Who seeks with a pure heart, finds. Disbelieving in God's existence has in my view, only in very rare cases something to do with people not finding evidence. It has to do with people rejecting God on other grounds.

ElShamah - Reason & Science: Defending ID and the Christian Worldview
Welcome to my library—a curated collection of research and original arguments exploring why I believe Christianity, creationism, and Intelligent Design offer the most compelling explanations for our origins. Otangelo Grasso
276 Re: My articles Thu Aug 11, 2022 6:42 am
Re: My articles Thu Aug 11, 2022 6:42 am
Otangelo
Admin
Yesterday, I was at Rebekah's Bread of Life channel, and asked the atheists, what they would do, if the God of the Bible would prove his existence to them. If they would worship Him, or just acknowledge His existence, and move on with their life as if he would not exist. First, they were questioning how God would do that and ranted about that for minutes. After I finally got them to answer my question, explaining that in my thought experiment, they should simply take it as a fact, that God was capable of doing that, they, hesitating, tried to evade again, asking which God of the Bible, there was Elohim, Jahweh, Elijah, etc. In the end, they admitted: The God of the Bible is evil, a genocidal monster, not worth our worship.
Who seeks with a pure heart, finds. Disbelieving in God's existence has in my view, only in very rare cases something to do with people not finding evidence. It has to do with people rejecting God on other grounds.
Who seeks with a pure heart, finds. Disbelieving in God's existence has in my view, only in very rare cases something to do with people not finding evidence. It has to do with people rejecting God on other grounds.
277 Re: My articles Tue Aug 16, 2022 8:22 pm
Re: My articles Tue Aug 16, 2022 8:22 pm
Otangelo
Admin
We celebrate and worship the Lord, which is our hope and savior.
Atheists have nothing to celebrate nor to worship, or hope for.
We have a positive case that we proclaim,
Atheists only lament and criticize what we believe, and only proclaim what they lack.
We have evidence pointing to the God of the Bible through natural and scriptural theology. They have no evidence that warrants a no-God world.
We have intellectual satisfaction. They have cognitive dissonance.
We look forward to our eternal destiny in heaven. They look forward to the grave.
We know our Lord lives, even if we cannot prove it. They confess ignorance.
We: 1.
Atheists: O.
I see no reason why I should become an atheist.

Atheists have nothing to celebrate nor to worship, or hope for.
We have a positive case that we proclaim,
Atheists only lament and criticize what we believe, and only proclaim what they lack.
We have evidence pointing to the God of the Bible through natural and scriptural theology. They have no evidence that warrants a no-God world.
We have intellectual satisfaction. They have cognitive dissonance.
We look forward to our eternal destiny in heaven. They look forward to the grave.
We know our Lord lives, even if we cannot prove it. They confess ignorance.
We: 1.
Atheists: O.
I see no reason why I should become an atheist.

278 Extreme genome repair, and remarkable morphogenesis by self-assembly point to design Sat Oct 29, 2022 10:30 am
Extreme genome repair, and remarkable morphogenesis by self-assembly point to design Sat Oct 29, 2022 10:30 am
Otangelo
Admin
Extreme genome repair, and remarkable morphogenesis by self-assembly point to design
https://reasonandscience.catsboard.com/t2061p275-my-articles#9626
Extreme Genome Repair (2009): If its naming had followed, rather than preceded, molecular analyses of its DNA, the extremophile bacterium Deinococcus radiodurans might have been called Lazarus. After shattering of its 3.2 Mb genome into 20–30 kb pieces by desiccation or a high dose of ionizing radiation, D. radiodurans miraculously reassembles its genome such that only 3 hr later fully reconstituted nonrearranged chromosomes are present, and the cells carry on, alive as normal 1
T. Devitt (2014): John R. Battista, a professor of biological sciences at Louisiana State University, showed that E. coli could evolve to resist ionizing radiation by exposing cultures of the bacterium to the highly radioactive isotope cobalt-60. “We blasted the cultures until 99 percent of the bacteria were dead. Then we’d grow up the survivors and blast them again. We did that twenty times,” explains Cox. The result were E. coli capable of enduring as much as four orders of magnitude more ionizing radiation, making them similar to Deinococcus radiodurans, a desert-dwelling bacterium found in the 1950s to be remarkably resistant to radiation. That bacterium is capable of surviving more than one thousand times the radiation dose that would kill a human. 2
Simple bacteria can restart their 'outboard motor' by hotwiring their own genes (2015):
Unable to move and facing starvation, the bacteria evolve a replacement flagellum - a rotating tail-like structure that acts like an outboard motor - by patching together a new genetic switch with borrowed parts. When an organism suffers a life-threatening mutation, it can rapidly rewire its genes. The remarkable speed with which old genes take on new tasks suggests that life has unexpected levels of genetic flexibility. In theory, the bacteria should have starved to death and effectively gone extinct. Yet over the course of a weekend, they managed to patch themselves back together with borrowed genes." Scientists made the discovery by accident while researching ways to use naturally occurring bacteria to improve the yield of crops. A microbe was engineered so that it could not make its ‘propeller-like' flagellum and forage for food. However, when a researcher accidentally left the immotile strain out on a lab bench, the team discovered the bacteria had evolved over just a few days. The new variety of bacteria had resurrected their flagella in the process.
Remarkably, this happened because the mutants had rewired a cellular switch, which normally controls nitrogen levels in the cell, to activate the flagellum. This rescued these bacteria, which faced certain death if they didn't move to new food sources. The bacteria being studied, Pseudomonas fluorescens, are among a group of bacteria scientists are researching for use in agriculture, as a kind of ‘plant probiotic'. These could help crops grow or fight off diseases, leading to higher yields. However, a key problem is that the bacteria lack resilience, as their positive effects can stop working after only a short period of time. Dr Jackson, a microbiologist at Reading, said: "Plant probiotics could make crops grow more reliably in the future, helping to feed the world's growing population. This new study shows that these bacteria are more resilient than previously thought, as they show a remarkable capacity to overcome catastrophic changes and find a way to survive. "This gives us crucial insights into how bacteria could survive and change, and the challenge now is to see if this occurs in their natural soil and plant environment." 3
K. Eric Drexler: Engines of Creation 2.0 ( 2006): The T4 phage, acts like a spring-loaded syringe and looks like something out of an industrial parts catalog. It can stick to a bacterium, punch a hole, and inject viral DNA (yes, even bacteria suffer infections). Like a conqueror seizing factories to build more tanks, this DNA then directs the cell’s machines to build more viral DNA and syringes. Like all organisms, these viruses exist because they are fairly stable and are good at getting copies of themselves made. Whether in cells or not, nanomachines obey the universal laws of nature. Ordinary chemical bonds hold their atoms together, and ordinary chemical reactions (guided by other nanomachines) assemble them. Protein molecules can even join to form machines without special help, driven only by thermal agitation and chemical forces. By mixing viral proteins (and the DNA they serve) in a test tube, molecular biologists have assembled working T4 viruses. The machinery of the T4 phage, for example, self-assembles from solution, apparently aided by a single enzyme.…self-assembling structures (…For a description of molecular self-assembly, including that of the T4 phage and the ribosome, see Chapter 36 of Lehninger’s Biochemistry 7)
This ability is surprising: imagine putting automotive parts in a large box, shaking it, and finding an assembled car when you look inside! Yet the T4 virus is but one of many self-assembling structures. (M. YANAGIDA, 1984: The virus particle contains more than 3,000 protein subunits of some 30 polypeptide species !!) Molecular biologists have taken the machinery of the ribosome apart into over fifty separate protein and RNA molecules, and then combined them in test tubes to form working ribosomes again. To see how this happens, imagine different T4 protein chains floating around in water. Each kind folds up to form a lump with distinctive bumps and hollows, covered by distinctive patterns of oiliness, wetness, and electric charge. Picture them wandering and tumbling, jostled by the thermal vibrations of the surrounding water molecules. From time to time two bounce together, then bounce apart. Sometimes, though, two bounce together and fit, bumps in hollows, with sticky patches matching; they then pull together and stick. In this way protein adds to protein to make sections of the virus, and sections assemble to form the whole. 4
E. V. Koonin, the logic of chance, page 376: Breaking the evolution of the translation system into incremental steps, each associated with a biologically plausible selective advantage is extremely difficult even within a speculative scheme let alone experimentally. Speaking of ribosomes, they are so well structured that when broken down into their component parts by chemical catalysts (into long molecular fragments and more than fifty different proteins) they reform into a functioning ribosome as soon as the divisive chemical forces have been removed, independent of any enzymes or assembly machinery – and carry on working. Design some machinery that behaves like this and I personally will build a temple to your name! 5
AlphaFold (2020) What a protein does largely depends on its unique 3D structure. Figuring out what shapes proteins fold into is known as the “protein folding problem”, and has stood as a grand challenge in biology for the past 50 years. In a major scientific advance, the latest version of our AI system AlphaFold has been recognized as a solution to this grand challenge by the organizers of the biennial Critical Assessment of protein Structure Prediction (CASP). 6
Comment: Imagine the engineering effort that it would take for protein engineers to produce nanomachines that would need no nano arms and nano hands to assemble complex nanomachines, but design parts that would be able to assemble on their own just by shaking them, like motors, bearings, and moving parts coming together randomly, and then self-assemble into a fully operational nano-machine. The engineers would need to know the single individual forces and how they would interact with the forces from the other parts. The problem becomes even more apparent when we consider that one of the forces that influence proteins is for example Van der Waals forces which operate based on quantum mechanical principles. R. W. Newberry (2019): The dominant contributors to protein folding include the hydrophobic effect and conventional hydrogen bonding, along with Coulombic interactions and van der Waals interactions. Human technology and advance is far from being able to design this. What a feat would THAT be!
