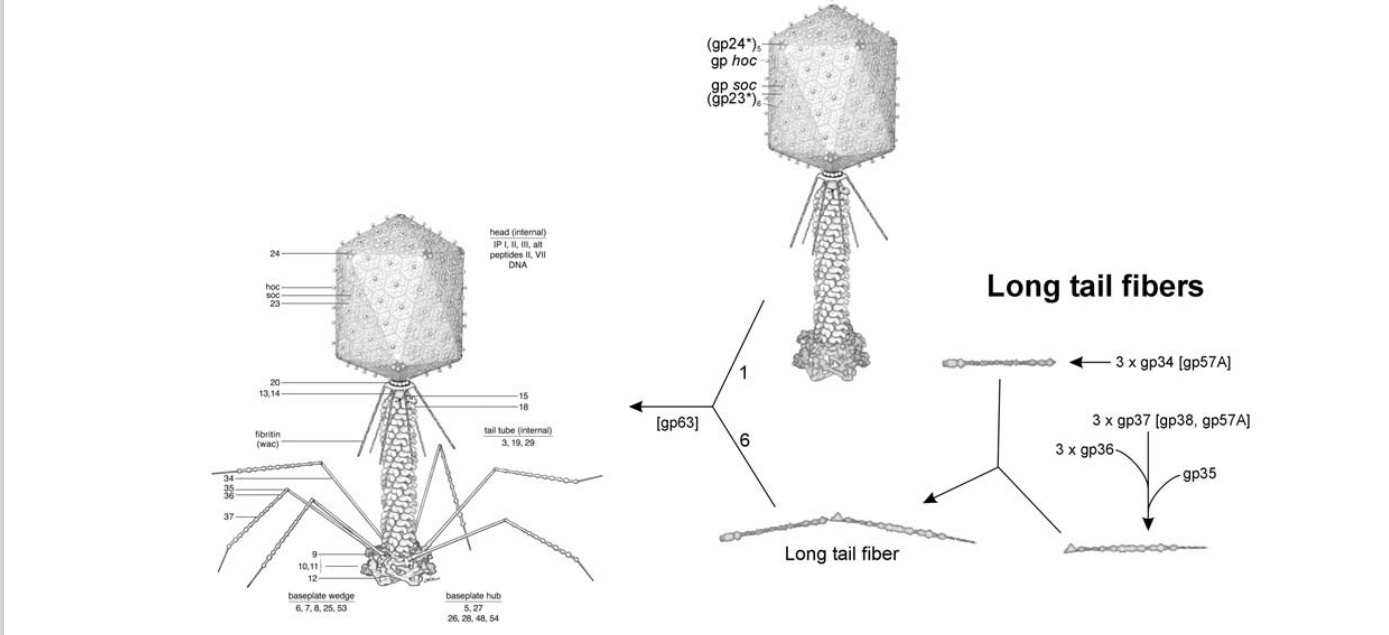
Head: It is elongated and hexagonal in shape. Possesses a prismoid structure. It is surrounded by an envelope called a capsid.
Capsid: It is produced by identical protein subunits called capsomeres. It contains around 2000 capsomeres.
Genetic material: It is 50 nm long and can be either DNA or RNA. The structure of genetic material can be linear or circular. It is tightly packed inside the head.
Neck: It is also called a collar, which connects the head and tail. It possesses a circular plate-like structure.
Tail: It resembles a hollow tube. A tail is surrounded by a protein sheath.
Sheath: It is composed of around 144 protein subunits. The sheath of the bacteriophage is highly contractile. It contains 24 rings.
Base plate: It is hexagonal in shape. The base plate is present at a distal end.
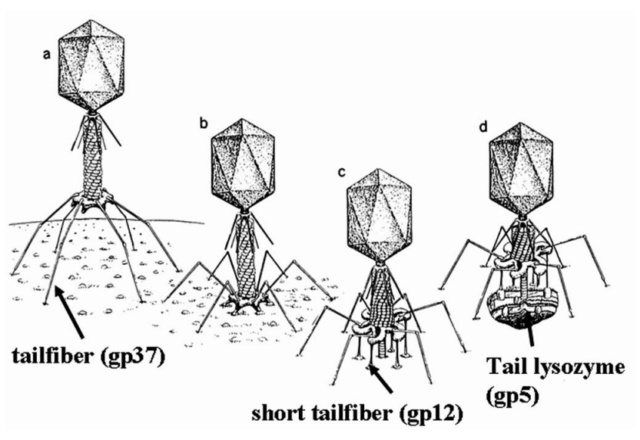
Tail fibers: These are attached to the base plate. It appears long and thread-like filaments. Tail fibers induce host specificity, or they are host-specific. They are generally found 6 in number. Size: 130x2nm
Spikes: It is also called a tail pin. Spikes recognize the receptor sites of the host cell.
Michael Rossmann, Purdue - T4 Bacteriophage Assembly
https://vimeo.com/10700469
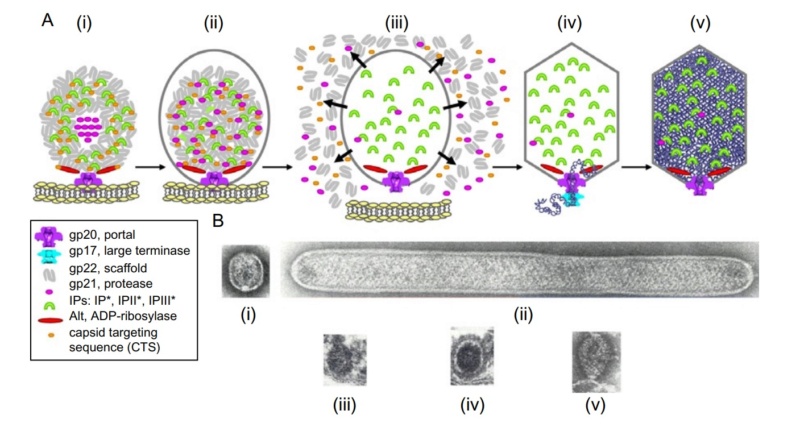
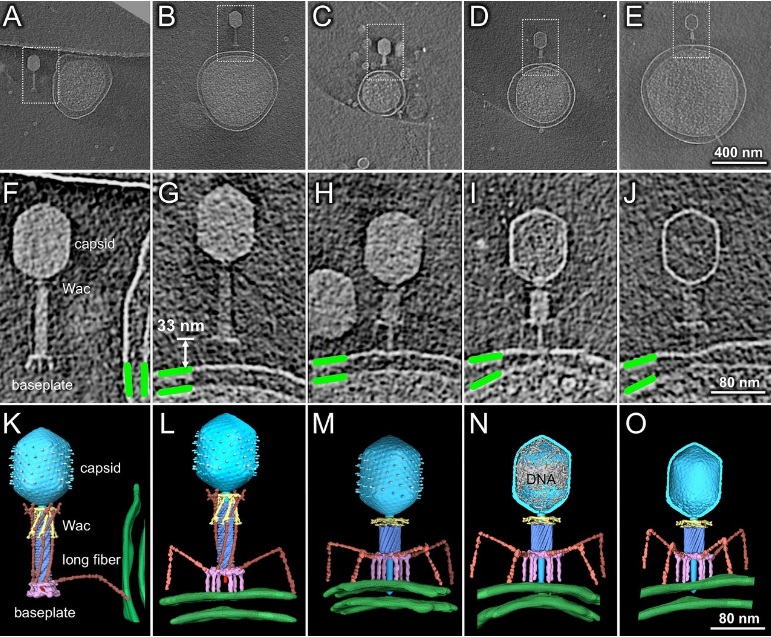
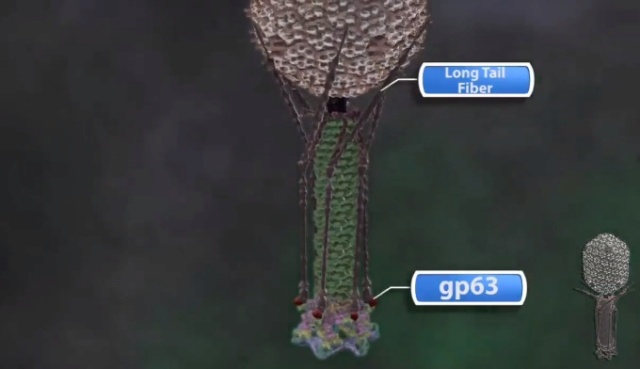
Bo Hu (2015): The T4 virion comprises a capsid containing a 170-kb dsDNA genome, a collar region that displays short whiskers called whisker antigen control (Wac) (also known as fibritin), a contractile tail with a complex baseplate that harbors short tail fibers (STFs), and a set of side or long tail fibers (LTFs) (Fig. 1D). After recognition of a host, the tail transmits a signal to the head for genome ejection and provides the channel through which the DNA moves. 28
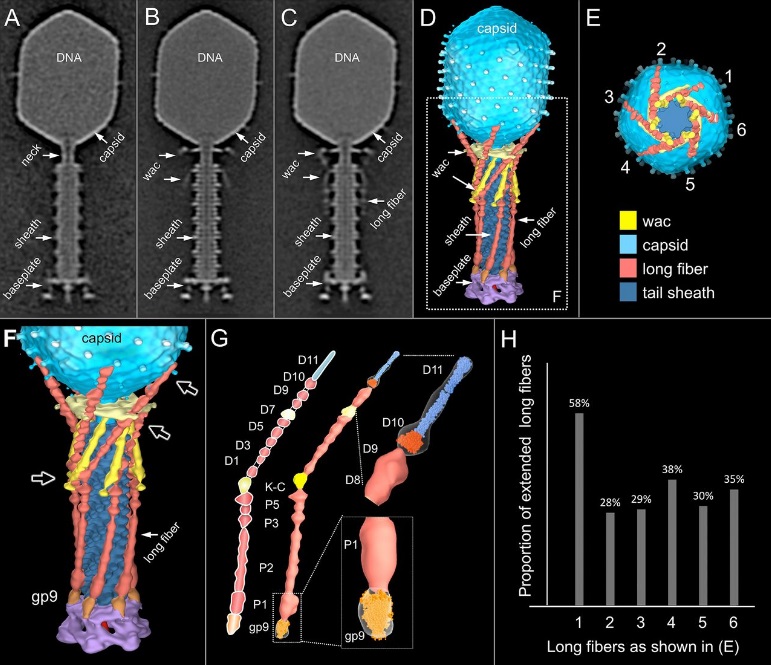
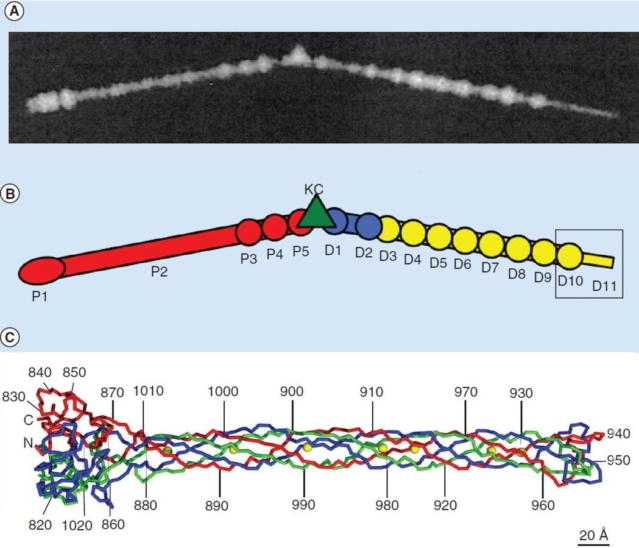
Comparative structural analysis of infective T4 virions.
Asymmetric 3D reconstructions are shown as central slices of Wac-minus
(A), fiberless (X4E mutant)
(B), and WT
(C) virions. In the absence of Wac, LTFs are seen only as short stubs emanating from the baseplate due to their flexibility when extended. A 3D reconstruction of WT T4
(D), enlarged in F, highlighting the six (orange) LTFs attached (open arrows) to the whiskers and collar of the sixfold symmetrical Wac (yellow) and the fivefold symmetrical capsid vertex.
