https://reasonandscience.catsboard.com/t1554-the-rubisco-enzymes-amazing-evidence-of-design
Although RubisCO has been intensively studied, its evolutionary origins and rise as Nature’s most dominant carbon dioxide (CO2)-fixing enzyme still remain in the dark. 4
Rubisco is the most important enzyme on the planet. virtually all the organic carbon in the biosphere derives ultimately from the carbon dioxide that this enzyme fixes from the atmosphere. Without it, advanced life would not be possible. And we would not be able to debate our origins. All inquiry and quest about if we are ultimately the result of a powerful creator, or just random natural chemical reactions and emerging properties of lifeless matter, if biodiversity is due to evolution, or an intelligent designer, is second to the inquiry of how Rubisco came to be. Rubisco has a complex structure, functioning and synthesis process, many cell compartments, enzymes, proteins, and pathways are involved and required to assemble it, the unfinished sub-units require co and post-translational modifications, specific proteins that help like assembly robots in the manufacturing process, sophisticated pathways, and mechanisms of protein import and targeting in chloroplasts through large multi-protein translocon complexes in the stroma, and advanced protein communication and information systems. All this is of bewildering complexity, where dozens of individual interconnected and finely tuned parts are required, a web of interlocked extremely complex advanced molecular machines where if one is missing, nothing goes, that defy the intelligence of the best scientists for decades to find out their structure, mechanisms, and functions. Could all this be due to natural processes?
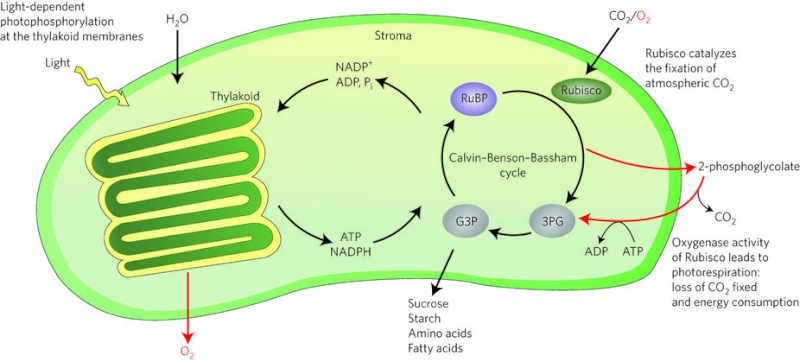
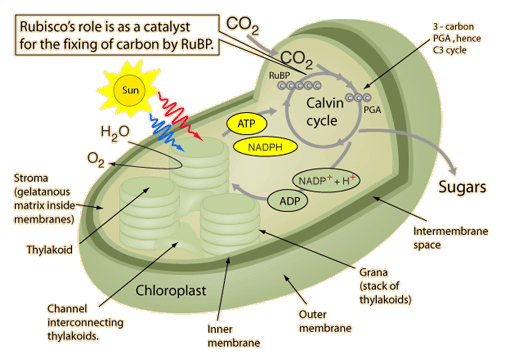
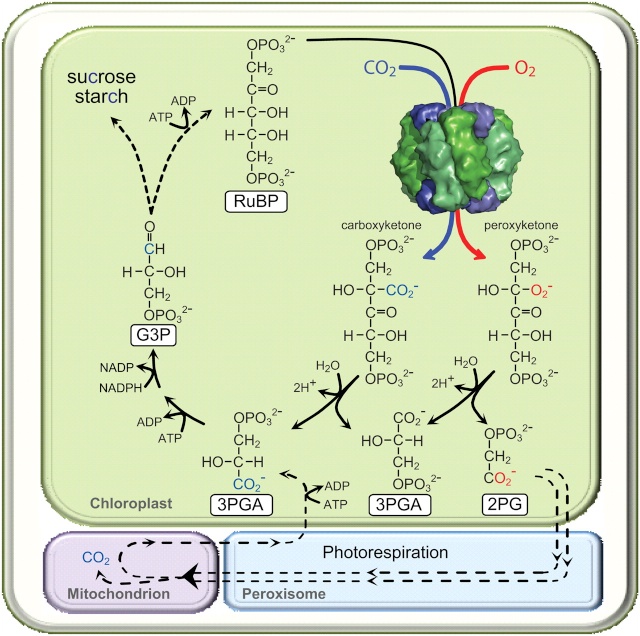
RuBisCO is a multi-subunit plant protein essential to photosynthesis. It catalyzes the primary chemical reaction by which inorganic carbon enters the biosphere. In the C3 pathway, RuBisCO is responsible for initiating the first step in carbon dioxide fixation, a process by which atmospheric carbon dioxide is converted by plants to energy-rich molecules such as glucose. This step of the Calvin Cycle plays a crucial role in providing energy for the cell.