1. Rodrigo S. Galhardo: Extreme Genome Repair 2012 Apr 4.
2. T. Devitt: In the lab, scientists coax E. coli to resist radiation damage March 17, 2014
3. WEEKEND EVOLUTION: BACTERIA 'HOTWIRE THEIR GENES' TO FIX A FAULTY MOTOR 26 February 2015
4. K. Eric Drexler: Engines of Creation 2.0 ( 2006)
5. E. V. Koonin, The logic of chance (2012), page 376
6. AlphaFold: a solution to a 50-year-old grand challenge in biology November 30, 2020
7. https://archive.org/details/biochemistrymole00lehn_0/mode/2up?view=theater&q=biochemistry%2C+lehninger
https://reasonandscience.catsboard.com/t2061p275-my-articles#9626
Extreme Genome Repair (2009): If its naming had followed, rather than preceded, molecular analyses of its DNA, the extremophile bacterium Deinococcus radiodurans might have been called Lazarus. After shattering of its 3.2 Mb genome into 20–30 kb pieces by desiccation or a high dose of ionizing radiation, D. radiodurans miraculously reassembles its genome such that only 3 hr later fully reconstituted nonrearranged chromosomes are present, and the cells carry on, alive as normal 1
T. Devitt (2014): John R. Battista, a professor of biological sciences at Louisiana State University, showed that E. coli could evolve to resist ionizing radiation by exposing cultures of the bacterium to the highly radioactive isotope cobalt-60. “We blasted the cultures until 99 percent of the bacteria were dead. Then we’d grow up the survivors and blast them again. We did that twenty times,” explains Cox. The result were E. coli capable of enduring as much as four orders of magnitude more ionizing radiation, making them similar to Deinococcus radiodurans, a desert-dwelling bacterium found in the 1950s to be remarkably resistant to radiation. That bacterium is capable of surviving more than one thousand times the radiation dose that would kill a human. 2
Simple bacteria can restart their 'outboard motor' by hotwiring their own genes (2015):
Unable to move and facing starvation, the bacteria evolve a replacement flagellum - a rotating tail-like structure that acts like an outboard motor - by patching together a new genetic switch with borrowed parts. When an organism suffers a life-threatening mutation, it can rapidly rewire its genes. The remarkable speed with which old genes take on new tasks suggests that life has unexpected levels of genetic flexibility. In theory, the bacteria should have starved to death and effectively gone extinct. Yet over the course of a weekend, they managed to patch themselves back together with borrowed genes." Scientists made the discovery by accident while researching ways to use naturally occurring bacteria to improve the yield of crops. A microbe was engineered so that it could not make its ‘propeller-like' flagellum and forage for food. However, when a researcher accidentally left the immotile strain out on a lab bench, the team discovered the bacteria had evolved over just a few days. The new variety of bacteria had resurrected their flagella in the process.
Remarkably, this happened because the mutants had rewired a cellular switch, which normally controls nitrogen levels in the cell, to activate the flagellum. This rescued these bacteria, which faced certain death if they didn't move to new food sources. The bacteria being studied, Pseudomonas fluorescens, are among a group of bacteria scientists are researching for use in agriculture, as a kind of ‘plant probiotic'. These could help crops grow or fight off diseases, leading to higher yields. However, a key problem is that the bacteria lack resilience, as their positive effects can stop working after only a short period of time. Dr Jackson, a microbiologist at Reading, said: "Plant probiotics could make crops grow more reliably in the future, helping to feed the world's growing population. This new study shows that these bacteria are more resilient than previously thought, as they show a remarkable capacity to overcome catastrophic changes and find a way to survive. "This gives us crucial insights into how bacteria could survive and change, and the challenge now is to see if this occurs in their natural soil and plant environment." 3
K. Eric Drexler: Engines of Creation 2.0 ( 2006): The T4 phage, acts like a spring-loaded syringe and looks like something out of an industrial parts catalog. It can stick to a bacterium, punch a hole, and inject viral DNA (yes, even bacteria suffer infections). Like a conqueror seizing factories to build more tanks, this DNA then directs the cell’s machines to build more viral DNA and syringes. Like all organisms, these viruses exist because they are fairly stable and are good at getting copies of themselves made. Whether in cells or not, nanomachines obey the universal laws of nature. Ordinary chemical bonds hold their atoms together, and ordinary chemical reactions (guided by other nanomachines) assemble them. Protein molecules can even join to form machines without special help, driven only by thermal agitation and chemical forces. By mixing viral proteins (and the DNA they serve) in a test tube, molecular biologists have assembled working T4 viruses. The machinery of the T4 phage, for example, self-assembles from solution, apparently aided by a single enzyme.…self-assembling structures (…For a description of molecular self-assembly, including that of the T4 phage and the ribosome, see Chapter 36 of Lehninger’s Biochemistry 7)
This ability is surprising: imagine putting automotive parts in a large box, shaking it, and finding an assembled car when you look inside! Yet the T4 virus is but one of many self-assembling structures. (M. YANAGIDA, 1984: The virus particle contains more than 3,000 protein subunits of some 30 polypeptide species !!) Molecular biologists have taken the machinery of the ribosome apart into over fifty separate protein and RNA molecules, and then combined them in test tubes to form working ribosomes again. To see how this happens, imagine different T4 protein chains floating around in water. Each kind folds up to form a lump with distinctive bumps and hollows, covered by distinctive patterns of oiliness, wetness, and electric charge. Picture them wandering and tumbling, jostled by the thermal vibrations of the surrounding water molecules. From time to time two bounce together, then bounce apart. Sometimes, though, two bounce together and fit, bumps in hollows, with sticky patches matching; they then pull together and stick. In this way protein adds to protein to make sections of the virus, and sections assemble to form the whole. 4
E. V. Koonin, the logic of chance, page 376: Breaking the evolution of the translation system into incremental steps, each associated with a biologically plausible selective advantage is extremely difficult even within a speculative scheme let alone experimentally. Speaking of ribosomes, they are so well structured that when broken down into their component parts by chemical catalysts (into long molecular fragments and more than fifty different proteins) they reform into a functioning ribosome as soon as the divisive chemical forces have been removed, independent of any enzymes or assembly machinery – and carry on working. Design some machinery that behaves like this and I personally will build a temple to your name! 5
AlphaFold (2020) What a protein does largely depends on its unique 3D structure. Figuring out what shapes proteins fold into is known as the “protein folding problem”, and has stood as a grand challenge in biology for the past 50 years. In a major scientific advance, the latest version of our AI system AlphaFold has been recognized as a solution to this grand challenge by the organizers of the biennial Critical Assessment of protein Structure Prediction (CASP). 6
Comment: Imagine the engineering effort that it would take for protein engineers to produce nanomachines that would need no nano arms and nano hands to assemble complex nanomachines, but design parts that would be able to assemble on their own just by shaking them, like motors, bearings, and moving parts coming together randomly, and then self-assemble into a fully operational nano-machine. The engineers would need to know the single individual forces and how they would interact with the forces from the other parts. The problem becomes even more apparent when we consider that one of the forces that influence proteins is for example Van der Waals forces which operate based on quantum mechanical principles. R. W. Newberry (2019): The dominant contributors to protein folding include the hydrophobic effect and conventional hydrogen bonding, along with Coulombic interactions and van der Waals interactions. Human technology and advance is far from being able to design this. What a feat would THAT be!
1. Rodrigo S. Galhardo: Extreme Genome Repair 2012 Apr 4.
2. T. Devitt: In the lab, scientists coax E. coli to resist radiation damage March 17, 2014
3. WEEKEND EVOLUTION: BACTERIA 'HOTWIRE THEIR GENES' TO FIX A FAULTY MOTOR 26 February 2015
4. K. Eric Drexler: Engines of Creation 2.0 ( 2006)
5. E. V. Koonin, The logic of chance (2012), page 376
6. AlphaFold: a solution to a 50-year-old grand challenge in biology November 30, 2020
7. https://archive.org/details/biochemistrymole00lehn_0/mode/2up?view=theater&q=biochemistry%2C+lehninger

279 Re: My articles Mon Nov 07, 2022 6:31 am
Re: My articles Mon Nov 07, 2022 6:31 am
Otangelo
Admin
God is the ultimate, and fundamental being, over all.
He glorifies Himself through those that he saved through Christ's suffering on the cross, giving them loving grace, forgiveness for his sins, and eternal life.
But he also glorifies himself over evildoers and those that deny him.
Because he will display His justice to them.
5 Moses 32.35: Vengeance is mine, and recompense, for the time when their foot shall slip; for the day of their calamity is at hand, and their doom comes swiftly.’
Justice is only served when unrighteousness is punished accordingly, and righteously. By judging the world at the end of the day, God will display that he is just, that he is light, and no darkness in Him. All evil will come to light and judged, and justice will be established, and God as the holy, governing his creation in righteousness and justice, confirmed.
The result will be:
Philippians 2:10-11 New Century Version (NCV)
so that every knee will bow to the name of Jesus— everyone in heaven, on earth, and under the earth. And everyone will confess that Jesus Christ is Lord and bring glory to God the Father.
That means, all those that will be condemned, will agree that their condemnation is justified and just. They may curse God in their pain in all eternity, but it will not be because they are angry for thinking they were judged in unrighteousness, but because of their pain.
God also displays His power by being the creator of the micro, and the macro. For knowing each atom, each molecule in our body, and knowing each star by name. He knew how to create life, and is lord over every detail of His creation.
The ribosome is analogous to a human-made 3D printer
Vault particles, made by a 3d Polyribosome nano-printer
https://reasonandscience.catsboard.com/t2112-vault-particles-made-by-a-3d-polyribosome-nano-printer
It is the translation machinery in every cell, that translates the message of DNA, and synthesizes proteins, the molecular machines in the cell. It polymerizes amino acids, or joins one to another, forming long amino acid chains, that afterward fold into 3D, to become functional. Each exercises its specific task and function in the cell.