(G) Domain structure of an LTF (4) aligned with our cryo-ET reconstruction. For clarity, not all subdomains are named on the figure. Subdomains P1–P5 correspond to a trimer of gp34, K-C to gp35, and part of gp36, and subdomains D1–D11 in the distal half-fiber are comprised of trimeric gp36 and gp37. Crystal structures of gp9 (PDB ID code 1S2E) and the gp37 fiber tip (PDB ID code 2XGF) are fitted into the density map. Classification of each LTF reveals the symmetry mismatch viewed along the tail sheath axis, where fiber 1 is set to interact with a capsid edge
(H) Classification was also used to evaluate the density of each LTF and thus to estimate the distribution of extended fibers on a WT virion. Also, see Figs. S1–S3.
Hari charan (2020): This overall structure is necessary the way the phages deliver their payload of genetic material into bacteria. Once on the surface of a bacterium, the tube portion contracts, and the phage acts like microscopic hypodermic needle, literally injecting the genetic material into the bacterium.
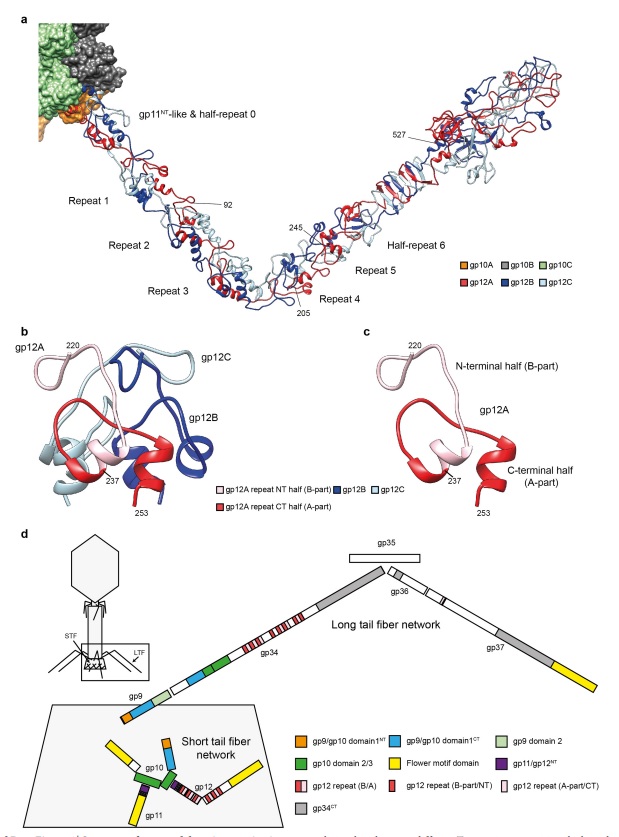
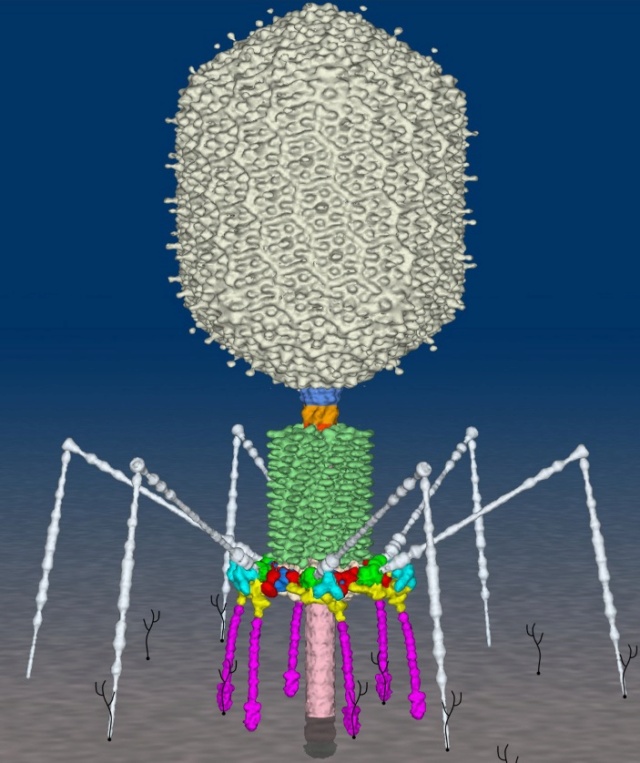
P. G. Leiman (2003): Bacteriophage T4 is one of the most complex viruses. More than 40 different proteins form the mature virion, which consists of a protein shell encapsidating a 172-kbp double-stranded genomic DNA, a ‘tail,’ and fibers, attached to the distal end of the tail. The fibers and the tail carry the host cell recognition sensors and are required for attachment of the phage to the cell surface. The tail also serves as a channel for delivery of the phage DNA from the head into the host cell cytoplasm. The tail is attached to the unique ‘portal’ vertex of the head through which the phage DNA is packaged during head assembly. Similar to other phages, and also herpes viruses, the unique vertex is occupied by a dodecameric portal protein, which is involved in DNA packaging. Bacteriophage T4 is a double-stranded DNA (dsDNA) tailed virus that infects Escherichia coli (figure below).

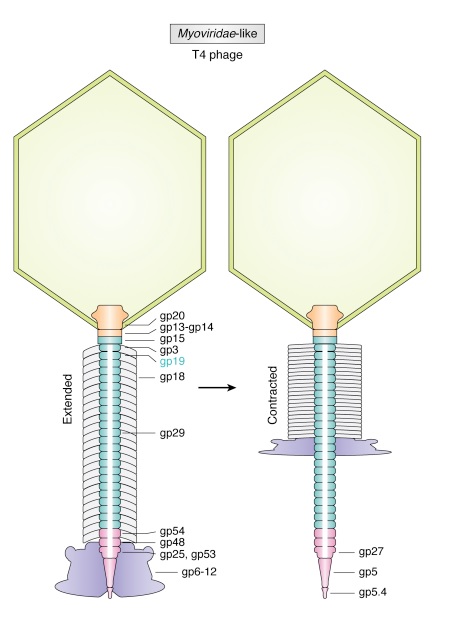
Structure of bacteriophage T4. The proteins comprising the virion are labeled with their corresponding gene number or name.
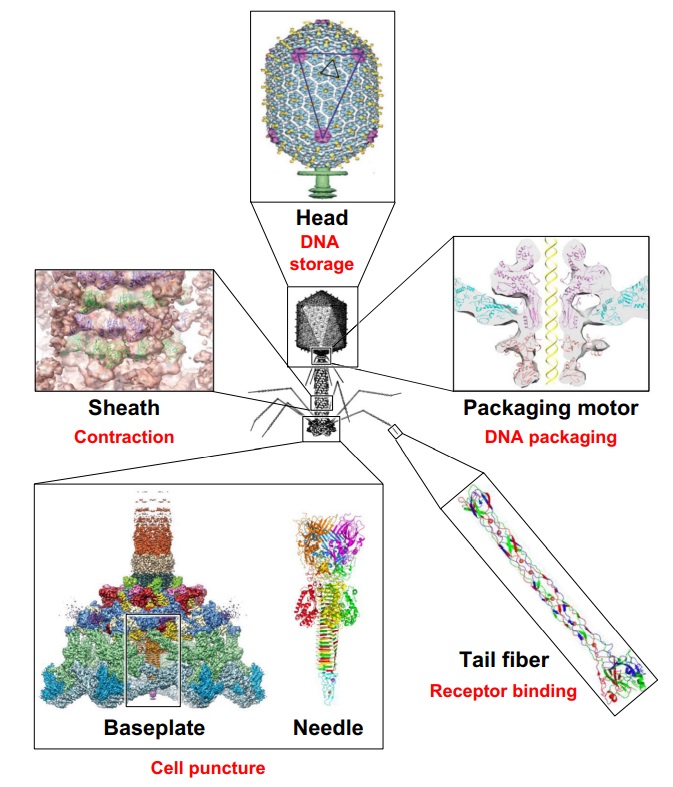
Bacteriophage T4 with detailed structures of the head, the packaging motor, the sheath, the tail fiber and the baseplate with the needle
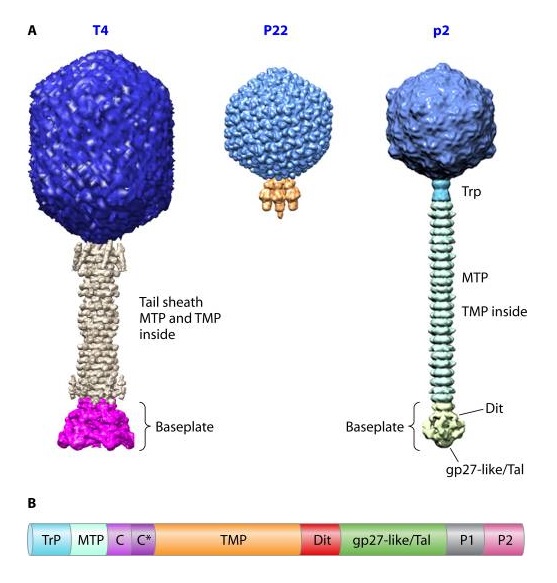
It is one of the most complex viruses, with a genome that contains 274 open reading frames out of which more than 40 encode structural proteins. The mature virus, or ‘virion,’ consists of a prolate head with hemiicosahedral ends encapsidating the genomic DNA; a cocylindrical contractile tail, terminated with a baseplate; and six fibers attached to the baseplate. The head, tail, and fibers assemble via independent ordered pathways and join together to form a mature virus particle. Unlike animal viruses, infection of host cells by tailed bacteriophages is highly efficient – only one bacteriophage T4 particle is required, in general, to infect a host cell. Upon infection, the phage shuts down host-specific nucleic acid and protein syntheses, thus ensuring production of only its own components in amounts sufficient to assemble up to 200 progeny virus particles per infected cell. The efficiency of the infection process and the large genome of bacteriophage T4, in which only half of the genes are necessary for proliferation on E. coli, contribute to the diversity of the phages from the T4-like family, a subgroup of Myoviridae. These phages propagate on a wide range of bacterial hosts that grow in diverse environments.