https://reasonandscience.catsboard.com/t2164-the-calvin-benson-cycle
Rubisco is also the most abundant enzyme on earth. It is present in every plant and photosynthetic organism, from the smallest cyanobacteria and plankton to palm trees and giant sequoias. Rubisco is a complex composed of eight large subunits and eight small subunits
Synthesized RuBisCO does not have a fully functional active site. It needs to be activated by a CO2 molecule that carbamylates its catalytic Lys to bind Mg2+ that completes the activation process... The carboxylation involves at least four, perhaps five discrete steps and at least three transition states;
The origin of these highly specific, regulated, and coordinated steps, which are essential for the activation of Rubisco, is best explained through a planning mind, which all set it up. Natural mechanisms are extremely unlikely to be capable to produce these sophisticated metabolic multistep pathways and assembly lines to make Rubisco in the first place. No wonder, that no mainstream scientific papers are able to provide compelling evolutionary scenarios. As long as the enzyme is not fully functional, nothing goes, and ultimately, advanced life on earth would not be possible. How did the correct insertion of the correct metal cation Mg2+ surrounded by three H2O/OH molecules emerge? Trial and error? The genome needs the right information in order to get the right materials, the right shape and quantity of each subunit co-factors and metal clusters, how to position them at the right active site, and how to mount these parts in the right order. That seems to me only being explained in a compelling manner by the wise planning of a super-intelligent engineer, which knew how to invent and build this highly sophisticated and complex machine and make it fully functional right from scratch. A stepwise unguided emergence seems to be extremely unlikely. This mechanism seems to be the result of intelligence, which sets it all up through power, will, and information.
The eight large subunits of Rubisco are coded by the chloroplast DNA and the eight small subunits by nuclear DNA. The small subunit of Rubisco and all the other Calvin cycle enzymes are encoded by nuclear genes and must be transported and travel to the chloroplast site after their synthesis in the cytosol.
https://reasonandscience.catsboard.com/t2165-pathways-and-mechanisms-of-protein-import-and-targeting-in-chloroplasts
The precursor forms of these stromal proteins contain an N-terminal stromal-import sequence. This transit peptide allows transfer of the small subunits synthesized in the cytosol through the chloroplast envelope translocon complexes into the plastid. These are highly complex molecular gates in the chloroplast inner and outer membrane, which filter which molecules go in. After the unfolded precursor enters the stromal space, it binds transiently to a stromal Hsc70 chaperone and the N-terminal sequence is cleaved.