The marvel of the ribosome is, it has a pocket, called the inner core of the peptidyl transferase center. Researchers reported:
" We identified a single functional group with crucial importance for peptide bond catalysis— namely, the ribose 20'-OH at A2451. This ribose 20 group needs to maintain hydrogen donor characteristics in order to promote effective amide bond formation.
My comment: A precise, minutely orchestrated arrangement of just two main players, the interaction of ribose 2'-OH at position A2451, and the 2’ hydroxyl of the P site substrate A76 is pivotal in orienting substrates in the active site for optimal catalysis, and play a key role in polypeptide bond formation. The ribosome promotes the reaction of the amino acid condensation by properly orienting the reaction substrates. Key in the reaction is the presence of a proton shuttling group. The observed 100-fold reduction in the reaction rate by mutation of the P-site A76 20-OH group is an indication of this group's activity during the peptidyl transfer reaction.
The positioning of all substrates, transition states, and ribosomal residues contributing to the concerted redistribution of charges must be tightly controlled to achieve efficient transpeptidation. This 2'-OH renders almost full catalytic power. These data highlight the unique functional role of the A2451 2'-OH for peptide bond synthesis among all other functional groups at the ribosomal peptidyl transferase active site.
Protein synthesis is VITAL for all life. If that arrangement was not right from the beginning, no life. How did that state of affairs originate? The peptidyl transferase center is often claimed to be the product of the RNA world since it is not made of proteins using amino acids, but RNA. It has a size of almost 3000 RNAs. How did that form prebiotically, if RNA chains over 40 monomer units break down? If RNA is highly unstable, and ribose, depending on the temperatures, can disintegrate in days, usually in 40 days or so?
I go in-depth into the topic of the prebiotic origin of RNA in my recent book: On the Origin of Life and Virus World by means of an Intelligent Designer: The Factory Maker, Paley's Watchmaker Argument 2.0
https://www.amazon.com/-/pt/dp/B0BJ9RYMD6/ref=sr_1_1?__mk_pt_BR=%C3%85M%C3%85%C5%BD%C3%95%C3%91&crid=1487RO32KGIY&keywords=otangelo%20grasso&qid=1665878930&sprefix=otangelo%20grasso%2Caps%2C271&sr=8-1
He is also Lord over the macro world.
Eric Metaxas: Is atheism dead? page 55
Where We Are in the Universe Before we move from this chapter on the fine-tuning of Earth to our next chapter on the fine-tuning of the entire universe, we should touch on what lies between them. Because the very placement of Earth within the universe is an example of fine-tuning. This is probably even harder for us to comprehend than the idea that Jupiter’s and Saturn’s existence is crucial to our existence. But even the position of our solar system within our galaxy— the Milky Way—is vital to the existence of life here on Earth. Our solar system is located on the inner edge of the Orion Arm of our galaxy, about twenty-six thousand light-years from the center. Science now understands that this is crucial to life on Earth in several ways. If we were closer to the galaxy’s center, the radiation hitting us would be far greater, because there are many more stars in the galaxy’s center than out here on the spiral arms where we exist. So at the center there are more “active galactic nucleus outbursts” (AGNs), as well as more supernovae and more gamma ray bursts. That would make life here impossible. We would also be far more likely to be hit by comets, which are more numerous. Gonzalez and Richards call where we are in our solar system the “Galactic Habitable Zone,” meaning that it is the ideal location for a planet like ours to form and support life. But if we were farther out from the center, there would be other problems. Stars farther out are orbited by planets significantly smaller than Earth, so as we have said, that would mean no atmosphere capable of supporting life. Neither would they be able to sustain plate tectonics, which is another element absolutely crucial to life as we know it that we will touch on in Chapter Five. The authors even say that our galaxy is better suited for life than 98 percent of the other galaxies near us. For one thing, it is shaped like a spiral. Stars in elliptical galaxies have less-ordered orbits, like bees flying around a hive, so they are more likely to visit their galaxy’s dangerous central regions. They’re also more likely to pass through interstellar clouds at disastrously high speeds. So in many ways our galaxy —a late-type, metal-rich, spiral galaxy with orderly orbits and comparatively little danger between spiral arms—just happens to be that rare galaxy perfectly suited for life, and our placement within that galaxy also happens to be perfectly suited for life. What shall we make of any of this? Science now tells us that all of these varied parameters are not merely helpful for life on Earth, but are inescapably necessary for it? Can we face that our existence looks like nothing less than a mathematical impossibility? It is as though the more clearly we see these things, the more difficult they are to take in.
https://3lib.net/book/18063091/2dbdee
God is glorified by displaying his love, grace, and mercy through Jesus Christ, who did not fear the cross but obeyed the father, humbly carried it, and permitted it to be nailed on to it like a criminal, naked, and with shame, without having committed any crime or sin.
God is glorified in each YouTube atheist who lies, claiming that there is no evidence for His existence, and calls Him evil, because he judges the world. On judgment day, they will know better, and bow and give Him glory.
God is glorified in His creation. A true master of wisdom, sublime intelligence, and power, far above our comprehension, creating the universe, galaxies, stars, and everything fine-tuned, from the universe to the earth, with creatures of incredible beauty, no human artist comes even close to creating such beauty. Creating atoms with the right laws, sizes, masses, charges, and forces. He created the molecular world, unimagined a hundred years ago, with incredibly ingenious masterfully engineered solutions, arranging atoms and fine-tuning molecules like the nucleobases to permit the right hydrogen bonding to form the Watson Crick ladder, and the DNA information molecule, which is life essential.
God is worthy of worship, glory, praise, and Lordship. You believe? You are beloved and will be rewarded according to your faith. You disbelieve? God will display His glory on you, and one day, you will bow as well.
It's time to take the right decision now. Tomorrow, it could be too late.
How you can get Saved!
https://reasonandscience.catsboard.com/t2501-how-you-can-get-saved
He glorifies Himself through those that he saved through Christ's suffering on the cross, giving them loving grace, forgiveness for his sins, and eternal life.
But he also glorifies himself over evildoers and those that deny him.
Because he will display His justice to them.
5 Moses 32.35: Vengeance is mine, and recompense, for the time when their foot shall slip; for the day of their calamity is at hand, and their doom comes swiftly.’
Justice is only served when unrighteousness is punished accordingly, and righteously. By judging the world at the end of the day, God will display that he is just, that he is light, and no darkness in Him. All evil will come to light and judged, and justice will be established, and God as the holy, governing his creation in righteousness and justice, confirmed.
The result will be:
Philippians 2:10-11 New Century Version (NCV)
so that every knee will bow to the name of Jesus— everyone in heaven, on earth, and under the earth. And everyone will confess that Jesus Christ is Lord and bring glory to God the Father.
That means, all those that will be condemned, will agree that their condemnation is justified and just. They may curse God in their pain in all eternity, but it will not be because they are angry for thinking they were judged in unrighteousness, but because of their pain.
God also displays His power by being the creator of the micro, and the macro. For knowing each atom, each molecule in our body, and knowing each star by name. He knew how to create life, and is lord over every detail of His creation.
The ribosome is analogous to a human-made 3D printer
Vault particles, made by a 3d Polyribosome nano-printer
https://reasonandscience.catsboard.com/t2112-vault-particles-made-by-a-3d-polyribosome-nano-printer
It is the translation machinery in every cell, that translates the message of DNA, and synthesizes proteins, the molecular machines in the cell. It polymerizes amino acids, or joins one to another, forming long amino acid chains, that afterward fold into 3D, to become functional. Each exercises its specific task and function in the cell.
The marvel of the ribosome is, it has a pocket, called the inner core of the peptidyl transferase center. Researchers reported:
" We identified a single functional group with crucial importance for peptide bond catalysis— namely, the ribose 20'-OH at A2451. This ribose 20 group needs to maintain hydrogen donor characteristics in order to promote effective amide bond formation.
My comment: A precise, minutely orchestrated arrangement of just two main players, the interaction of ribose 2'-OH at position A2451, and the 2’ hydroxyl of the P site substrate A76 is pivotal in orienting substrates in the active site for optimal catalysis, and play a key role in polypeptide bond formation. The ribosome promotes the reaction of the amino acid condensation by properly orienting the reaction substrates. Key in the reaction is the presence of a proton shuttling group. The observed 100-fold reduction in the reaction rate by mutation of the P-site A76 20-OH group is an indication of this group's activity during the peptidyl transfer reaction.
The positioning of all substrates, transition states, and ribosomal residues contributing to the concerted redistribution of charges must be tightly controlled to achieve efficient transpeptidation. This 2'-OH renders almost full catalytic power. These data highlight the unique functional role of the A2451 2'-OH for peptide bond synthesis among all other functional groups at the ribosomal peptidyl transferase active site.
Protein synthesis is VITAL for all life. If that arrangement was not right from the beginning, no life. How did that state of affairs originate? The peptidyl transferase center is often claimed to be the product of the RNA world since it is not made of proteins using amino acids, but RNA. It has a size of almost 3000 RNAs. How did that form prebiotically, if RNA chains over 40 monomer units break down? If RNA is highly unstable, and ribose, depending on the temperatures, can disintegrate in days, usually in 40 days or so?
I go in-depth into the topic of the prebiotic origin of RNA in my recent book: On the Origin of Life and Virus World by means of an Intelligent Designer: The Factory Maker, Paley's Watchmaker Argument 2.0
https://www.amazon.com/-/pt/dp/B0BJ9RYMD6/ref=sr_1_1?__mk_pt_BR=%C3%85M%C3%85%C5%BD%C3%95%C3%91&crid=1487RO32KGIY&keywords=otangelo%20grasso&qid=1665878930&sprefix=otangelo%20grasso%2Caps%2C271&sr=8-1
He is also Lord over the macro world.