The amazing bacteriophage DNA packaging motor
The bacteriophage DNA injection machine
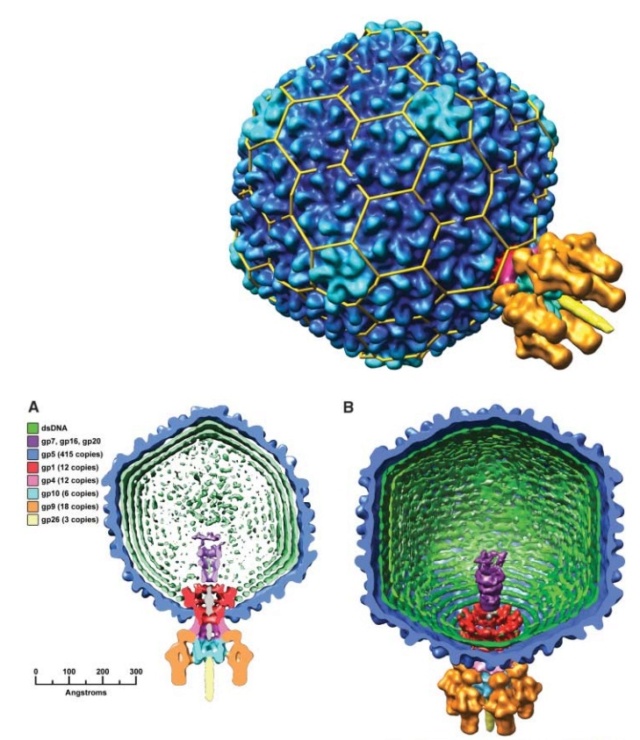
Bacteriophage T4 head structure
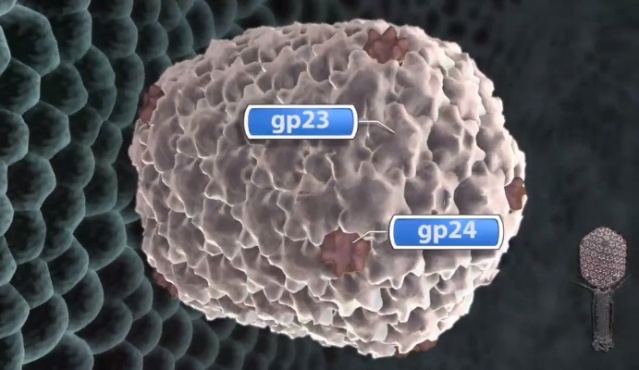
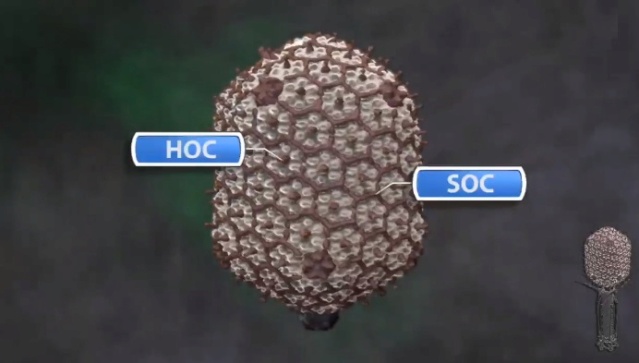
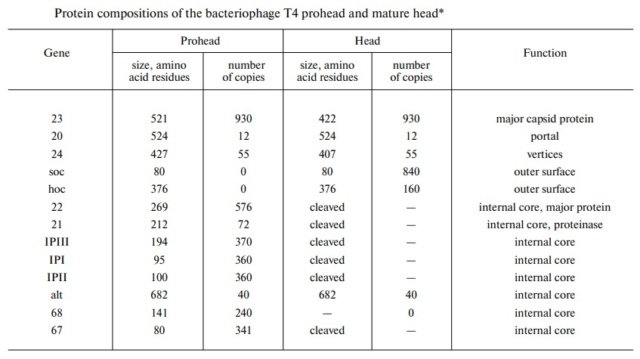
M.YANAGIDA (1984): There are three classes of lattice proteins, namely, the major coat protein (gp23), soc, and hoe. Six molecules of gp23 and one molecule of hoe produce a hexamer and a center unit, respectively, Six molecules of soc form six bridges (as ellipsoidal particles in Fig. 2). Thus the repeating unit of the head lattice consists of "hexamer+a center unit+six bridges," or type "(6+ 1)+6". This model contains 840 subunits for hexamers, 60 subunits for pentamers (at vertices), 900 bridges, and 140 center units, that is, 900 gp23, 900 soc, and 140 hoc molecules. About 1,000 gp23, 1,000 soc, and 150 hoc were reported to be present in the capsid from chemical analysis, in agreement with the ultrastructural analysis. 27
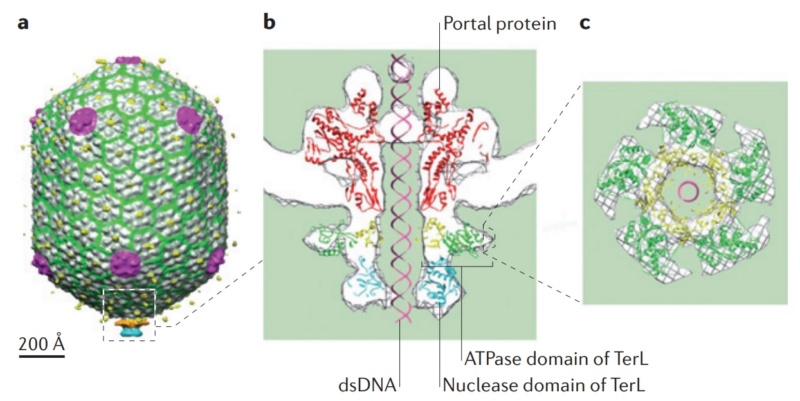
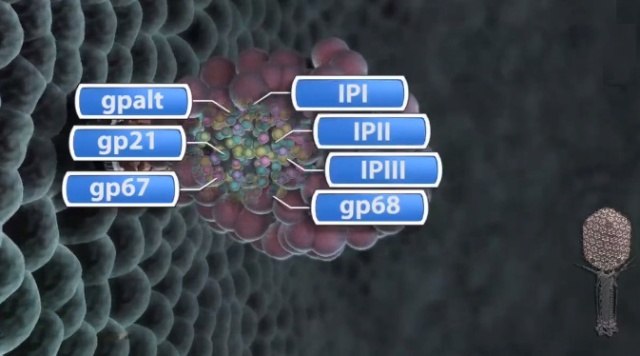
The head of bacteriophage T4 is composed of more than 3000 polypeptide chains of at least 12 kinds of protein and a 172-kbp dsDNA chromosome, which comprises 102% of the unique region of about 169 kbp.. The shell has icosahedral ends and a cylindrical equatorial midsection with a unique portal vertex where the phage tail is attached. 9
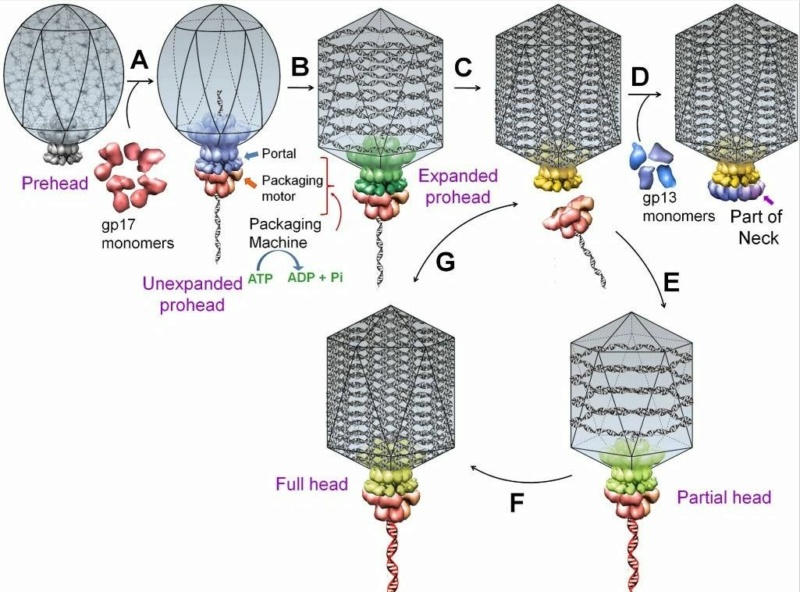
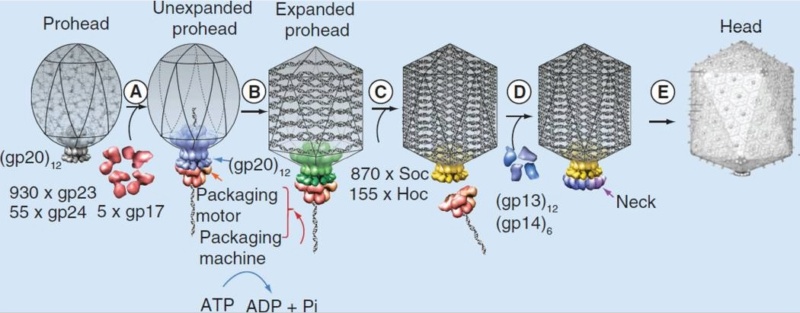
Moh Lan Yap (2015): The mature head encapsidates 172 kbp dsDNA. The head is first assembled as an empty capsid that is subsequently packaged with DNA by an ATP-dependent packaging machine. This machine binds to the same special pentameric vertex that is later occupied by the phage tail. The head is prolate, meaning that it has two icosahedral ends and a cylindrical mid-section. The geometrical organization (expressed as triangulation numbers) of the ends and mid-section are based on planar hexagonal grids. The capsid is composed of 930 post-translationally modified monomers, or 155 hexamers of the major protein, gene product 23 (gp23*, where the * signifies post-translational cleavage). The presence of proteins, homologous to the major capsid protein, which form pentamers as opposed to hexamers is a frequent solution to the formation of the pentameric vertices in icosahedral viruses. The portal protein has multiple roles. It initiates head assembly, genome packaging and serves as the genome gatekeeper to prevent leakage of the packaged DNA. Two accessory proteins, Hoc (highly antigenic outer capsid protein) and Soc (small outer capsid protein) attach to the capsid surface (Figure below).
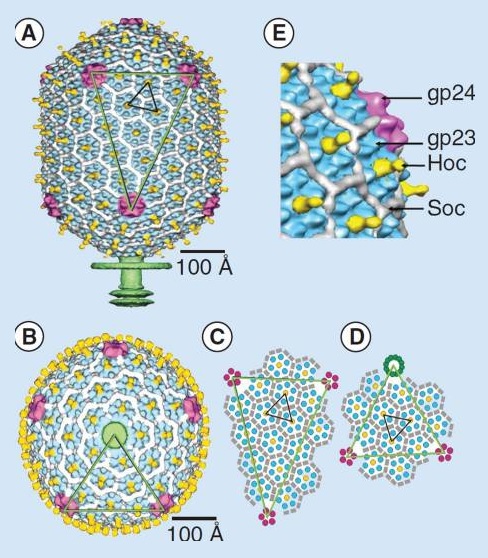
The structure of the bacteriophage T4 head
The symmetry of the gp23* major capsid protein shell is characterized by triangulation numbers Tend = 13 laevo and Tmid = 20. The facet triangles are shown in green and the basic triangles are shown in black.
(A) Shaded surface representation of the cryo-electron microscopy reconstruction viewed perpendicular to the fivefold axis. gp23* is shown in blue, gp24* is in magenta, Soc is in white, Hoc is in yellow and the tail is in green.
(B) View of the reconstruction along the fivefold axis with the portal vertex toward the observer; the tail has been cut away at the level shown by the black arrow in (A). Proteins are colored as described for (A).
(C) Schematic representation of the distribution of proteins in the elongated midsection facet.
(D) Schematic representation of an end-cap facet. Proteins are colored as described for (A) except the Soc molecules are shown as gray rectangles. (E) A closer look of the distribution of proteins on the head.