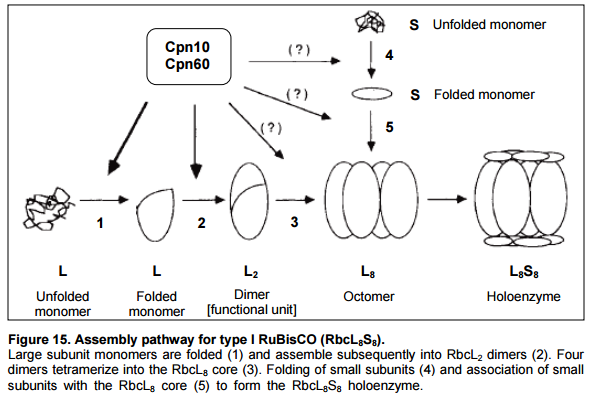
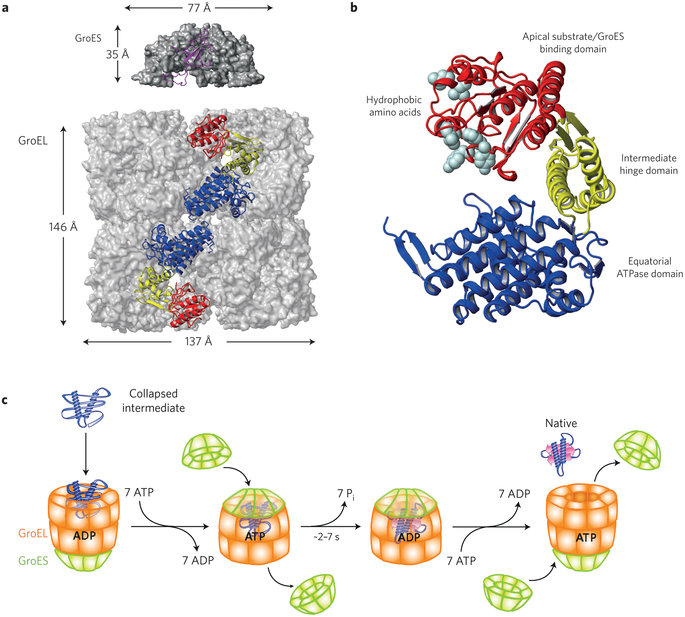
Folding of the small and large Rubisco subunit proteins is mediated by the amazing GroEL–GroES chaperonin system. Protein folding mediated by chaperonins is the process by which newly synthesized polypeptide chains acquire the three-dimensional structures necessary for biological function. For many years, protein folding was believed to occur spontaneously. But it has become apparent that large proteins frequently fail to reach native state, forming nonfunctional aggregates instead. They need the aid of these sophisticated barrel-shaped proteins.
https://reasonandscience.catsboard.com/t1437-chaperones?highlight=chaperones
That raises interesting questions: How should and could natural non-guided natural mechanisms foresee the necessity of chaperones in order to get a specific goal, that is the right precise 3-dimensional folding resulting in functional proteins to make living organisms? The nonliving matter has no natural " drive " or purpose or goal to become living. The make of proteins to create life, however, is a multistep process of many parallel acting complex metabolic pathways and production-line like processes to make proteins and other life-essential products like nucleotides, amino acids, lipids, carbohydrates, etc. The right folding of proteins is just one of several other essential processes in order to get a functional protein. But a functional protein by its own has no function unless correctly embedded in the right assembly sequence and order at the right functional place.
Eight S subunits combine with the eight L subunits to yield the active rubisco enzyme. At least three chloroplast outer membrane proteins, including a receptor that binds the stromal-import sequence and a translocation channel protein, and five inner-membrane proteins are known to be essential for directing proteins to the stroma. Import into the stroma depends on ATP hydrolysis catalyzed by a stromal Hsc70 chaperone. Chloroplasts cannot generate an electrochemical gradient (proton-motive force) across their inner membrane. Thus protein import into the chloroplast stroma is powered solely by ATP hydrolysis. Within the stroma, the S-subunits undergo further posttranslational modification (transit peptide cleavage, Met-1 aN- methylation) prior to assembly into final L8S8 Rubisco complexes. How did natural evolutionary processes find out how to do it? Trial and error?