Eric Metaxas: Is atheism dead? page 55
Where We Are in the Universe Before we move from this chapter on the fine-tuning of Earth to our next chapter on the fine-tuning of the entire universe, we should touch on what lies between them. Because the very placement of Earth within the universe is an example of fine-tuning. This is probably even harder for us to comprehend than the idea that Jupiter’s and Saturn’s existence is crucial to our existence. But even the position of our solar system within our galaxy— the Milky Way—is vital to the existence of life here on Earth. Our solar system is located on the inner edge of the Orion Arm of our galaxy, about twenty-six thousand light-years from the center. Science now understands that this is crucial to life on Earth in several ways. If we were closer to the galaxy’s center, the radiation hitting us would be far greater, because there are many more stars in the galaxy’s center than out here on the spiral arms where we exist. So at the center there are more “active galactic nucleus outbursts” (AGNs), as well as more supernovae and more gamma ray bursts. That would make life here impossible. We would also be far more likely to be hit by comets, which are more numerous. Gonzalez and Richards call where we are in our solar system the “Galactic Habitable Zone,” meaning that it is the ideal location for a planet like ours to form and support life. But if we were farther out from the center, there would be other problems. Stars farther out are orbited by planets significantly smaller than Earth, so as we have said, that would mean no atmosphere capable of supporting life. Neither would they be able to sustain plate tectonics, which is another element absolutely crucial to life as we know it that we will touch on in Chapter Five. The authors even say that our galaxy is better suited for life than 98 percent of the other galaxies near us. For one thing, it is shaped like a spiral. Stars in elliptical galaxies have less-ordered orbits, like bees flying around a hive, so they are more likely to visit their galaxy’s dangerous central regions. They’re also more likely to pass through interstellar clouds at disastrously high speeds. So in many ways our galaxy —a late-type, metal-rich, spiral galaxy with orderly orbits and comparatively little danger between spiral arms—just happens to be that rare galaxy perfectly suited for life, and our placement within that galaxy also happens to be perfectly suited for life. What shall we make of any of this? Science now tells us that all of these varied parameters are not merely helpful for life on Earth, but are inescapably necessary for it? Can we face that our existence looks like nothing less than a mathematical impossibility? It is as though the more clearly we see these things, the more difficult they are to take in.
https://3lib.net/book/18063091/2dbdee
God is glorified by displaying his love, grace, and mercy through Jesus Christ, who did not fear the cross but obeyed the father, humbly carried it, and permitted it to be nailed on to it like a criminal, naked, and with shame, without having committed any crime or sin.
God is glorified in each YouTube atheist who lies, claiming that there is no evidence for His existence, and calls Him evil, because he judges the world. On judgment day, they will know better, and bow and give Him glory.
God is glorified in His creation. A true master of wisdom, sublime intelligence, and power, far above our comprehension, creating the universe, galaxies, stars, and everything fine-tuned, from the universe to the earth, with creatures of incredible beauty, no human artist comes even close to creating such beauty. Creating atoms with the right laws, sizes, masses, charges, and forces. He created the molecular world, unimagined a hundred years ago, with incredibly ingenious masterfully engineered solutions, arranging atoms and fine-tuning molecules like the nucleobases to permit the right hydrogen bonding to form the Watson Crick ladder, and the DNA information molecule, which is life essential.
God is worthy of worship, glory, praise, and Lordship. You believe? You are beloved and will be rewarded according to your faith. You disbelieve? God will display His glory on you, and one day, you will bow as well.
It's time to take the right decision now. Tomorrow, it could be too late.
How you can get Saved!
https://reasonandscience.catsboard.com/t2501-how-you-can-get-saved
280 Elaborated Tunnel Architectures in Enzyme Systems point to a designed setup Wed Nov 09, 2022 8:12 am
Elaborated Tunnel Architectures in Enzyme Systems point to a designed setup Wed Nov 09, 2022 8:12 am
Otangelo
Admin
Elaborated Tunnel Architectures in Enzyme Systems point to a designed setup
RNA and DNA belong to the four basic building blocks of life. They are complex macromolecules made of three constituents: the base, the backbone, which is the ribose five-carbon sugar, and phosphate, the moiety which permits DNA polymerization and catenation of monomers, to become polymers. The nucleobases are divided into pyrimidine and purines. These bases must be made in complex biosynthesis pathways in the cell, requiring several molecular machines, and enzymes, that perform the gradual, stepwise operations to yield the nucleobases, which, in the end, are handed over for further processing. Pyrimidines, one of the two classes, require 7 enzymes, of which Carbamoyl phosphate synthase II is the first in the production line.
In bacteria, a single enzyme supplies carbamoyl phosphate for the synthesis of arginine and pyrimidines. The bacterial enzyme has three separate active sites, spaced along a tunnel nearly 100 Å long. Bacterial carbamoyl phosphate synthetase provides a vivid illustration of the channeling of unstable reaction intermediates between active sites. This reaction consumes two molecules of ATP: One provides a phosphate group and the other energizes the reaction. The need for this channel exists to efficiently translocate reactive gaseous molecules that can either be toxic to the cell or are reactive intermediates that need to be delivered to complete a coupled reaction.
Comment: Consider that no lifeform exists that does not use DNA and RNA. Therefore, the synthesis of these molecules is a prerequisite for life. The origin of this metabolic pathway can therefore not be explained through evolution. Either it was design or random nonguided fortunate events.
Tunnel Architectures in Enzyme Systems that Transport Gaseous Substrates
Derinkuyu Underground City in Cappadocia, Turkey, is one of the deepest and most fascinating multilevel subterranean cities, excavated in tunnel systems. Specifically constructed, elaborated Air ducts ensure fresh oxygen supply, and the oxygen ratio inside never changes no matter at what level one is in. Such systems are always engineering marvels, and must be precisely calculated, and constructed. Remarkably, some proteins act similarly and exist in molecular biological systems.
Ruchi Anand (2021): Tunnels connect the protein surface to the active site or one active site with the others and serve as conduits for the convenient delivery of molecules. Tunnels transferring small molecules such as N2, CH4, C2H6, O2, CO, NH3, H2, C2H2, NO, and CO2 are termed gaseous tunnels. Conduits that have a surface-accessible connection and can accept gases from the surroundings are named external gaseous (EG) tunnels. Whereas, buried gaseous tunnels that do not emerge to the surface are named internal gaseous (IG) tunnels. In some cases, the tunnels can be performed, permanently visible within the protein structure such that the natural breathing motions in proteins do not alter the tunnel dimensions to the extent that the radius of the gaseous tunnel falls below the minimum threshold diameter, e.g., carbamoyl phosphate synthetase (CPS) has a preformed tunnel. In contrast, it can be transient such that the tunnel diameter is not sufficiently wide enough to allow the incoming molecule to pass through it or certain constrictions in the tunnel block its delivery. This could be either to control the frequency of molecules traveling across or to coordinate and facilitate coupled reaction rates. Another possible scenario of transient tunnel formation is one in which the tunnel is nonexistent in the apo state, and only upon significant conformational change, under appropriate cues, is the tunnel formed. In several cases transient tunnels require intermediate/substrate-induced conformational changes in the tunnel residues to open up for the transport of the incoming molecule, within the respective enzyme. These tunnels undergo enormous fluctuations and switch between open and close states. It is remarkable that the presence of these conduits, which are as long as 20−30 Å and even longer like 96 Å in CPS,6a run inside the protein body, forming pores that serve as highways for transport of these gaseous molecules. In several cases, an added level of tuning into the tunnel architecture is introduced by incorporating gating mechanisms into the EG and IG tunnel architectures.
Gates serve as checkpoints and vary from system to system; some are as simple as an amino acid blocking the path which moves out upon receiving appropriate cues such as the swinging door type in cytidine triphosphate synthase (CTP) and in others more complex arrangement of amino acids come together to form control units such as aperture gates, drawbridge, and shell type gates. These tunnels and their gates are connected via an active communication network that spans between distal centers and hence introduces both conformation and dynamic allostery into the protein systems. It is not uncommon to observe long-distance allosteric networks that can be dynamic in nature and transiently formed via the motion of loop elements, secondary structural rearrangements, or of entire domains.
EXTERNAL GASEOUS (EG) TUNNEL ARCHITECTURES
EG tunnels connect the bulk solvent with the active site of an enzyme. These tunnels are found in several enzymes that accept gaseous substrates to facilitate their delivery to the buried active site. A class of predominant gaseous substrates are alkanes such as methane and ethane gases that are oxidized aerobically or via anaerobic pathways. Recently, the crystal structure of the enzyme that anaerobically oxidizes ethane to ethylCoM from Candidatus Ethanoperedens thermophilum was determined, and named it ethylCoM reductase. The enzyme belongs to the broad methylCoM reductase superfamily, which oxidizes methane. The ethylCoM reductase has a 33 Å tunnel that runs across the length of the protein. Interestingly, the EG tunnel present in ethylCoM reductase has some very unique features. At the end of the tunnel, near the Ni-cofactor F430 active site, there are several residues that are post-translationally modified. Methylated amino acids, such as S-methylcysteine, 3-methylisoleucine, 2(S)-methylglutamine, and N2 -methylhistidine line the tunnel. It is likely that these residues tune the enzyme to select for ethane by creating a very hydrophobic environment and prevent similar-sized hydrophilic molecules such as methanol from reaching the active center. The larger hydrophobic alkanes are selected out via optimization of the tunnel diameter, which is fit to accommodate ethane. Another example of an alkane transporting tunnel exists in soluble methane monooxygenase (sMMO) that performs C− H functionalization by breaking the strongest C−H bond, among saturated hydrocarbons, in methane and aerobically oxidizes it to form methanol. In methanotrophs, these enzymes are tightly regulated, and the complex formation between the two proteins, hydroxylase MMOH and regulatory protein MMOB, is required for function. The EG tunnel formed in this system is very hydrophobic, and the diameter is such that it only allows for smaller gases such as methane and O2 to percolate into the di-Fe cluster harboring active site. In Methylosinus trichosporium OB3b, half of the tunnel is at the interface of the MMOH/MMOB complex, and another half of the tunnel is buried within MMOH, where the oxidation reaction is catalyzed. As an added control feature, the complex has multiple gates to regulate its function. Residues W308 and P215 guard the entrance of the substrate molecules and block the formation of the EG tunnel in the absence of the complex between MMOH and MMOB.