The rod-shaped Soc binds between two gp23* hexamers, thus forming a continuous mesh surrounding the hexameric gp23* on the capsid. Soc maintains the stability of the head under extreme environments. Hoc is an elongated molecule protruding from the center of gp23* hexamers. Its Ig-like domains, exposed on the outer surface of the head, may provide survival advantages to the phage. 21
V. V. Mesyanzhinov (2004): The head of phage T4, or capsid, is a prolate icosahedron elongated along a fivefold axis and is composed of more than 1500 protein subunits encoded by at least twelve genes (Table 1).In total, the mature T4 capsid contains 930 subunits of gp23* (* indicates a protein proteolytically processed during capsid maturation) and 55 subunits of gp24*. Pentamers of gp24* occupy eleven vertices of the icosahedron, and gp20 forms a unique portal vertex required for DNA packaging and subsequent attachment of the tail. The T4 capsid shell is decorated on the outside with gphoc (highly antigenic outer capsid protein) and gpsoc (small outer capsid protein). The latter two proteins enhance head stability. 23
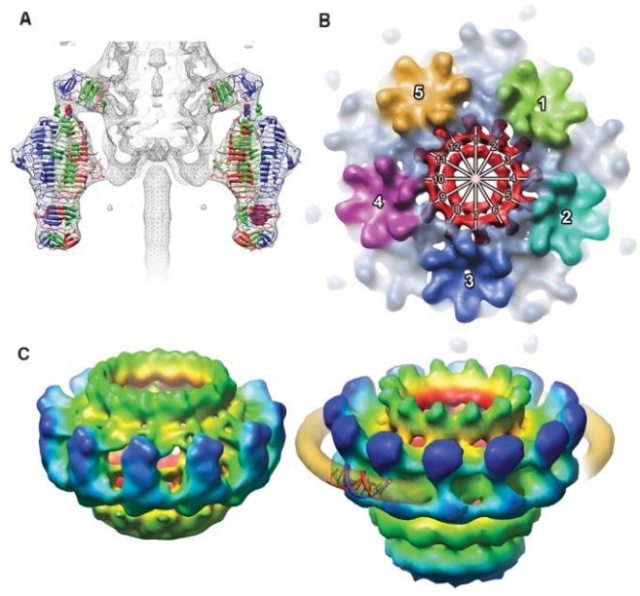
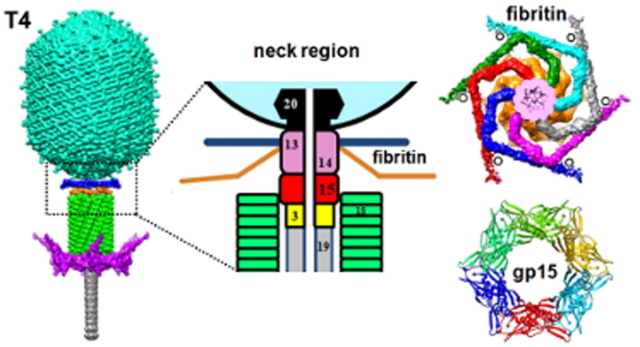
The Molecular Architecture of the Bacteriophage T4 Neck
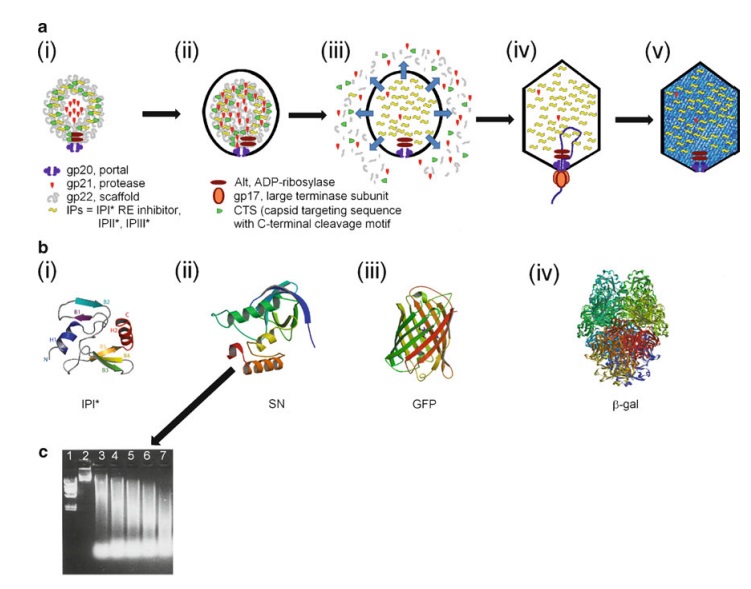
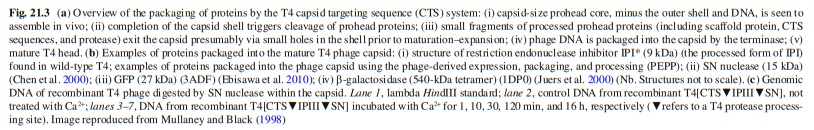
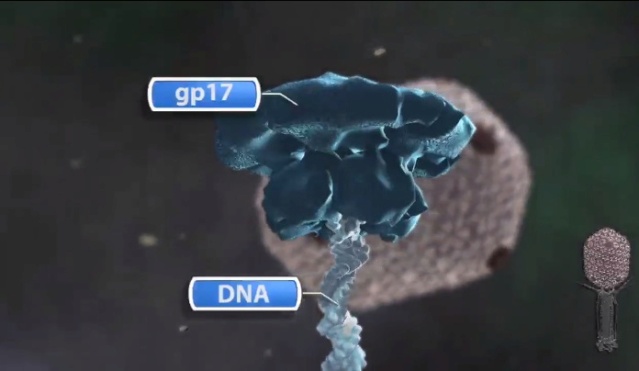
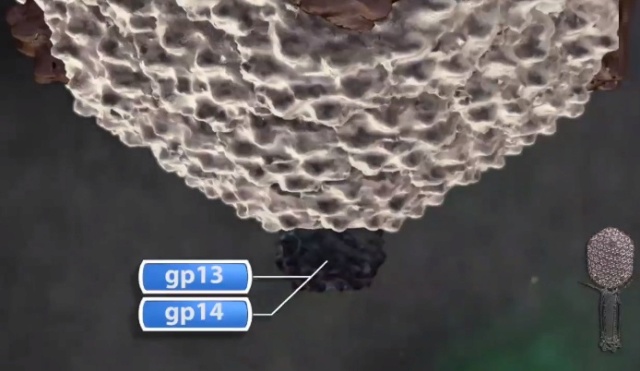
Andrei Fokine et.al. (2013): The T4 head and tail are assembled via independent pathways. Assembly of the T4 head is a complex process that includes a number of intermediate stages. The head assembly is initiated by the dodecamer (A dodecamer (protein) is a protein complex with 12 protein subunits.) of the portal protein, gp20 (gp, gene product). First, a head precursor, called the prohead, is assembled, which is subsequently processed by a scaffold-associated protease. Then the phage genomic DNA is packaged into the capsid through the portal vertex by an ATP-driven motor composed of five gp17 molecules. Upon completion of the DNA packaging, the head assembly is finalized by attachment of several copies of the gp13 and gp14 proteins to the portal vertex. Monomers of gp13 and gp14 have a size of 309 and 256 amino acid (aa) residues, respectively. The gp13–gp14 complex seals the portal vertex and creates a site for attachment of the independently assembled tail. Mutant phages lacking these proteins produce heads that are unable to bind tails and lose their DNA.
The T4 tail assembly begins with the baseplate formation and proceeds with polymerization of the tail tube and the contractile sheath. The tail tube is formed by gp19 molecules (163 aa residues). The length of the tube is controlled by a mechanism involving the “tape-measure protein”, gp29. The elongation of the tail tube is terminated by attachment of the hexamer of the 175-residue tail tube terminator protein, gp3, which binds to the last row of gp19 subunits (probably also to gp29) and stabilizes the tail tube. The T4 tail tube is used as a scaffold for the polymerization of the contractile sheath. The gp18 sheath molecules (659 aa residues) assemble around the tube in the form of a six-start helix. The T4 tail assembly is completed by the hexamer of the tail terminator protein, gp15 (the monomer is 272 aa residues long), which binds to the top† of the tail. Contraction of the tail during infection is associated with a substantial rearrangement of the gp18 subunits and results in shortening of the sheath to less than one-half of its original length. 22
Coat, scaffolding, and portal proteins are encoded by P22 genes 5, 8, and 1, respectively. In the absence of scaffolding protein, P22 coat protein assembles into TZ4 and TZ7 icosahedral shells as well as “spiral” structures, and all of these lack the essential portal protein and at least one protein required for DNA injection. 10
Lei Sun et al., (2015):The portal structure probably dates back to a time when self-replicating microorganisms were being established on Earth. 11
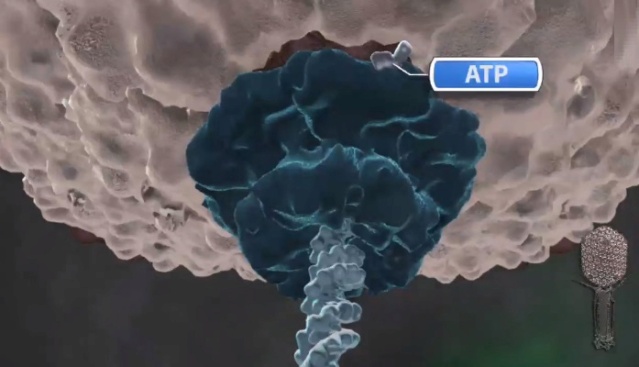
Amy D. Migliori (2014): Recent structural studies of the bacteriophage T4 packaging motor have led to a proposed mechanism wherein the gp17 motor protein translocates DNA by transitioning between extended and compact states, orchestrated by electrostatic interactions between complimentarily charged residues across the interface between the N- and C-terminal subdomains. 2
They are the most numerous biological entity on earth, with an estimated number of 10^31 tailed phages in the biosphere. They are arguably very ancient as a group, with some estimates placing their ancestors before the divergence of the Bacteria from the Archaea and Eukarya

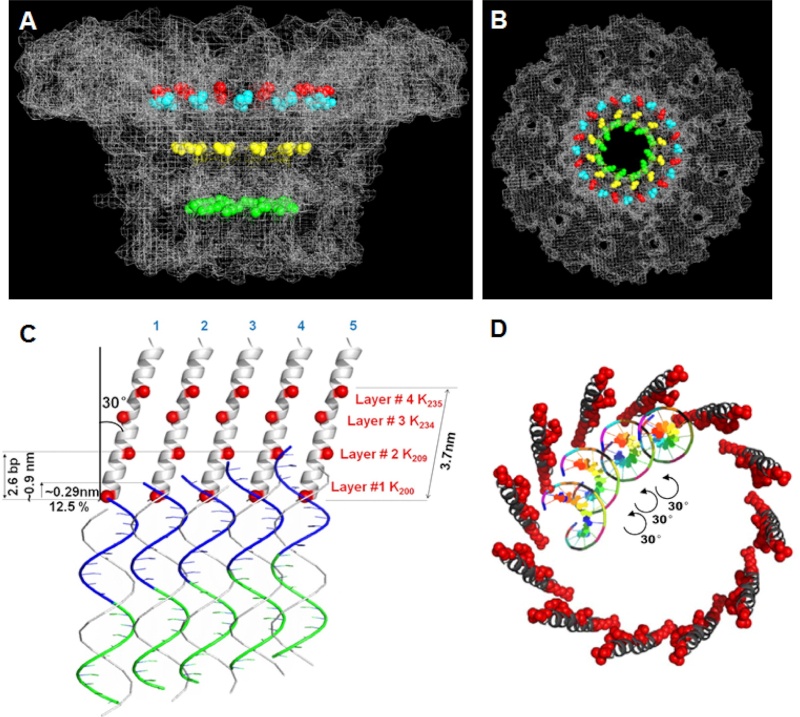
(a) 3D density map of T4 portal protein assembly at 3.6 Å resolution with each subunit color-coded. Shown is the top view (left)
and side view (right).
(b) Ribbon diagram of the gp20 atomic model with each subunit color-coded. Shown is the top view (left)
and side view (right).
(a) Charge distribution on the outer surface of dodecameric gp20. Blue and red colours correspond to 10 kT e− positive and negative potential,
respectively.
(b) Charge distribution on the inner surface of dodecameric gp20. (c) Ribbon drawing of the gp20 monomer structure with each
domain colour-coded.