In order to make and assemble Rubisco, at least 25 parts, most of the essential and irreducible, are directly involved in Rubisco function, activation, and synthesis:
https://reasonandscience.catsboard.com/t1554-the-rubisco-enzymes-amazing-evidence-of-design#3899
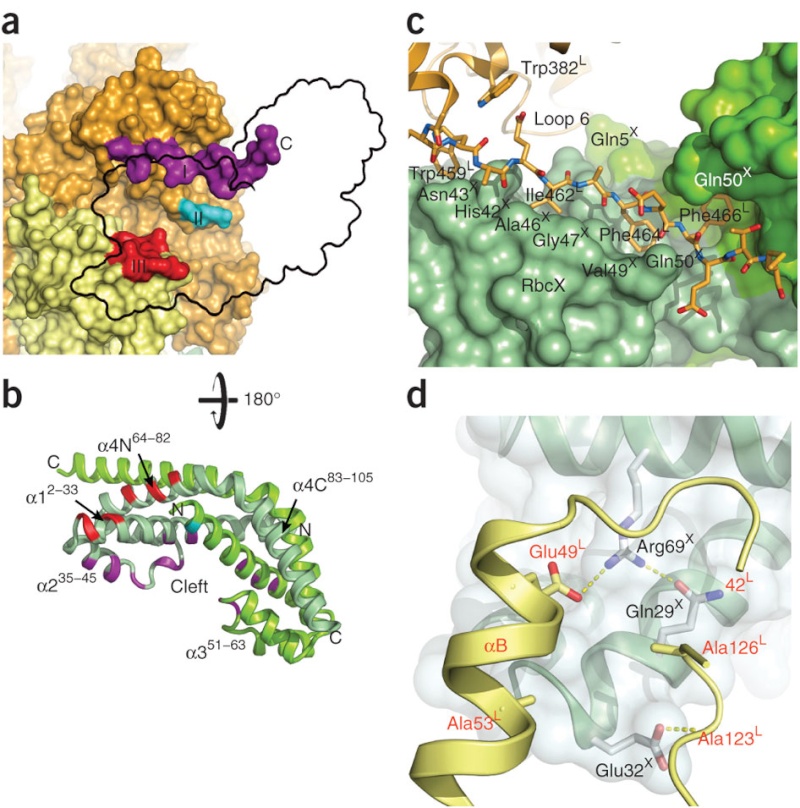
Could these parts, proteins enzymes, etc. have evolved separately and gradually? What about the RbcX Assembly Chaperone, specifically used as an assembly tool of Rubisco? What about the barrel-shaped GroEL/GroES chaperonins which perform their function with extremely impressive simplicity and elegance, namely helping over 100 different proteins to get into their correct shape and form, essential for function? ( in our case, helping the Rubisco RbcL subunits to get their proper shape )
These chaperone systems are themselves made of proteins which also require the assistance of chaperones to correctly fold and to maintain integrity once folded. Chaperones for chaperones in fact. The very simplest of cells that we know of have these systems in place.
Or how do proponents of evolution explain how natural selection would have favored the emergence of Hsp70 chaperones, central components of the cellular network, proteins which assist a large variety of protein folding processes in the cell by the transient association of their substrate-binding domain with short hydrophobic peptide segments within their substrate proteins? That is in our case, their function of which was to prevent a still-useless rubisco small subunit from folding outside the chloroplast? They are made, used during the synthesis process, and once Rubisco assembly has finished, these enzymes are discarded. This is very much a factory-like production and assembly-line process, using fully automatized and programmed nano-robot like molecular machines, namely enzymes. Most parts, if missing, render 1. the assembly of Rubisco impossible, and 2. Rubisco useless. Many parts, if missing render it not fully functional and defective. Besides the enzymes that have used in other biological systems, there would be no reason to make them unless all other parts were there too, and the assembly instructions of Rubisco. As an analogy, if you had to make the implementation of a car factory, why would you make the assembly chain of a piston, if you do not have all the precise instructions to make 1. the car as a whole, and 2. the instructions of the precise shape and the materials required for the piston in particular, and how to mount it in the motor? That's precisely what happens in the cell. Evolution has no consciousness, and no foresight nor intelligence. But precisely that is required for PLANNING and make of blueprints. I cannot create a machine, without the precise drawing and project information in advance, which is required to make 1. the assembly tools 2. the subparts 2. the whole machine.