Comment: This demonstrates and exemplifies how in many cases, single monomers have important functions, and changing them through mutations can remove the function of the entirety of the enzyme.
Upon complexation, a conformational change is triggered, and these residues move out of the path, opening the passage for the entire tunnel. When the upper gating residues move upon MMOB/MMOH complex formation, another residue F282 right near the active site also concomitantly undergoes a shift, allowing methane and oxygen to access the di-Fe center. MMOH also has an alternative secondary hydrophilic passage, accessible only when MMOB/MMOH complex dissociates which allows the polar methanol product to be released through it. The gating residues, F282 in the hydrophobic EG tunnel and E240 in the hydrophilic passage, switch between open and close states alternately upon binding/unbinding of MMOB and hence opens one of the two tunnels at a time. This regulates the flow of substrates and products and avoids overoxidation of methanol by releasing it through the hydrophilic passage prior to the entry of substrates in the active site via the hydrophobic EG tunnel.
One of the most common gaseous substrates for which several examples of tunneling enzymes exist is oxygen (O2). It is used in several important oxidation reactions for the generation of essential pathway intermediates and also is a key transport gas in cells. Interestingly in several cases, oxygen is transported to the desired site via molecular tunnels, perhaps to modulate its flow. There are two types of tunnel architectures that are prevalent: first, where there is a main tunnel connected to several subsidiary tunnels, and second, those with fewer tunnels but with stringent gating controls. For instance, soybean lipoxygenase-1 is an example of a multitunnel system that has eight EG tunnels, out of which the one that is formed by hydrophobic residues, such as L496, I553, I547, and V564, has the highest throughput and is identified as the main gaseous tunnel for delivering O2 to the reaction center. It catalyzes the stereospecific peroxidation of linoleic acid via forming a pentadienyl radical intermediate. Under oxygen-deficient conditions, the intermediate escapes from the active site to the bulk and forms four products, i.e., 13S-, 13R-, 9S-, and 9R-hydroperoxy-octadecadienoic acid, in equal distributions. However, under ambient O2 conditions, the EG tunnel delivers O2 efficiently into the active site which has a properly positioned and oriented radical intermediate. Here, O2 is delivered by the EG tunnel such that it stereo- and regiospecifically attacks the radical intermediate to yield 13S-hydroperoxy-octadecadienoic acid as a major product with ∼90% yield. It has also been shown that when the EG tunnel residue L496 is mutated to a bulky tryptophan, it opens up a new gaseous tunnel for O2 delivery, where it attacks at the different side of the pentadienyl intermediate, preferring the formation of 9S- and 9Rproducts. This example showed the importance of the gaseous tunnel in determining the stereo- and regiospecificity for product formation
INTERNAL GASEOUS (IG) TUNNEL ARCHITECTURES
While the EG tunnels transport gases and have pores that are accessible to the surface, there is another class of tunnels formed within the core of the enzyme system, buried in the body of the protein, called the IG tunnels.
Question: How could these tunnels be the product of evolutionary pressures, requiring long periods of time, if, in case the tunnel that protects the toxic intermediates is not instantiated from the beginning, the products would leak, and eventually kill the cell? This is an all-or-nothing business, where these tunnels had to be created right from the start, fully set up and developed.
These systems generally have the tunnel connecting two reactive centers, and the product of one reaction is transported to the second active site. In some cases, an IG tunnel network, instead of leading to another active site, can also lead to the lipid membrane so as to directly access the active site of membrane-bound enzymes. The substrate is generated within one of the active centers and is in the limiting amount as well as it could be toxic or unstable in the presented environment. Therefore, to ensure it reaches the destination reaction center, nature has devised strategies by constructing IG tunnels which, in several instances, are transient tunnels that only form upon entry of substates and have much more controlled and complex gating architectures. 57
Comment: This is truly fascinating evidence of intended design for important functions: To direct gases to where they are needed to perform a reaction.
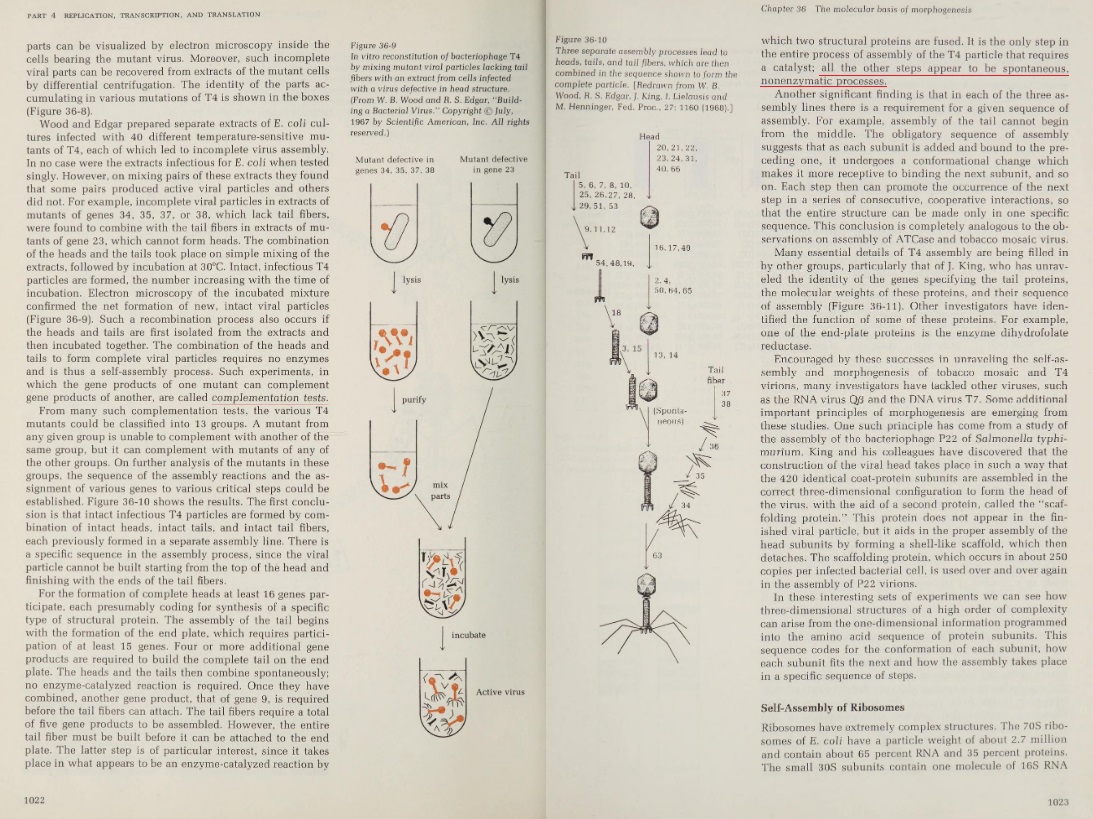
Image description: The structure of carbamoyl phosphate synthetase
The small subunit that contains the active site for the hydrolysis of glutamine is shown in green. The N-terminal domain of the large subunit that contains the active site for the synthesis of carboxy phosphate and carbamate is shown in red. The C-terminal domain of the large subunit that contains the active site for the synthesis of carbamoyl phosphate is shown in blue. The two molecular tunnels for the translocation of ammonia and carbamate are shown in yellow dotted lines 56
Nucleotide metabolism: By evolution?
G. Caetano-Anollés (2013): The origin of metabolism has been linked to abiotic chemistries that existed in our planet at the beginning of life. While plausible chemical pathways have been proposed, including the synthesis of nucleobases, ribose and ribonucleotides, the cooption of these reactions by modern enzymes remains shrouded in mystery. Pathways of nucleotide biosynthesis, catabolism, and salvage originated ∼300 million years later by concerted enzymatic recruitments and gradual replacement of abiotic chemistries. The simultaneous appearance of purine biosynthesis and the ribosome probably fulfilled the expanding matter-energy and processing needs of genomic information. 59
Comment: These are assertions, clearly not based on scientific data and observations, but ad-hoc conclusions that lack evidence.
RNA and DNA belong to the four basic building blocks of life. They are complex macromolecules made of three constituents: the base, the backbone, which is the ribose five-carbon sugar, and phosphate, the moiety which permits DNA polymerization and catenation of monomers, to become polymers. The nucleobases are divided into pyrimidine and purines. These bases must be made in complex biosynthesis pathways in the cell, requiring several molecular machines, and enzymes, that perform the gradual, stepwise operations to yield the nucleobases, which, in the end, are handed over for further processing. Pyrimidines, one of the two classes, require 7 enzymes, of which Carbamoyl phosphate synthase II is the first in the production line.
In bacteria, a single enzyme supplies carbamoyl phosphate for the synthesis of arginine and pyrimidines. The bacterial enzyme has three separate active sites, spaced along a tunnel nearly 100 Å long. Bacterial carbamoyl phosphate synthetase provides a vivid illustration of the channeling of unstable reaction intermediates between active sites. This reaction consumes two molecules of ATP: One provides a phosphate group and the other energizes the reaction. The need for this channel exists to efficiently translocate reactive gaseous molecules that can either be toxic to the cell or are reactive intermediates that need to be delivered to complete a coupled reaction.