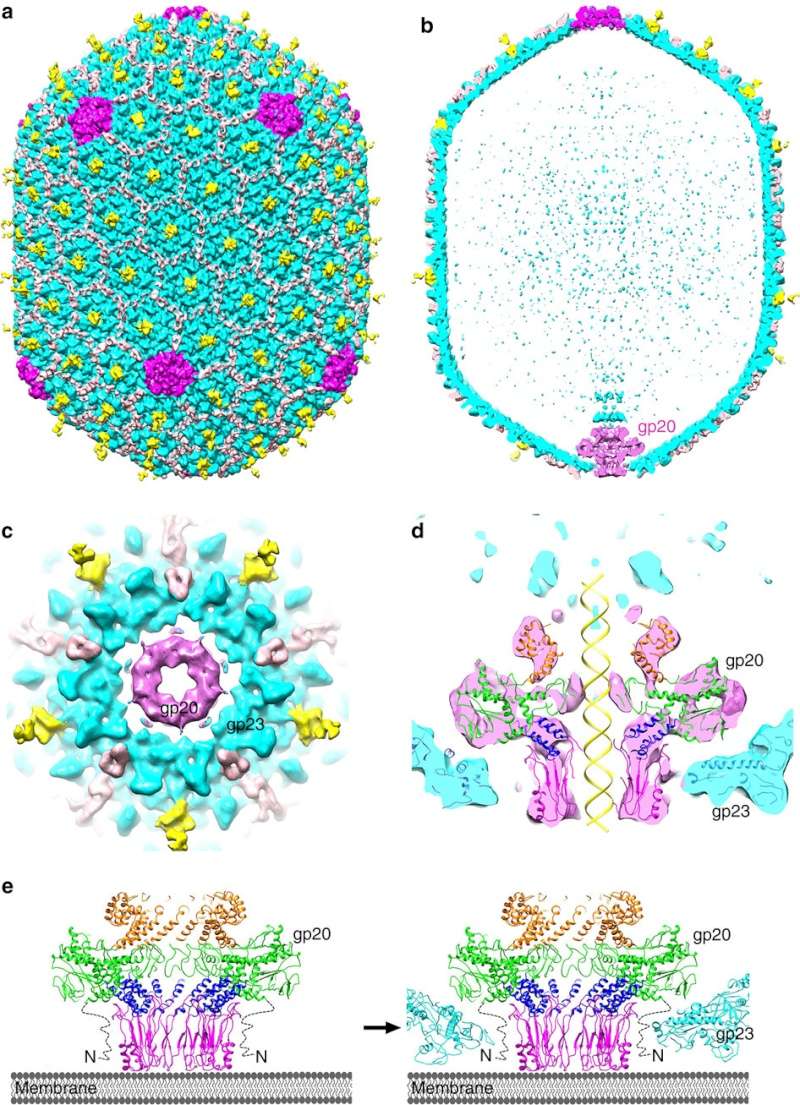
(a,b) Cryo-EM density map of the T4 prolate head (gp23: cyan; gp24:magenta; Soc: pink; Hoc: yellow).
(c) Bottom view of the prolate head, showing the gap between gp20 and the capsid. (d) Fit of the gp20 and gp23
structures into the cryo-EM map of the T4 prolate head. (e) A model of the T4 head assembly. A dodecameric portal is
assembled on the inner membrane of E. coli with the assistance of the phage-coded chaperone gp40 and the E. coli chaperone YidC58.
The portal assembly acts as an initiator for head assembly, leading to co-polymerization of the major capsid protein gp23
and scaffolding proteins.
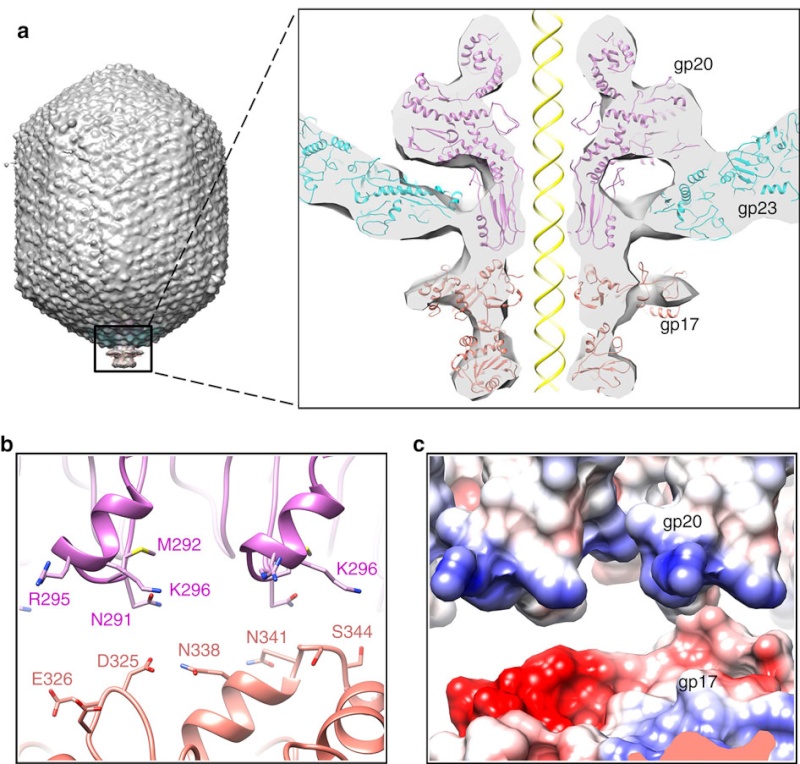
(a) Fitting of the T4 portal protein (purple) and gp17 (tan) into the 35 Å cryo-EM reconstruction of the procapsid+gp17
(EMD-1572 accession number).
(b) Residues involved in the interaction between gp20 (purple) and gp17 (tan) are shown
as sticks.
(c) The surface charge of gp20 and gp17 around the interface area showing electrostatic interactions. The view
orientation is the same as in panel (b).

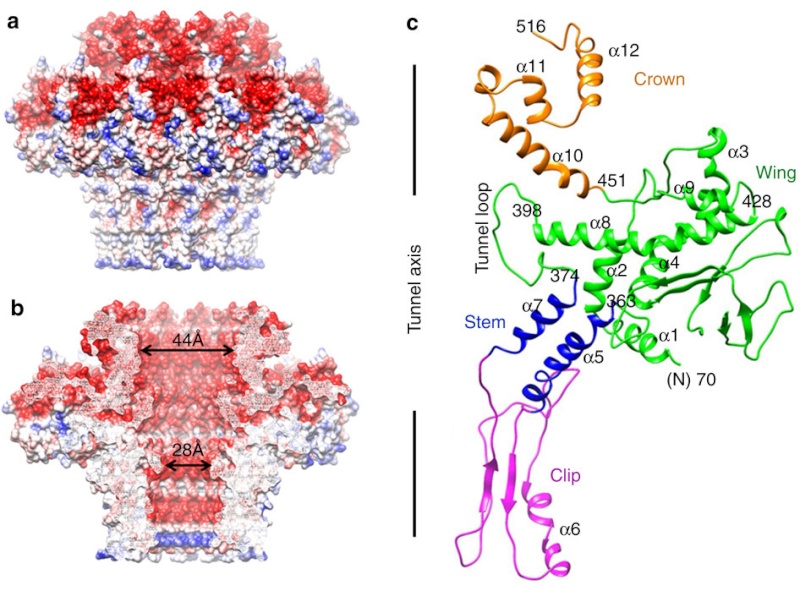
The different portal protein subunits with their wing, stem, clip and crown domains are coloured green, blue, purple and orange, respectively.
Phyiscsworld ( 2014): The molecular motor that folds and packs DNA into a virus is at its most efficient when the DNA shows some self-repulsion. That is the surprising finding of researchers based in the US – it was previously thought that such repulsion would act as an obstacle in the packing process. The team also found that pausing the motor and allowing it to relax increased the rate of the whole packaging process. In addition to providing new insights into how viruses function, the work could benefit biotechnologies that enclose long polymers into nanoscale devices. 6
After invading its host cell, a virus reprogrammes the cell's nucleus to duplicate it.
Question: How was the virus programmed to re-program the cell's nucleus? trial and error? Had the function of reprogramming not have to be fully operating since the beginning, otherwise, the virus would not be able to replicate.
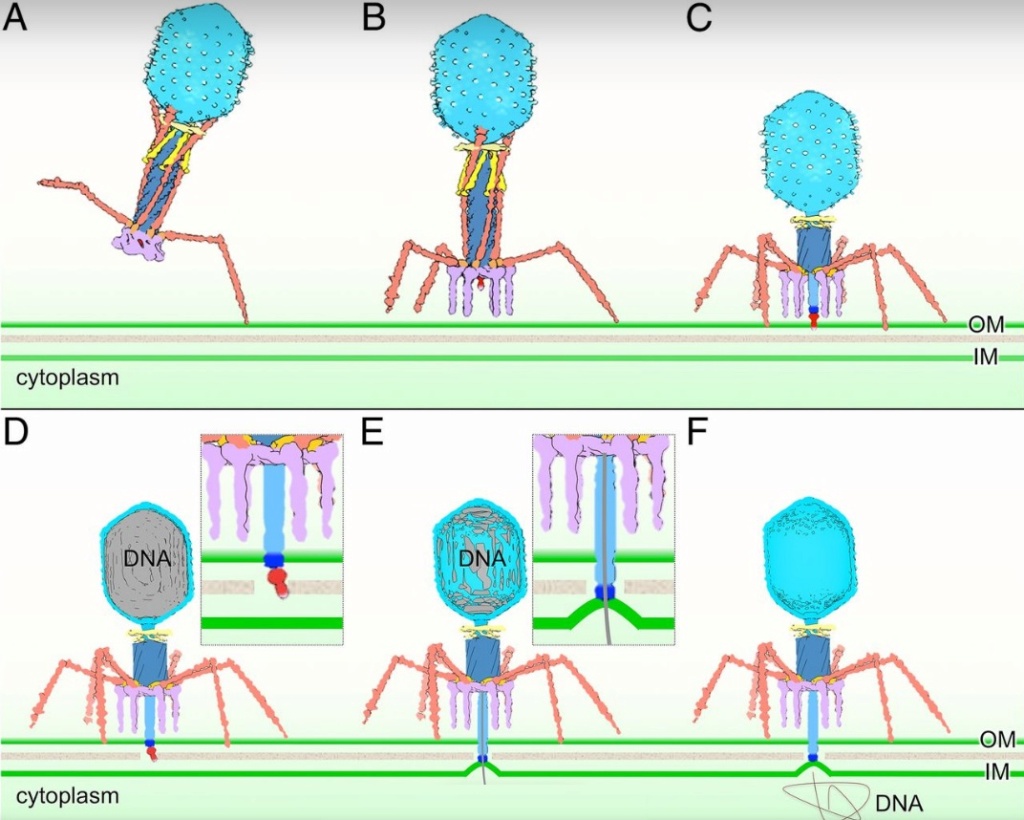
As it replicates, a strand of DNA is pulled from an infected host cell and squeezed into a protein shell – known as a prohead – which then carries the DNA to infect other cells. In some species, the prohead is produced first, leaving only a small hole at one end through which a powerful molecular motor pushes the DNA in and then packs it at very high densities.
Question: How did it emerge the function to pack the DNA at very high densities? trial and error?
The motor has to overcome three forces: the electrostatic self-resistance that comes into play because DNA is negatively charged; the mechanical resistance of DNA to bending; and the entropic resistance of DNA to be crowded on itself.
Question: How did the motor emerge this function of overcoming the three forces? trial and error?
The tail structure
Phys.Org ( 2016): To infect bacteria, most bacteriophages employ a 'tail' that stabs and pierces the bacterium's membrane to allow the virus's genetic material to pass through. The most sophisticated tails consist of a contractile sheath surrounding a tube akin to a stretched coil spring at the nanoscale. When the virus attaches to the bacterial surface, the sheath contracts and drives the tube through it. All this is controlled by a million-atom baseplate structure at the end of the tail. Phages are widely distributed on the planet. They accompany bacteria everywhere - in the soil, water, hot springs, algal bloom, animal intestines etc - and have a dramatic impact on the diversity of bacterial populations, including for example, the microbiome of the human gut. 18
Petr G.Leiman (2006): Bacteriophage T4 has one of the most complex tails of all studied phages. The T4 tail is composed of ∼400 polypeptide chains that form the tube, the contractile sheath around the tube, and the baseplate that terminates both of them. 26
Petr G Leiman (2006): Bacteriophage T4 has one of the most complex tails of all studied phages. It is composed of w400 polypeptide chains that form the tube, the contractile sheath around the tube, and the baseplate that terminates both of them. https://pubmed.ncbi.nlm.nih.gov/16554069/
The tail, fibers, and infection process
Phages from the Myoviridae family have exceptionally complex, contractile tails. Bacteriophage T4 devotes 25 kbp of its genome to tail assembly, which is comparable with the size of the entire adenovirus genome (36 kbp). Products of at least 22 genes are involved in tail assembly (table 3),
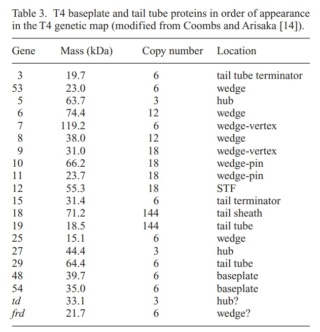
which include a phage-encoded chaperone that participates in folding of the long and short tail fibers (table 2).