How do proponents of evolution explain how natural selection would have favored a protein complex the function of which was to prevent a still-useless Rubisco small subunit from folding outside the chloroplast? Before it evolved a way to get the protein inside, there would be no benefit from keeping it unfolded outside. How could blind chance ‘know’ it needed to cause large subunit polypeptides to fold ‘correctly’ and to keep them from clumping? It could not ‘anticipate’ the ‘correct’ confirmation before the protein became useful. And evolution would need to be clever indeed to chemically modify something not yet useful so that it could be folded ‘correctly’ when even the ‘correctly’ folded polypeptide would not yet become useful.
Only a designer would know why it would be necessary
to produce a specialized protease, target it to the chloroplast, and program it to clip off the targeting sequence of the small subunit at just the right place. And what about the assembly of a collection of meaningless rubisco parts in just one certain way? In order to design a sophisticated set of tools to make something else useful in the future that had, as yet, no function, evolution (as ‘designer’) would have had to have detailed knowledge of the future usefulness of the protein it was so cleverly engineering. If evolution managed to generate any one of these chaperone protein complexes (and it would not), it would still be useless for generating rubisco unless all the other chaperones were also present. Without any one of them, the sixteen-unit complex could not be generated.
That totally destroys the Evolution Theory: How should and could natural nonguided natural mechanisms foresee the necessity of chaperones in order to get a specific goal, that is the right precise 3-dimensional folding resulting in functional proteins to make living organisms? The nonliving matter has no natural " drive " or purpose or goal to become living. The make of proteins to create life, however, is a multistep process of many parallel acting complex metabolic pathways and production-line like processes to make proteins and other life-essential products like nucleotides, amino acids, lipids, carbohydrates, etc. The right folding of proteins is just one of several other essential processes in