Comment: Consider that no lifeform exists that does not use DNA and RNA. Therefore, the synthesis of these molecules is a prerequisite for life. The origin of this metabolic pathway can therefore not be explained through evolution. Either it was design or random nonguided fortunate events.
Tunnel Architectures in Enzyme Systems that Transport Gaseous Substrates
Derinkuyu Underground City in Cappadocia, Turkey, is one of the deepest and most fascinating multilevel subterranean cities, excavated in tunnel systems. Specifically constructed, elaborated Air ducts ensure fresh oxygen supply, and the oxygen ratio inside never changes no matter at what level one is in. Such systems are always engineering marvels, and must be precisely calculated, and constructed. Remarkably, some proteins act similarly and exist in molecular biological systems.
Ruchi Anand (2021): Tunnels connect the protein surface to the active site or one active site with the others and serve as conduits for the convenient delivery of molecules. Tunnels transferring small molecules such as N2, CH4, C2H6, O2, CO, NH3, H2, C2H2, NO, and CO2 are termed gaseous tunnels. Conduits that have a surface-accessible connection and can accept gases from the surroundings are named external gaseous (EG) tunnels. Whereas, buried gaseous tunnels that do not emerge to the surface are named internal gaseous (IG) tunnels. In some cases, the tunnels can be performed, permanently visible within the protein structure such that the natural breathing motions in proteins do not alter the tunnel dimensions to the extent that the radius of the gaseous tunnel falls below the minimum threshold diameter, e.g., carbamoyl phosphate synthetase (CPS) has a preformed tunnel. In contrast, it can be transient such that the tunnel diameter is not sufficiently wide enough to allow the incoming molecule to pass through it or certain constrictions in the tunnel block its delivery. This could be either to control the frequency of molecules traveling across or to coordinate and facilitate coupled reaction rates. Another possible scenario of transient tunnel formation is one in which the tunnel is nonexistent in the apo state, and only upon significant conformational change, under appropriate cues, is the tunnel formed. In several cases transient tunnels require intermediate/substrate-induced conformational changes in the tunnel residues to open up for the transport of the incoming molecule, within the respective enzyme. These tunnels undergo enormous fluctuations and switch between open and close states. It is remarkable that the presence of these conduits, which are as long as 20−30 Å and even longer like 96 Å in CPS,6a run inside the protein body, forming pores that serve as highways for transport of these gaseous molecules. In several cases, an added level of tuning into the tunnel architecture is introduced by incorporating gating mechanisms into the EG and IG tunnel architectures.
Gates serve as checkpoints and vary from system to system; some are as simple as an amino acid blocking the path which moves out upon receiving appropriate cues such as the swinging door type in cytidine triphosphate synthase (CTP) and in others more complex arrangement of amino acids come together to form control units such as aperture gates, drawbridge, and shell type gates. These tunnels and their gates are connected via an active communication network that spans between distal centers and hence introduces both conformation and dynamic allostery into the protein systems. It is not uncommon to observe long-distance allosteric networks that can be dynamic in nature and transiently formed via the motion of loop elements, secondary structural rearrangements, or of entire domains.
EXTERNAL GASEOUS (EG) TUNNEL ARCHITECTURES
EG tunnels connect the bulk solvent with the active site of an enzyme. These tunnels are found in several enzymes that accept gaseous substrates to facilitate their delivery to the buried active site. A class of predominant gaseous substrates are alkanes such as methane and ethane gases that are oxidized aerobically or via anaerobic pathways. Recently, the crystal structure of the enzyme that anaerobically oxidizes ethane to ethylCoM from Candidatus Ethanoperedens thermophilum was determined, and named it ethylCoM reductase. The enzyme belongs to the broad methylCoM reductase superfamily, which oxidizes methane. The ethylCoM reductase has a 33 Å tunnel that runs across the length of the protein. Interestingly, the EG tunnel present in ethylCoM reductase has some very unique features. At the end of the tunnel, near the Ni-cofactor F430 active site, there are several residues that are post-translationally modified. Methylated amino acids, such as S-methylcysteine, 3-methylisoleucine, 2(S)-methylglutamine, and N2 -methylhistidine line the tunnel. It is likely that these residues tune the enzyme to select for ethane by creating a very hydrophobic environment and prevent similar-sized hydrophilic molecules such as methanol from reaching the active center. The larger hydrophobic alkanes are selected out via optimization of the tunnel diameter, which is fit to accommodate ethane. Another example of an alkane transporting tunnel exists in soluble methane monooxygenase (sMMO) that performs C− H functionalization by breaking the strongest C−H bond, among saturated hydrocarbons, in methane and aerobically oxidizes it to form methanol. In methanotrophs, these enzymes are tightly regulated, and the complex formation between the two proteins, hydroxylase MMOH and regulatory protein MMOB, is required for function. The EG tunnel formed in this system is very hydrophobic, and the diameter is such that it only allows for smaller gases such as methane and O2 to percolate into the di-Fe cluster harboring active site. In Methylosinus trichosporium OB3b, half of the tunnel is at the interface of the MMOH/MMOB complex, and another half of the tunnel is buried within MMOH, where the oxidation reaction is catalyzed. As an added control feature, the complex has multiple gates to regulate its function. Residues W308 and P215 guard the entrance of the substrate molecules and block the formation of the EG tunnel in the absence of the complex between MMOH and MMOB.
Comment: This demonstrates and exemplifies how in many cases, single monomers have important functions, and changing them through mutations can remove the function of the entirety of the enzyme.
Upon complexation, a conformational change is triggered, and these residues move out of the path, opening the passage for the entire tunnel. When the upper gating residues move upon MMOB/MMOH complex formation, another residue F282 right near the active site also concomitantly undergoes a shift, allowing methane and oxygen to access the di-Fe center. MMOH also has an alternative secondary hydrophilic passage, accessible only when MMOB/MMOH complex dissociates which allows the polar methanol product to be released through it. The gating residues, F282 in the hydrophobic EG tunnel and E240 in the hydrophilic passage, switch between open and close states alternately upon binding/unbinding of MMOB and hence opens one of the two tunnels at a time. This regulates the flow of substrates and products and avoids overoxidation of methanol by releasing it through the hydrophilic passage prior to the entry of substrates in the active site via the hydrophobic EG tunnel.
One of the most common gaseous substrates for which several examples of tunneling enzymes exist is oxygen (O2). It is used in several important oxidation reactions for the generation of essential pathway intermediates and also is a key transport gas in cells. Interestingly in several cases, oxygen is transported to the desired site via molecular tunnels, perhaps to modulate its flow. There are two types of tunnel architectures that are prevalent: first, where there is a main tunnel connected to several subsidiary tunnels, and second, those with fewer tunnels but with stringent gating controls. For instance, soybean lipoxygenase-1 is an example of a multitunnel system that has eight EG tunnels, out of which the one that is formed by hydrophobic residues, such as L496, I553, I547, and V564, has the highest throughput and is identified as the main gaseous tunnel for delivering O2 to the reaction center. It catalyzes the stereospecific peroxidation of linoleic acid via forming a pentadienyl radical intermediate. Under oxygen-deficient conditions, the intermediate escapes from the active site to the bulk and forms four products, i.e., 13S-, 13R-, 9S-, and 9R-hydroperoxy-octadecadienoic acid, in equal distributions. However, under ambient O2 conditions, the EG tunnel delivers O2 efficiently into the active site which has a properly positioned and oriented radical intermediate. Here, O2 is delivered by the EG tunnel such that it stereo- and regiospecifically attacks the radical intermediate to yield 13S-hydroperoxy-octadecadienoic acid as a major product with ∼90% yield. It has also been shown that when the EG tunnel residue L496 is mutated to a bulky tryptophan, it opens up a new gaseous tunnel for O2 delivery, where it attacks at the different side of the pentadienyl intermediate, preferring the formation of 9S- and 9Rproducts. This example showed the importance of the gaseous tunnel in determining the stereo- and regiospecificity for product formation
INTERNAL GASEOUS (IG) TUNNEL ARCHITECTURES
While the EG tunnels transport gases and have pores that are accessible to the surface, there is another class of tunnels formed within the core of the enzyme system, buried in the body of the protein, called the IG tunnels.
Question: How could these tunnels be the product of evolutionary pressures, requiring long periods of time, if, in case the tunnel that protects the toxic intermediates is not instantiated from the beginning, the products would leak, and eventually kill the cell? This is an all-or-nothing business, where these tunnels had to be created right from the start, fully set up and developed.
These systems generally have the tunnel connecting two reactive centers, and the product of one reaction is transported to the second active site. In some cases, an IG tunnel network, instead of leading to another active site, can also lead to the lipid membrane so as to directly access the active site of membrane-bound enzymes. The substrate is generated within one of the active centers and is in the limiting amount as well as it could be toxic or unstable in the presented environment. Therefore, to ensure it reaches the destination reaction center, nature has devised strategies by constructing IG tunnels which, in several instances, are transient tunnels that only form upon entry of substates and have much more controlled and complex gating architectures. 57
Comment: This is truly fascinating evidence of intended design for important functions: To direct gases to where they are needed to perform a reaction.
Image description: The structure of carbamoyl phosphate synthetase
The small subunit that contains the active site for the hydrolysis of glutamine is shown in green. The N-terminal domain of the large subunit that contains the active site for the synthesis of carboxy phosphate and carbamate is shown in red. The C-terminal domain of the large subunit that contains the active site for the synthesis of carbamoyl phosphate is shown in blue. The two molecular tunnels for the translocation of ammonia and carbamate are shown in yellow dotted lines 56
Nucleotide metabolism: By evolution?