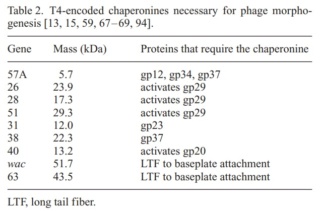
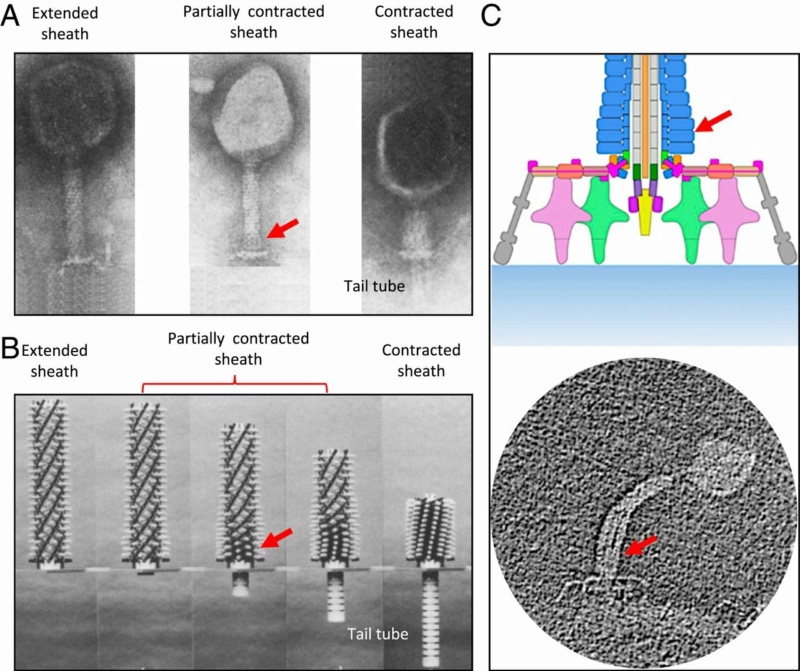
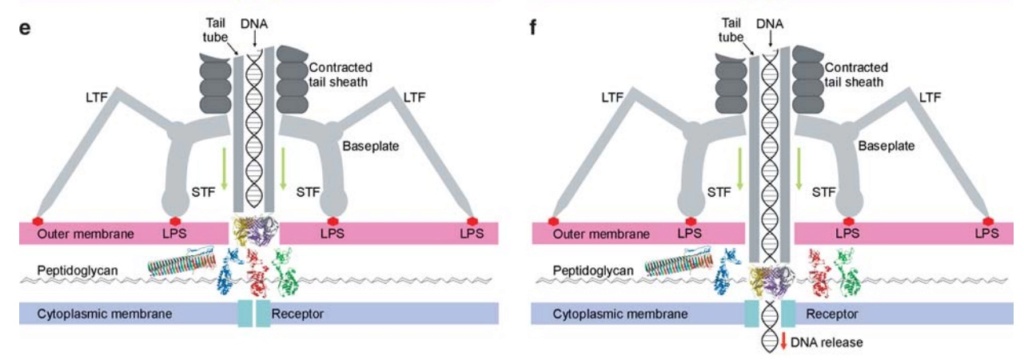
The bacteriophage T4 tail is composed of two concentric protein cylinders, at one end of which is the baseplate and fibers. The inner cylinder, called the tail tube, is built of 144 copies of gp19. The tail tube has a 40 Å-diameter channel for DNA passage from the head to the infected cell. The outer cylinder, called the tail sheath, tightly envelopes the 90 Å-diameter tail tube and has a width of about 210 Å. It is composed of 144 copies of gp18. The subunits comprising each cylinder form a six-start helix with a pitch of 41 Å and a righthanded twist angle of 17°. The helix has a length of 984 Å and contains 24 repeats. During infection, the phage recognizes an E. coli bacterium using its long tail fibers (LTFs) connected to the baseplate. The phage then anchors the baseplate to the lipopolysaccharide cell surface receptors using the short tail fibers (STF), which are initially assembled under the baseplate. This event triggers a hexagon-to-star conformational change in the baseplate and causes an irreversible contraction of the tail sheath, releasing about 25 kcal/ mol of energy per gp18 monomer. During this process, the gp18 hexamers flatten, rotate, and expand radially, resulting in a decrease of their thickness by 26 Å and an increase of the twist angle by 15°. The contracted tail sheath has a length of only 360 Å and a width of 270 Å. The tail tube does not change its length during sheath contraction. As a result, almost half of the tube protrudes out of the contracted tail sheath and the baseplate.
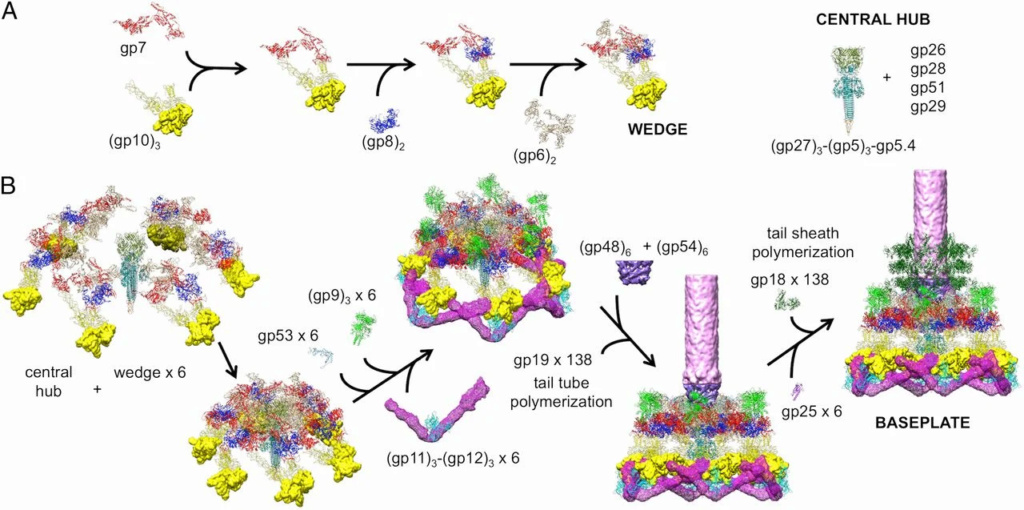
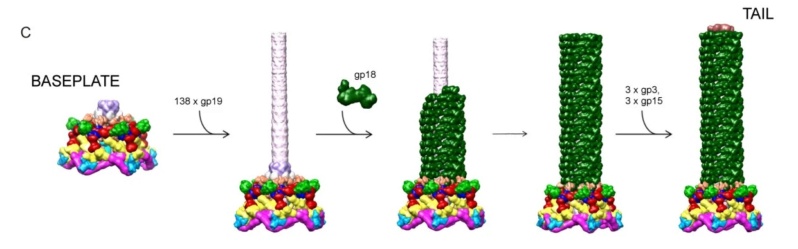
The sheath can be caused to contract by exposing the phage to 3 M urea. Nevertheless, the DNA is not released until the tail tube tip binds to a cytoplasmic membrane receptor common to enteric bacteria, suggesting that tail contraction does not cause the release of DNA. The interaction of the tail tube tip with the cytoplasmic membrane involves creation of a channel for DNA passage. During DNA transfer from the capsid into the cell, the membrane remains virtually undamaged since the transfer requires a proton motive force across the membrane. The assembly pathway of the bacteriophage T4 tail is regulated by ordered sequential interactions of proteins rather than sequential gene expression. The baseplate, a remarkably complex multiprotein structure, is assembled first. It is composed of about 150 subunits of at least 16 different gene products, many of which are oligomeric (table 3). These proteins form six independently assembled wedges that join together around the central hub with the help of the trimeric proteins (gp9) and (gp12). Each wedge is assembled by sequential interactions of the seven protein oligomers: (gp11), (gp10), (gp7), (gp8), (gp6), gp53, and gp25. The baseplate hub is formed by (gp5), (gp27), gp29 and, probably, gp28. The assembly of the baseplate is completed with the attachment of six copies of gp48 and six copies of gp54 to the external interface between the wedges and the hub. The latter proteins serve as a starting point for polymerization of gp19 to form the tail tube, which is terminated with gp3. The tail tube serves as a scaffold for polymerization of the tail sheath around it. During this process, gp18 stores energy in its conformation (possibly by ATP hydrolysis), making the non-contracted T4 tail a stretched spring. The length of the tail tube is controlled by the ruler protein, gp29, which also participates in assembly of the central part of the baseplate. The length of the tail sheath is determined by the length of the tube. The assembly of the tail is completed by attachment of a gp15 hexamer to the last ring of the tail sheath. The baseplate is a dome-shaped object. The hollow tail tube stems from the center of the baseplate. 15

V. V. Mesyanzhinov (2004): Products of at least 22 genes are involved in assembly of the T4 phage tail (Table above) that uses the energy of the sheath contraction for DNA ejection into the host cell. The assembly pathway of the tail is based on strictly ordered sequential interactions of proteins. The baseplate is a remarkably complex multiprotein structure of the tail that serves as a control unit of virus infection. The baseplate is composed of ~150 subunits of at least 16 different gene products, many of which are oligomeric, and assembled from six identical wedges that surround a central hub. The T4 gp11 (the short tail fiber connecting protein), gp10, gp7, gp8, gp6, gp53, and gp25 combine sequentially to built up a wedge. The central hub is formed by gp5, gp27, and gp29 and probably gp26 and gp28. Assembly of the baseplate is completed by attaching gp9 and gp12 forming the short tail fibers, and also gp48 and gp54 that are required to initiate polymerization of the tail tube, a channel for DNA ejection that is constructed of 138 copies of gp19. The length of the tail tube is probably determined by the “ruler protein” or template, gp29. The tail tube serves as a template for assembly of 138 copies of gp18 that form the contractile tail sheath. In the absence of the tail tube, gp18 assembles into long polysheaths with a structure similar in several aspects to the contracted state. Both the tail tube and the tail sheath have helical symmetry. Assembled tail sheath represents a metastable supramolecular structure, and sheath contraction is an irreversible process. During contraction the length of tail sheath decreases from 980 to 360 Å and its outer diameter increases from 210 to 270 Å. The assembly of the tail is completed by a gp15 hexamer that binds to the last gp18 ring of the tail sheath. The assembled tail associates with the head after DNA packaging. Then six gpwac (fibritin) molecules attach to the neck of the virion forming a ring embracing it (“collar”) and thin filaments protruding from the collar (“whiskers”) that help with attachment of the phage particle to other fibrous proteins, the long tail fibers. 23
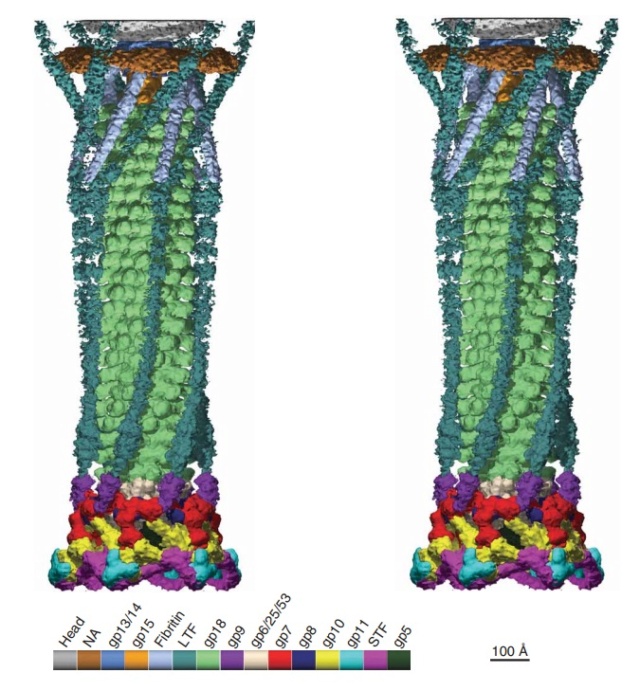
Stereo diagram of bacteriophage T4 showing the extended tail, the LTFs, the neck and a small part of the capsid. NA indicates that the density has not been assigned to a specific gene product.