order to get a functional protein. But a functional protein on its own has no function unless correctly embedded in the right assembly sequence and order at the right functional place." that's precisely the problem of evolution. there is no foresight. So why would evolution produce an assembly chaperone enzyme to make rubisco? You don't make a robot for an assembly line if the end product is not known.
(RuBisCO) is a multi-subunit plant protein essential to photosynthesis. It catalyzes the primary chemical reaction by which inorganic carbon enters the biosphere. In the C3 pathway, RuBisCO is responsible for initiating the first step in carbon dioxide fixation, a process by which atmospheric carbon dioxide is converted by plants to energy-rich molecules such as glucose. In chemical terms, it catalyzes the carboxylation of ribulose-1,5-bisphosphate (also known as RuBP).3 This step of the Calvin Cycle plays a crucial role in providing energy for the cell. It is the most important enzyme on the planet — virtually all the organic carbon in the biosphere derives ultimately from the carbon dioxide that this enzyme fixes from the atmosphere. 3 Rubisco is also the most abundant enzyme on earth. It is present in every plant and photosynthetic organism, from the smallest cyanobacteria and plankton to palm trees and giant sequoias. Rubisco is a complex composed of eight large subunits and eight small subunits 2
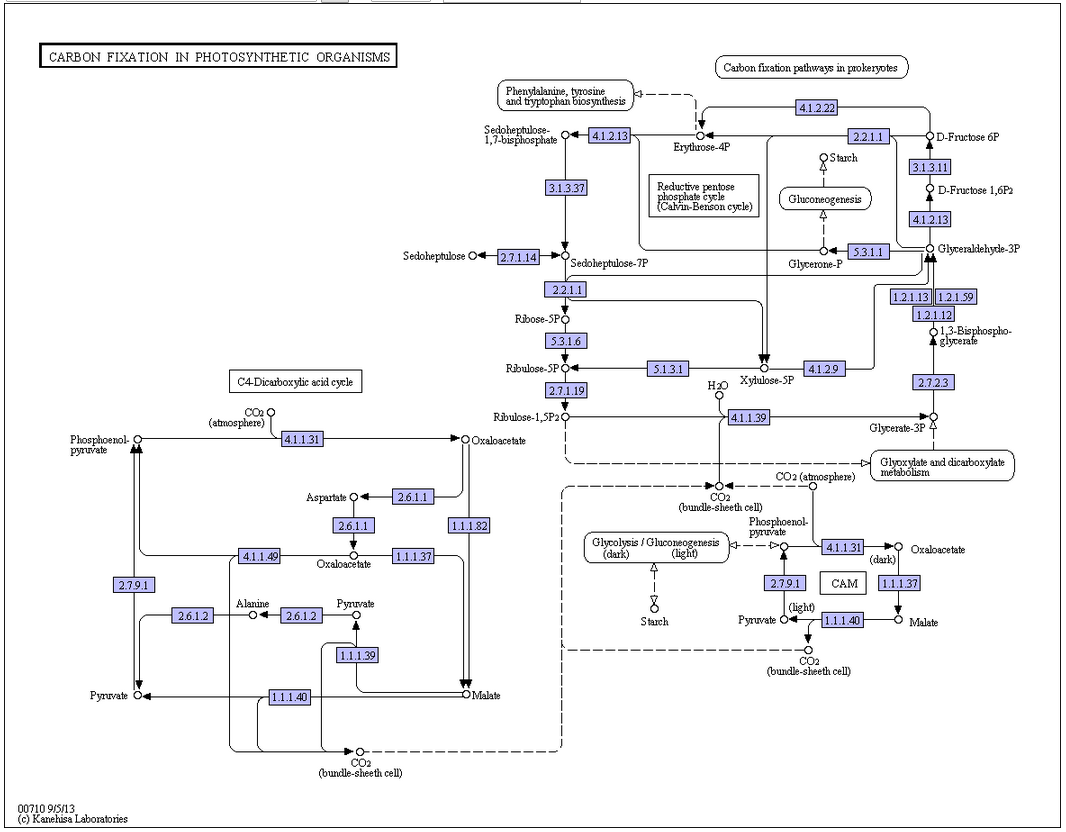
RuBisCO vs alternative carbon fixation pathways
While many autotrophic bacteria and archaea fix carbon via the reductive acetyl CoA pathway, the 3-hydroxypropionate cycle, or the reverse Krebs cycle, these pathways are relatively smaller contributors to global carbon fixation than that catalyzed by RuBisCO. Phosphoenolpyruvate carboxylase, unlike RuBisCO, only temporarily fixes carbon. Reflecting its importance, RuBisCO is the most abundant protein in leaves, accounting for 50% of soluble leaf protein in C3 plants (20–30% of total leaf nitrogen) and 30% of soluble leaf protein in C4 plants (5–9% of total leaf nitrogen).
Structure
In plants, algae, cyanobacteria, and phototrophic and chemoautotrophic proteobacteria, the enzyme usually consists of two types of protein subunit, called the large chain (L) and the small chain (S).