G. Caetano-Anollés (2013): The origin of metabolism has been linked to abiotic chemistries that existed in our planet at the beginning of life. While plausible chemical pathways have been proposed, including the synthesis of nucleobases, ribose and ribonucleotides, the cooption of these reactions by modern enzymes remains shrouded in mystery. Pathways of nucleotide biosynthesis, catabolism, and salvage originated ∼300 million years later by concerted enzymatic recruitments and gradual replacement of abiotic chemistries. The simultaneous appearance of purine biosynthesis and the ribosome probably fulfilled the expanding matter-energy and processing needs of genomic information. 59
Comment: These are assertions, clearly not based on scientific data and observations, but ad-hoc conclusions that lack evidence.

281 Re: My articles Mon Nov 14, 2022 4:24 am
Re: My articles Mon Nov 14, 2022 4:24 am
Otangelo
Admin
I think that in recent years, atheists are more and more becoming aware that they are losing the battle in regard to teleological arguments. The force of the evidence is too strong to be rationally denied. That is especially true in regard to the origin and fine-tuning of the universe, the origin of the laws of physics, the origin of complex instructional information, and the irreducibility of biological systems.
One of the adopted tactics by those that have decided, that God cannot be permitted to give even the tiniest step into the door, is shifting from strong to weak atheism, in order to remove the burden of proof from their shoulders.
Another tactic in order not to look silly has been to avoid discussing design altogether, arguing about moral issues, and accusing the God of the Bible of being immoral, as Dawkins did famously in his book: The God delusion.
One of the questions raised and argued about recently, that I have seen, is: God is evil because he created evil. Since he had the foresight and knew that in this world, Adam and Eve would sin, and sin would enter the world, and since he did not prevent it, he is ultimately responsible for evil, therefore he is evil.
But is that so? What are the answers? Is God like a surgeon that permitted pain to achieve a greater good? Did he permit evil to display what goodness is, contrasting it with evil? Did he permit it in order to display his redemptive power, and put in practice his plan of salvation, demonstrating his love, and willingness to be graceful and merciful?
But was that justified, considering, as Jesus said and foresaw, that most would walk on the highway to hell? Is, putting everything on balance, the amount of suffering not greater than the amount of joy and happiness, only enjoyed by those saved?
My standard answer to this is: How could or should I have the capacity, based on my limited knowledge and perspective, to judge God's plan, and intentions, on a holistic scale?
One of the adopted tactics by those that have decided, that God cannot be permitted to give even the tiniest step into the door, is shifting from strong to weak atheism, in order to remove the burden of proof from their shoulders.
Another tactic in order not to look silly has been to avoid discussing design altogether, arguing about moral issues, and accusing the God of the Bible of being immoral, as Dawkins did famously in his book: The God delusion.
One of the questions raised and argued about recently, that I have seen, is: God is evil because he created evil. Since he had the foresight and knew that in this world, Adam and Eve would sin, and sin would enter the world, and since he did not prevent it, he is ultimately responsible for evil, therefore he is evil.
But is that so? What are the answers? Is God like a surgeon that permitted pain to achieve a greater good? Did he permit evil to display what goodness is, contrasting it with evil? Did he permit it in order to display his redemptive power, and put in practice his plan of salvation, demonstrating his love, and willingness to be graceful and merciful?
But was that justified, considering, as Jesus said and foresaw, that most would walk on the highway to hell? Is, putting everything on balance, the amount of suffering not greater than the amount of joy and happiness, only enjoyed by those saved?
My standard answer to this is: How could or should I have the capacity, based on my limited knowledge and perspective, to judge God's plan, and intentions, on a holistic scale?
282 Re: My articles Tue Nov 29, 2022 5:06 am
Re: My articles Tue Nov 29, 2022 5:06 am
Otangelo
Admin
Hell does not exist, because God is evil, but because he is just. Likewise, people do not go to hell because God is evil, but because man is sinful, and deserves to be punished for his evil deeds. A righteous God cannot let sin and evil remain unpunished, otherwise, he would have created an unjust world.
The made-up invented God in Islam is unjust. He forgives sins without punishment. Imagine: There is a rich man, that had his entire family kidnapped for a ransom. The rich man paid with all his belongings, but the kidnappers tortured and killed his entire family. Then they are cached and brought to the judge. The judge asks the (now) poor man: What shall I do with the perpetrators that annihilated your family, and ruined your life?
He says: Punish them accordingly, with the hardest punishment permitted by law. The criminals however say to the judge: Sir, please forgive us, be merciful, we are repenting for what we did. We are so sorry. Then the judge says: Ok, I attend to your demand. You can go.
How would the victim feel? Obviously, outraged and even worse than before. A price has to be paid in order for justice to be maintained. The principle: evil demands punishment is the foundation of justice and is untradeable. A holy and just God cannot let pass evil unpunished. It does not matter, if just a "harmless" lie, or perpetuating an unjust war for the sake of one's own greatness, glory, and power, at the cost of millions of deaths. Sin is sin, and demands punishment. A holy God cannot let sin go, unpunished. That is the principle applied in the Bible, by the God of the Bible. That's what distinguishes Jahweh from Allah. The two cannot be conciliated. One is a different God than the other. Claiming that the God of Islam is the same deity as the God of the Bible is silly.
Unbelievers complain that this world is unjust, and not the best possible world. On what ground?
The made-up invented God in Islam is unjust. He forgives sins without punishment. Imagine: There is a rich man, that had his entire family kidnapped for a ransom. The rich man paid with all his belongings, but the kidnappers tortured and killed his entire family. Then they are cached and brought to the judge. The judge asks the (now) poor man: What shall I do with the perpetrators that annihilated your family, and ruined your life?
He says: Punish them accordingly, with the hardest punishment permitted by law. The criminals however say to the judge: Sir, please forgive us, be merciful, we are repenting for what we did. We are so sorry. Then the judge says: Ok, I attend to your demand. You can go.
How would the victim feel? Obviously, outraged and even worse than before. A price has to be paid in order for justice to be maintained. The principle: evil demands punishment is the foundation of justice and is untradeable. A holy and just God cannot let pass evil unpunished. It does not matter, if just a "harmless" lie, or perpetuating an unjust war for the sake of one's own greatness, glory, and power, at the cost of millions of deaths. Sin is sin, and demands punishment. A holy God cannot let sin go, unpunished. That is the principle applied in the Bible, by the God of the Bible. That's what distinguishes Jahweh from Allah. The two cannot be conciliated. One is a different God than the other. Claiming that the God of Islam is the same deity as the God of the Bible is silly.
Unbelievers complain that this world is unjust, and not the best possible world. On what ground?
283 Re: My articles Sat May 27, 2023 6:45 pm
Re: My articles Sat May 27, 2023 6:45 pm
Otangelo
Admin
Maybe you have heard the claim that the more science advances, the less God can hide in a gap of knowledge. Thats hogwash. The contrary is the case. The more science advances, the less naturalism can hide in the gaps of knowledge. See the state of knowledge a hundred years ago, and compare it to now:
Static eternal universe ---> Expanding finite-age universe
Ignorance of fine-tuning ---> knowledge of extensive fine-tuning
Cells are simple protoplasm, and to get life is a pretty simple pathway ---> naturalistic OOL is next-to-if, not impossible-to-explain
Biological systems are probably complicated ---> biological evolution of information-based and directed machines are next-to, if not -impossible-to-explain by unguided natural means.
The truth is, the more science advances, the more it points to an intelligent mind being the best explanation for all phenomena.
Unguided random events are implausible to the extreme.
Static eternal universe ---> Expanding finite-age universe
Ignorance of fine-tuning ---> knowledge of extensive fine-tuning
Cells are simple protoplasm, and to get life is a pretty simple pathway ---> naturalistic OOL is next-to-if, not impossible-to-explain
Biological systems are probably complicated ---> biological evolution of information-based and directed machines are next-to, if not -impossible-to-explain by unguided natural means.
The truth is, the more science advances, the more it points to an intelligent mind being the best explanation for all phenomena.
Unguided random events are implausible to the extreme.
284 Cellular Communication: The amazing size of Information Exchange in the Human Body Mon Aug 07, 2023 6:33 am
Cellular Communication: The amazing size of Information Exchange in the Human Body Mon Aug 07, 2023 6:33 am
Otangelo
Admin
"Cellular Communication: The amazing size of Information Exchange in the Human Body"
We have 37 trillion cells in our human body that communicate with each other, like a huge WWW.
The Human Genetic code information content is about 6 billion bits. The Epigenetic code information content is about ten times larger:≈ 60 billion bits, Sum, total, 66 billion bits.
To find the total estimated information content of the 37 trillion cells in the human body, we can multiply the 66 billion bits by the number of cells:
Total estimated information content = 66 billion bits × 37 trillion cells = 2.44 quintillion bits.
Now, let's compare this to the daily global internet traffic: Daily internet traffic ≈ 8 quintillion bits (8 x 10^18 bits)
Comparing the two values:
Total information content of human cells ≈ 2.44 quintillion bits (2.44 x 10^18 bits)
Daily internet traffic ≈ 8 quintillion bits (8 x 10^18 bits)
The daily global internet traffic (8 quintillion bits) is roughly three times larger than the estimated total information content within the 37 trillion cells in the human body (2.44 quintillion bits).
So the information content in the cells of 3 people is equal to all information exchange on the worldwide web, per day !!
Each cell contains about 2,3 billion proteins, and molecular machines. 20 thousand genes, which can be expressed in different ways, giving rise to 6 million protein species, through the spliceosome, which can splice the same gene over 300 times, giving rise to different gene products. The information can be read forward, or backward, with different reading frames. Life, in special complex life, is permeated with communication systems, that are employed
Each cell contains over 100 different epigenetic languages and codes, that work in an interdependent fashion together. These languages crosstalk with each other, dozens of them. Furthermore:
Wide-ranging conversations among various organelles respond to cell stress and provide quality control for mitochondria, membranes, and the production of proteins. Elaborate signaling inside neurons provides precise transport of materials along the axon and allows complex decision-making to take place among multiple compartments in dendrites.