The cell-puncturing device of bacteriophage T4
ScienceDaily (2016): To infect bacteria, most bacteriophages employ a 'tail' that stabs and pierces the bacterium's membrane to allow the virus's genetic material to pass through. The most sophisticated tails consist of a contractile sheath surrounding a tube akin to a stretched coil spring at the nanoscale. When the virus attaches to the bacterial surface, the sheath contracts and drives the tube through it. All this is controlled by a million-atom baseplate structure at the end of the tail. EPFL scientists have now shown, in atomic detail, how the baseplate coordinates the virus's attachment to a bacterium with the contraction of the tail's sheath.
ScienceDaily (2002): The viral machine works as follows: The virus uses its long-tail fibers to recognize its host and to send a signal back to the baseplate. Once the signal is received, the short-tail fibers help anchor the baseplate into the cell surface receptors. As the virus sinks down onto the surface, the baseplate undergoes a change — shifting from a hexagon to a star-shaped structure. At this time, the whole tail structure shrinks and widens, bringing the internal pin-like tube in contact with the outer membrane of the E. coli cell. As the tail tube punctures the outer and inner membranes of the E. coli cell, the virus' DNA is injected through the tail tube into the host cell. 17
P. G. Leiman (2003): Phages are widely distributed on the planet. They accompany bacteria everywhere -- in the soil, water, hot springs, algal bloom, animal intestines etc -- and have a dramatic impact on the diversity of bacterial populations, including for example, the microbiome of the human gut. The entire baseplate-tail-tube complex consists of one million atoms, making up 145 chains of 15 different proteins. The scientists were also able to identify a minimal set of molecular components in the baseplate that work together like miniature gears to control the activity of the virus's tail. These components, and the underlying functional mechanism, are the same across many viruses and even bacteria that use similar tail-like structures to inject toxins into neighboring cells. 15
One of the most remarkable features of the baseplate is the spike, or needle, along the axis of the dome. The crystal structure of the gp5-gp27 complex (fig. below) can be fitted into the baseplate map so that the needle density is occupied by the C-terminal domain of gp5. The gp27 trimer forms a channel suitable for the passage of a dsDNA and serves as an extension of the tail tube (figs below).
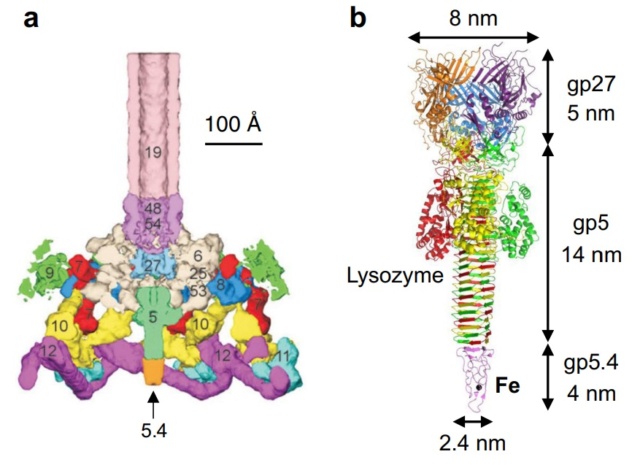
a Baseplate of bacteriophage T4
b gp5–gp27–gp5.4

Structure of the gp5-gp27 complex.
(a) Ribbon stereo diagram. The three gp5 monomers are colored red, green, and blue. The three gp27 monomers are colored yellow, gray, and purple. The K+ ion within gp5C is shown in pink. The (PO4)– is hidden behind the lysozyme domain.
(b) The structure of the gp27 monomer with its four domains colored cyan, pink, light green, and gold along the polypeptide chain.
(c) Top view of the gp27 cylinder shows that the cyan and green domains form a hexagonal torus
Gp5 consists of three domains: an N-terminal oligosaccharide binding-fold domain, the middle lysozyme domain, and the C-terminal triple b-helix domain (fig.above). The gp5 lysozyme domain has 43% sequence identity and a closely similar structure to the T4 lysozyme encoded by gene e (T4L).
Structure of the T4 baseplate
ScienceDaily (2002): The baseplate is the "nerve center" of the virus. When the long and short fibers attach to E. coli, the baseplate transmits this message to the tail, which contracts like a muscle. The baseplate both controls the needlepoint of the tail and the cutting enzyme that make a tiny, nanometer-sized hole through the cell wall of the E. coli. The viral DNA is then squeezed through the tail into the host. The E. coli, thus infected, starts to make only new phage particles and ultimately dies. 16
Moh Lan Yap (2016): T4 has a complex baseplate that is essential for assuring a highly efficient infection mechanism 25
M. I. Taylor (2016): Bacteriophages (viruses of bacteria) use a specialized organelle called a tail to deliver their genetic material and proteins across the cell envelope during infection. In phages with the most complex contractile tails, attachment to the host cell is accompanied by a substantial transformation of the tail structure: the external tail sheath contracts and drives a spike-tipped , rigid tube through the host cell membrane. Other macromolecular complexes, such as the type VI secretion system (T6SS), metamorphosis-associated contractile (MAC) arrays, R-type pyocins, Serratia antifeeding prophage, Photorhabdus virulence cassette and rhapidosomes, use a similar contractile sheath–rigid tube mechanism to breach the bacterial or eukaryotic cell envelope. The most complex part of these ‘contractile injection systems is the baseplate, which is responsible for coordinating host recognition or other environmental signals with sheath contraction. The T4 baseplate is currently thought to contain at least 15 different proteins with copy numbers ranging from 1 to 18. Assembly of the T4 baseplate involves two large independent intermediates: a hub and a wedge. Several phage and possibly host cell chaperones mediate the joining of six wedges to the hub, which is then followed by the attachment of receptor-binding fibres to this structure. The nascent baseplate initiates tube assembly and subsequent polymerization of the sheath in the extended high-energy state. The remarkable structure and transformation of the T4 tail and other contractile injection systems have received considerable attention. This is a 440,000-atom (not counting hydrogens) structure.
Overall structure of the T4 baseplate: The final atomic model is around 96% complete and contains 56,082 amino acid residues (9,886 unique). These amino acids belong to 145 polypeptide chains of 15 different proteins (gene products, gps) that comprise the baseplate (gp5, gp5.4, gp6, gp7, gp8, gp9, gp10, gp11, gp12, gp25, gp27, and gp53) and the proximal region of the tail tube (gp19, gp48 and gp54, although gp48 and gp54 could also be considered to be baseplate components). 19
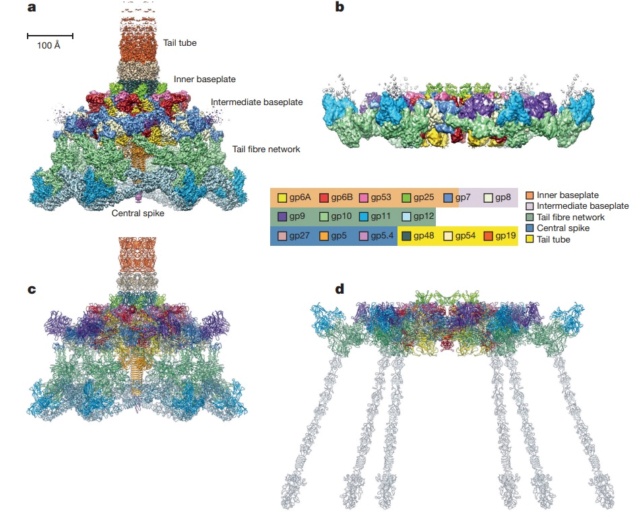
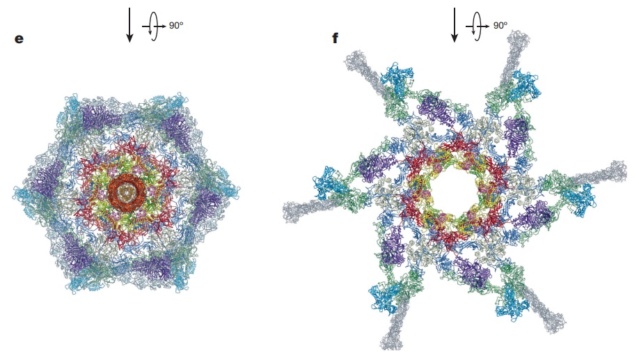
Maps and atomic models of T4 baseplate in pre- and post-attachment states.
a, b, Cryo-EM reconstructions of the pre- and post-attachment T4 baseplate, respectively. c–f, Atomic models of the two states with component proteins shown as ribbon diagrams. In d and f, the STFs (gp12 trimers) are displayed semi-transparently to indicate that they are not present in the refined model of the post-attachment baseplate.
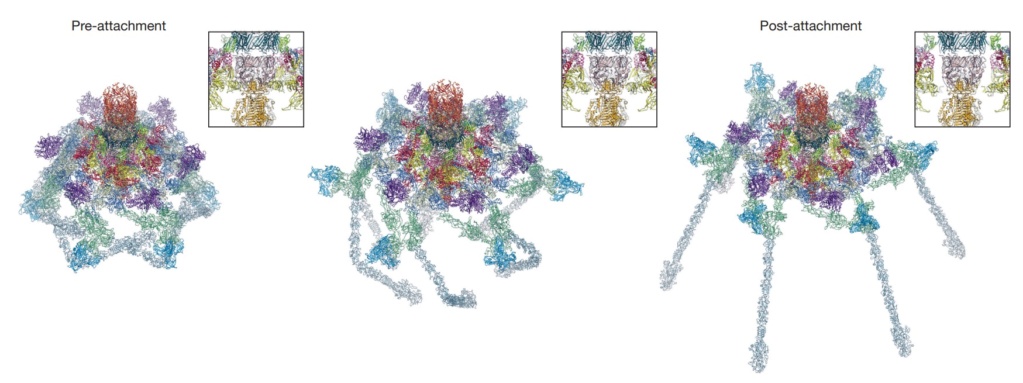
Conformational change of the T4 baseplate upon host cell attachment.
The left and right panels show the pre-and post-attachment structures that are derived from cryo-EM data. The middle image is a model of an intermediate with assumptions described in the text. The insets show close-up views of the central part of the baseplate demonstrating a release of the tail tube–central spike complex, whose position is kept unchanged throughout the transformation.