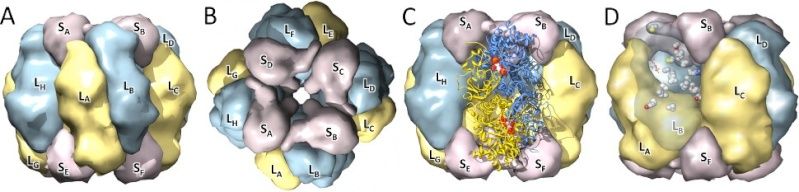
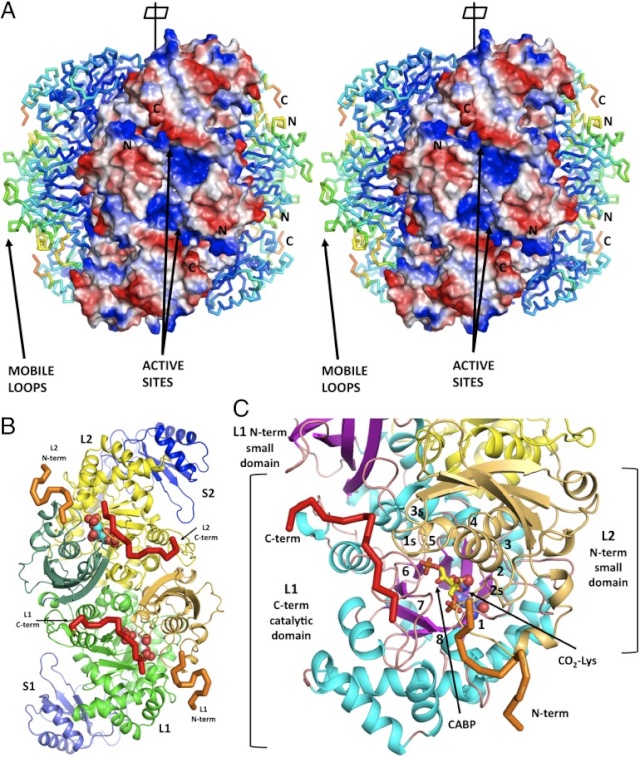
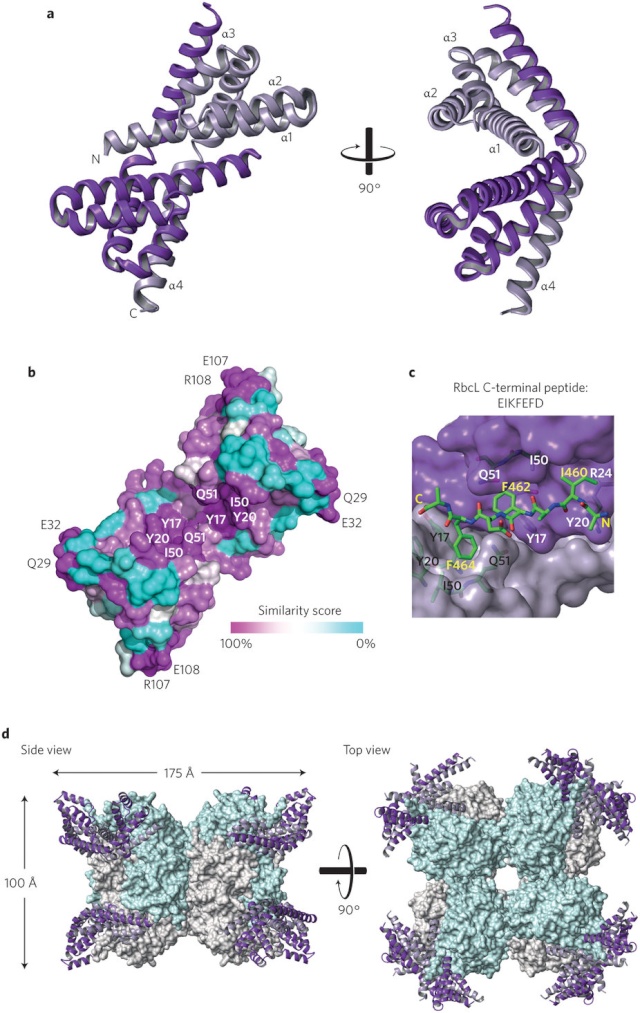
Structural view of the RubisCO enzyme. A) Front view, with the large subunits in blue/yellow and the small subunits in purple; B) Top view, with the central solvent channel visible; C) Front view, with the two large subunits (in ribbons) forming the catalytic dimer; D) Sites detected under functional divergence.
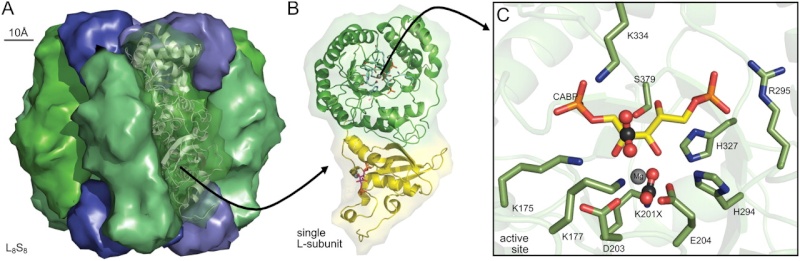
Conserved structural features of Rubisco.
A Spinach L8S8 Rubisco (Protein Data Bank 8RUC) drawn using Pymol to highlight arrangement of the S-subunits (blue) capping the catalytic core of four L2 subunits (green).
B Structural details for one L-subunit of an L2 pair highlighting one active site within the α/β-barrel of the C-terminal domain (green ribbons) and residues in the N-terminal domain (yellow ribbons) that contribute to the second active site in each L2.
C Arrangement of the conserved Rubisco active-site residues within a L-subunit C-terminal domain relative to carbamylated Lys-201 (K201X), bound Mg2+, and the six-carbon reaction intermediate mimic, 2-carboxyarabinitol 1,5-bisphosphate (CABP). The activating CO2 in K201X and the approximate positioning of substrate CO2 that binds to C-2 of the RuBP enediol are highlighted in ball-and-stick representations. Residues are numbered relative to spinach Rubisco. The conserved active site residues Glu-60 and Asn-123 from the N-terminal domain of the paired L-subunit are not shown.
From a structural point of view, all Rubisco enzymes comprise at least two large (L-) subunits. Despite there being as little as 30% amino acid identity between the different Rubisco forms, they all show a conserved L-subunit structure comprising an N-terminal domain (approximately 150 amino acids) and a larger C-terminal domain (approximately 320 amino acids) that forms an α/β-barrel . Paired L-subunits arrange head to tail to form a dimer (L2), with two active sites located at the L-L interface. The highly conserved catalytic residues predominantly reside within the α/β-barrel domain, with a few residues supplied by the N-terminal domain of the adjacent L-subunit.
Different organisms have diverse arrangements of the Rubisco L2 building blocks. Form I Rubiscos are the most abundant form found in plants, algae, and many photosynthetic bacteria. The L2 subunits in form I Rubiscos are arranged in an (L2)4 core, with two groups of four small (S-) subunits capping the L8 core to form an L8S8 molecule (Fig. A above). Although not strictly required for CO2 fixation, the S-subunits are essential for maximal activity and provide structural stability. Rubiscos classed as forms II and III lack S-subunits, containing only L-subunits arranged into L2 to (L2)5 complexes. These Rubiscos are primarily found in phototrophic proteobacteria, chemoautotrophs, dinoflagellates, and achaea.
Although the classification of Rubisco enzymes into forms I, II, and III is generally supported by sequence phylogenies, quaternary structures, and functional properties, there are exceptions.
The large-chain gene (rbcL) is part of the chloroplast DNA molecule in plants.
There are typically several related small-chain genes in the nucleus of plant cells, and the small chains are imported to the stromal compartment of chloroplasts from the cytosol by crossing the outer chloroplast membrane.
The enzymatically active RUBISCO substrate binding sites are located in the large chains that form dimers in which amino acids from each large chain contribute to the binding sites. A total of eight large-chains (= 4 dimers) and eight small chains assemble into a larger complex.
Magnesium ions (Mg2+) are needed for enzymatic activity. Correct positioning of Mg2+ in the active site of the enzyme involves addition of an "activating" carbon dioxide molecule (CO2) to a lysine in the active site (forming a carbamate). Formation of the carbamate is favored by an alkaline pH. The pH and the concentration of magnesium ions in the fluid compartment (in plants, the stroma of the chloroplast) increases in the light.
1) http://www.eplantscience.com/index/algae/photosynthesis/light_independent_reactions.php#aa
2) https://en.wikipedia.org/wiki/RuBisCO
3) https://en.wikipedia.org/wiki/RuBisCO
4. https://sci-hub.tw/https://www.sciencedirect.com/science/article/pii/S095816691730099X
Last edited by Otangelo on Wed Sep 28, 2022 5:19 am; edited 38 times in total



 4
4