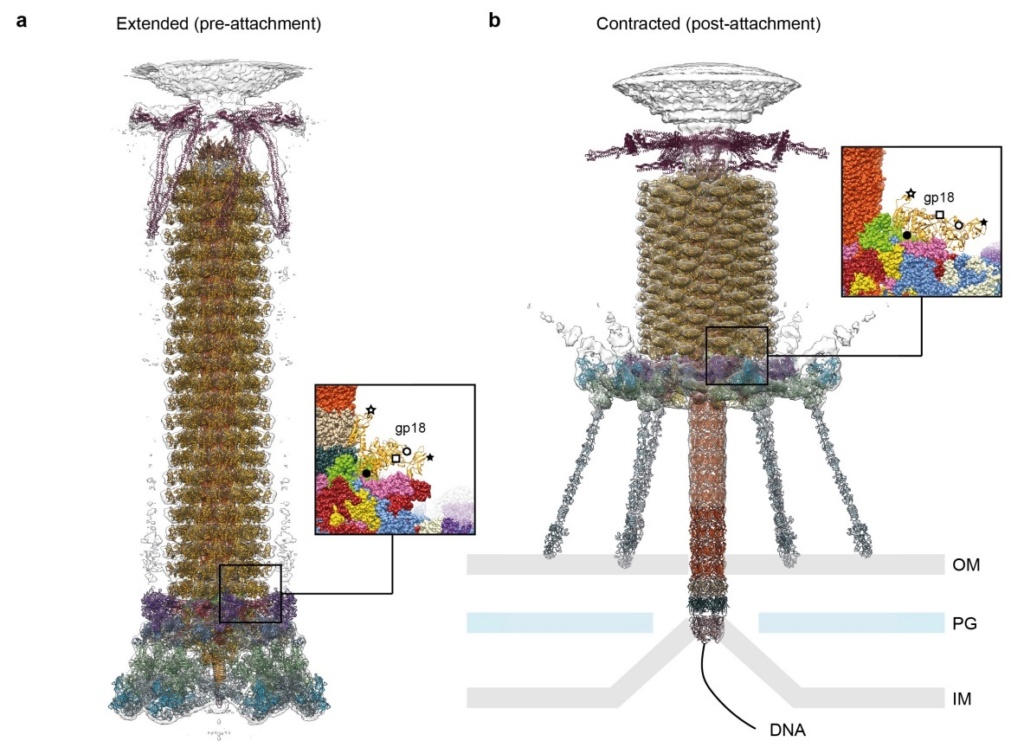
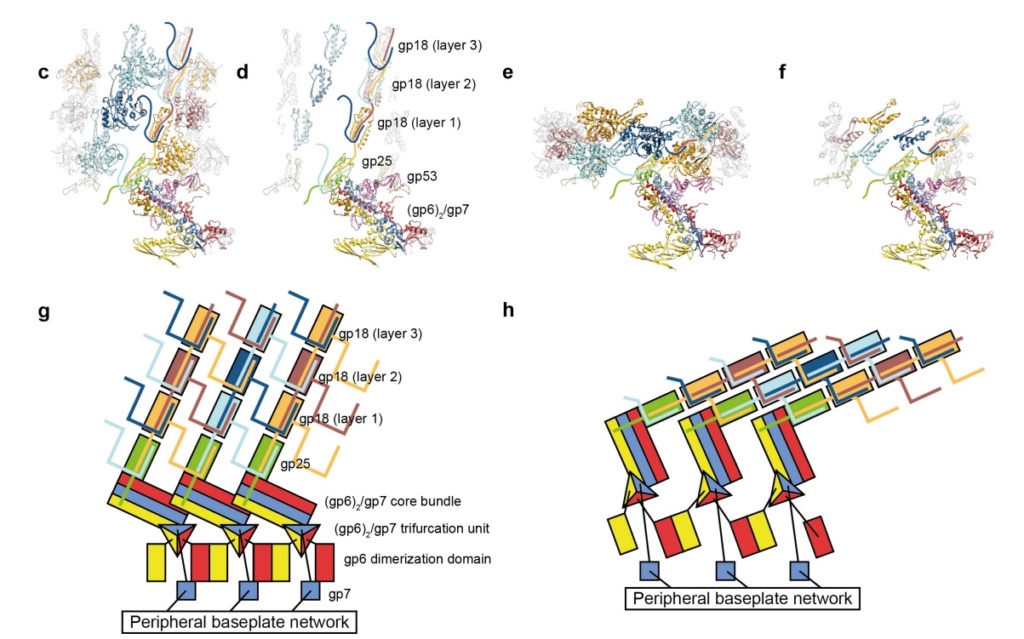
Model for baseplate-induced sheath contraction.
a, b, Pseudoatomic model of the complete T4 tail in the extended (pre-attachment) and contracted (post-attachment) conformations. The insets show a close-up view (labelled with a black box) of the position of the gp18 subunit on the baseplate in the extended and contracted conformations of the sheath. The white geometrical shapes label the same regions of the sheath subunit in both conformations.
c, Interaction of the two conserved domains of the gp18 sheath protein with the conserved components of the T4 baseplate wedge. Coloured lines indicate the putative topology of the N- and C-terminal gp18 extensions, as well as the gp25 C-terminal strand.
d, The same view as in c, but the external domain is now not shown for clarity to demonstrate the interaction of gp25-like sheath domains with each other and with gp25.
e, f, The same as c and d but in the contracted state.
g, h, Two diagrams demonstrating the motion of baseplate components that results in sheath contraction.
V. V. Mesyanzhinov (2004): Practically every assembled T4 particle is able to infect an E. coli host cell. The baseplate is the control center of the viral infectivity, and understanding of the baseplate structure, a multiprotein machine, is a challenging problem. Below we represent the data about baseplate proteins with known atomic structure.
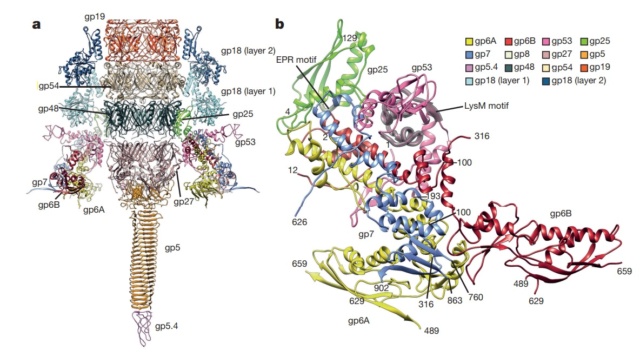
Structure of the conserved inner baseplate.
a, The minimal composition of a contractile injection system is derived from the T4 tail structure: the central spike complex (gp5, gp5.4, gp27), the conserved part of the wedge (gp6, gp7, gp25, gp53), the tail tube (gp19, gp48, gp54) and the conserved part of the sheath (gp18), some of which is modelled using the pyocin sheath14.
b, A close-up of the structure of the conserved region of the wedge consisting of the (gp6)2–gp7 heterotrimer, gp25 and gp53. The EPR motif of gp25 and the LysM motif of gp53 are highlighted with semi-transparent grey.
The tail tube & tail sheath terminators
Moh Lan Yap (2015): The polymerized tail tube and sheath are capped by the terminator proteins gp3 and gp15, respectively, to prevent depolymerization before the tail attaches to the head. Both gp3 and gp15 form hexameric rings that interact with the last row of gp19 and gp18 molecules. The central pore and the side surface of gp15 are negatively charged, whereas the top and the bottom surfaces are positively charged. The top and bottom surfaces interact with gp14 and gp3 proteins, respectively. The interaction between gp15 and gp18 is different in the extended and contracted (postinfection) conformations. In the contracted tail, the negatively charged side surface of the gp15 hexamer interacts with positively charged surfaces of the C-terminal domains of the gp18 molecules. These interactions help to maintain the integrity of the tail in its contracted form. The gp15 hexamer may have undergone a conformational change during the infection process, which might be propagated through gp14 and gp13, to the portal assembly to allow the release of the genomic DNA. 21
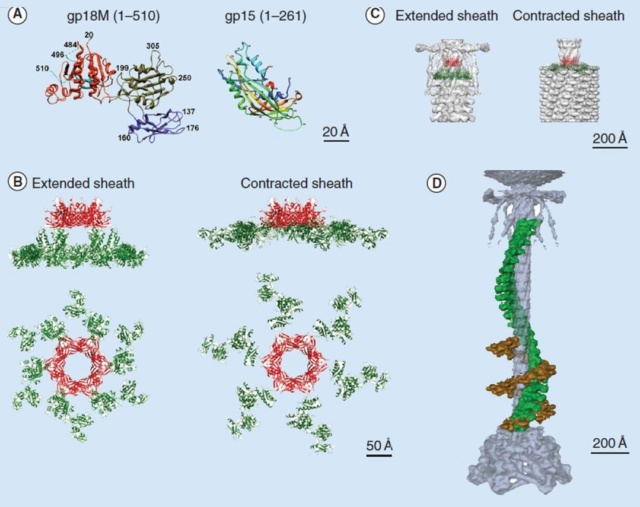
Crystal structures of gp18 and gp15
(A) Ribbon diagrams of gp18M and gp15 monomers. The three domains of gp18M are shown in blue (domain I), olive green (domain II) and orange red (domain III); the β-hairpin (residues 454–470) and the last 14 C-terminal residues are shown in cyan. Gp15 represented in rainbow color running from N terminus (blue) to C terminus (red). The broken lines indicate protein regions that are disordered in the crystals.
(B) Relative positions of the gp15 and gp18 molecules in the extended and contracted T4 tails, viewed from the side (upper) and from the top (lower) of the phage. A model of the entire gp18 molecule was created based on the crystal structure of gp18M and the prophage tail sheath protein LIN1278. The models of gp18 molecules belonging to the topmost ring of the contractile sheath are shown in green. The gp15 hexamer is shown in red.
(C) The gp15 and gp18 molecules are fitted into the cryo-electron microscopy reconstructions of the extended and contracted tails.
(D) A helical strand of gp18 in the extended (green) and contracted (brown) tail. The hexagonal baseplate, tail tube, whiskers and collar are shown in gray-blue.

Structure of the contracted T4 tail
(side view (a), with an inclination (b), cross-section (c), from the bottom (d)). Each protein or complex is labeled with their respective gene number and indicated by color: spring green, gp5; red, putative gp7; dark blue, gp8; green, gp9; yellow, putative gp10; cyan, gp11; magenta, gp12; salmon, gp19; sky blue, gp27; pink, putative gp48 or gp54; beige, gp6 + gp25 + gp53; orange, putative gp26. 23
1. http://www.pnas.org/content/111/42/15096.short
2. Anjeanette Roberts Celebrating 3.8 Billion Years of Bacteriophage October 22, 2015
3. http://www.ncbi.nlm.nih.gov/pubmed/22297528
4. https://creation.com/images/pdfs/tj/j22_1/j22_1_15-16.pdf
6. http://creation.com/did-god-make-pathogenic-viruses
7. Joseph W. Francis: The Organosubstrate of Life: A Creationist Perspective of Microbes and Viruses 2003
8. http://teaguesterling.com/dna/motor-protein.pdf
9. G. Leiman: Structure and morphogenesis of bacteriophage T4 P. 9 May 2003
10. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3109452/
11. Nicola Twilley: Inside the World of Viral Dark Matter February 6, 2015
12. Eric S Miller Bacteriophage T4 and its relatives 28 October 2010
13. Eric S Miller Bacteriophage T4 genome 2003 Mar;6
14. Vincent R. Racaniello: Principles of Virology, Volume 1: Molecular Biology 18 agosto 2015
15. Science Daily: How viruses infect bacteria: A tale of a tail May 18, 2016
16. Science Daily: New Understanding Of Complex Virus Nano-Machine For Cell Puncturing And DNA Delivery February 4, 2002
17. Science Daily: Study Reveals New Information On How Viruses Enter Cells February 7, 2002
18. Phys.Org: How viruses infect bacteria: A tale of a tail MAY 18, 2016
19. M. I. Taylor et.al. Structure of the T4 baseplate and its function in triggering sheath contraction 18 May 2016
20. Jaap Vergote Design and nature Jun 24, 2018
21. Moh Lan Yap: Structure and function of bacteriophage T4 2015 Aug 1
22. Andrei Fokine: The Molecular Architecture of the Bacteriophage T4 Neck 2013 Feb 19
23. V. V. Mesyanzhinov: Molecular Architecture of Bacteriophage T4 July 9, 2004
24. Fumio Arisaka:[url=https://aip.scitation.org/doi
25. Petr G. Leiman: Evolution of Bacteriophage Tails: Structure of T4 Gene Product 10 2006 May 5
26. Petr G.Leiman: Evolution of Bacteriophage Tails: Structure of T4 Gene Product 10 5 May 2006
27. MITSUHIRO YANAGIDA: MOLECULAR ORGANIZATION OF THE HEAD OF BACTERIOPHAGE Teven: UNDERLYING DESIGN PRINCIPLES 1984
28. Bo Hu: Structural remodeling of bacteriophage T4 and host membranes during infection initiation August 17, 2015
Last edited by Otangelo on Sat Oct 29, 2022 9:17 am; edited 117 times in total