We are also learning about how the primary cilium might function as a cell’s central control center, as sort of the “brain” of a cell, with its tubular structure used as an antenna for signaling. Long considered independent, the microtubule and actin network systems are now known to engage in functional cross-talk to drive essential cellular processes such as whole-cell migration, mitotic spindle positioning, or cell-wide organelle transport.
Cell-Cell interactions are performed in multicellular organisms through a sophisticated intercellular communication machinery. There are many ways like gap junctions and exosomes. But recently, it has been discovered that Cells talk and help each other via tiny tube networks. Cells are known to use intracellular microtubules, veritable nanotubular highways which direct proteins to their correct final destination inside of Cells. But, remarkably they are also used for intercellular communication, from Cells to Cells, and furthermore, for organelle Transport between Cells.
Just as humans use letters and words to create messages, cells release molecules called ligands into the environment to send messages to their neighbors—such as an instruction to differentiate or proliferate. The ligands bind to receptors on the surface of other cells, which interpret the message and trigger a set of chemical reactions to relay it to the correct molecules in the cell's interior.
One major communication channel is called the bone morphogenetic protein (BMP) pathway, which operates in nearly all tissues. This system uses many different ligands and receptors in various combinations.
1,894 ligand-receptor pairs—based on 642 ligands and 589 receptors— have been reported in the literature so far, and drew up a large-scale map of cell-to-cell. Most cells express tens or even hundreds of ligands and receptors, creating a highly connected signaling network made up of cell types that can communicate with each other through multiple ligand-receptor paths.
When biting into an apple, the body will immediately signal a complex sequence of messages and processes to break down the apple into energy and essential structural nutrients for cellular repair and replacement. That initial signal activates communication throughout the entire body, enabling metabolism to send support to every facet of the organisms function, be it mental, emotional or physical. Health and performance are completely dependent upon how efficient that signalling and communication process works.
Cells and tissues work together and respond to changes in the internal and external environment. Epigenetic mechanisms refer to heritable changes in gene expression that occur without changes in the DNA sequence. These mechanisms involve modifications to the structure of DNA and histone proteins that affect how genes are activated or silenced. Epigenetic languages enable cells to differentiate into various specialized cell types during development and play a vital role in maintaining cell identity and function throughout an organism's life. Epigenetic languages help determine which genes are active or inactive in specific cells, influencing processes such as cell growth, differentiation, and response to environmental cues.
Communication theory, specifies five stages for any communication system, regardless of its form: an information source, an encoder, a communication channel, a decoder (or receiver), and a user. The information source generates the information to be transmitted; the encoder transforms the information into a suitable message form for transmission over the communication channel; and the decoder performs the inverse operation of the encoder, or approximately so, for the user at the other end of the channel.
1. A living cell is an information-driven factory. The human body is like a huge city of interlinked factories, that work in a joint venture, together.
2. The information drives the operation in a manner analogous to how software in a computer drives computer hardware.
3. Computers are the product of deliberate intelligent action, not random processes.
We have 37 trillion cells in our human body that communicate with each other, like a huge WWW.
The Human Genetic code information content is about 6 billion bits. The Epigenetic code information content is about ten times larger:≈ 60 billion bits, Sum, total, 66 billion bits.
To find the total estimated information content of the 37 trillion cells in the human body, we can multiply the 66 billion bits by the number of cells:
Total estimated information content = 66 billion bits × 37 trillion cells = 2.44 quintillion bits.
Now, let's compare this to the daily global internet traffic: Daily internet traffic ≈ 8 quintillion bits (8 x 10^18 bits)
Comparing the two values:
Total information content of human cells ≈ 2.44 quintillion bits (2.44 x 10^18 bits)
Daily internet traffic ≈ 8 quintillion bits (8 x 10^18 bits)
The daily global internet traffic (8 quintillion bits) is roughly three times larger than the estimated total information content within the 37 trillion cells in the human body (2.44 quintillion bits).
So the information content in the cells of 3 people is equal to all information exchange on the worldwide web, per day !!
Each cell contains about 2,3 billion proteins, and molecular machines. 20 thousand genes, which can be expressed in different ways, giving rise to 6 million protein species, through the spliceosome, which can splice the same gene over 300 times, giving rise to different gene products. The information can be read forward, or backward, with different reading frames. Life, in special complex life, is permeated with communication systems, that are employed
Each cell contains over 100 different epigenetic languages and codes, that work in an interdependent fashion together. These languages crosstalk with each other, dozens of them. Furthermore:
Wide-ranging conversations among various organelles respond to cell stress and provide quality control for mitochondria, membranes, and the production of proteins. Elaborate signaling inside neurons provides precise transport of materials along the axon and allows complex decision-making to take place among multiple compartments in dendrites.
We are also learning about how the primary cilium might function as a cell’s central control center, as sort of the “brain” of a cell, with its tubular structure used as an antenna for signaling. Long considered independent, the microtubule and actin network systems are now known to engage in functional cross-talk to drive essential cellular processes such as whole-cell migration, mitotic spindle positioning, or cell-wide organelle transport.
Cell-Cell interactions are performed in multicellular organisms through a sophisticated intercellular communication machinery. There are many ways like gap junctions and exosomes. But recently, it has been discovered that Cells talk and help each other via tiny tube networks. Cells are known to use intracellular microtubules, veritable nanotubular highways which direct proteins to their correct final destination inside of Cells. But, remarkably they are also used for intercellular communication, from Cells to Cells, and furthermore, for organelle Transport between Cells.
Just as humans use letters and words to create messages, cells release molecules called ligands into the environment to send messages to their neighbors—such as an instruction to differentiate or proliferate. The ligands bind to receptors on the surface of other cells, which interpret the message and trigger a set of chemical reactions to relay it to the correct molecules in the cell's interior.
One major communication channel is called the bone morphogenetic protein (BMP) pathway, which operates in nearly all tissues. This system uses many different ligands and receptors in various combinations.
1,894 ligand-receptor pairs—based on 642 ligands and 589 receptors— have been reported in the literature so far, and drew up a large-scale map of cell-to-cell. Most cells express tens or even hundreds of ligands and receptors, creating a highly connected signaling network made up of cell types that can communicate with each other through multiple ligand-receptor paths.
When biting into an apple, the body will immediately signal a complex sequence of messages and processes to break down the apple into energy and essential structural nutrients for cellular repair and replacement. That initial signal activates communication throughout the entire body, enabling metabolism to send support to every facet of the organisms function, be it mental, emotional or physical. Health and performance are completely dependent upon how efficient that signalling and communication process works.
Cells and tissues work together and respond to changes in the internal and external environment. Epigenetic mechanisms refer to heritable changes in gene expression that occur without changes in the DNA sequence. These mechanisms involve modifications to the structure of DNA and histone proteins that affect how genes are activated or silenced. Epigenetic languages enable cells to differentiate into various specialized cell types during development and play a vital role in maintaining cell identity and function throughout an organism's life. Epigenetic languages help determine which genes are active or inactive in specific cells, influencing processes such as cell growth, differentiation, and response to environmental cues.
Communication theory, specifies five stages for any communication system, regardless of its form: an information source, an encoder, a communication channel, a decoder (or receiver), and a user. The information source generates the information to be transmitted; the encoder transforms the information into a suitable message form for transmission over the communication channel; and the decoder performs the inverse operation of the encoder, or approximately so, for the user at the other end of the channel.
1. A living cell is an information-driven factory. The human body is like a huge city of interlinked factories, that work in a joint venture, together.
2. The information drives the operation in a manner analogous to how software in a computer drives computer hardware.
3. Computers are the product of deliberate intelligent action, not random processes.

285 Unveiling the convergence, how science points to creation Tue Aug 08, 2023 7:55 am
Unveiling the convergence, how science points to creation Tue Aug 08, 2023 7:55 am
Otangelo
Admin
Unveiling the convergence, how science points to creation
In scientific discovery, moments of profound revelation have punctuated our understanding of the cosmos and life's origins. The journey began with the proclamation of the Big Bang theory in 1931, conceived by Georges Lemaître. This monumental theory unveiled the birth of the universe itself, a cataclysmic event that set the stage for everything that followed. Then, in a parallel avenue of inquiry, 2007 brought forth another groundbreaking notion—the Biological Big Bang model. Eugene Koonin's audacious proposal illuminated the grand transitions in biology, suggesting that life's progression, much like the universe's, was marked by sudden transformative leaps. In 2013, the words of Wilhelm Huck, a chemist from the Netherlands, said that a functioning cell must possess intricate correctness from the very outset, corroborating the concept of irreducible complexity and interdependence underscoring the astonishing complexity of life's origin and development and that life had to emerge all at once, in all its complexity, and not in a gradualistic fashion, like proposed by chemical evolution.
And now, in 2023, the James Webb Space Telescope delivers a revelation that amplifies this theme of swift emergence. Its gaze deep into the past has uncovered the Universe's early beginning, a crescendo of star formation bursts. The NASA article published on June 5th succinctly captures this newfound insight into the cosmos. It's a revelation that harmonizes with the overarching narrative—one of sudden emergence and creation. NASAs article from Jun 5, 2023 states: Early Universe Crackled With Bursts of Star Formation, Webb Shows
Remarkably, this pace of discovery harmonizes with an ancient account—a narrative of creation that transcends centuries. The echo of Genesis resonates anew as the scientific narrative and the scriptural testimony align. Science, with its probing inquiry, and the words of the Bible, rooted in faith, converge on a shared revelation. The culmination of knowledge and spiritual understanding bears witness to the profundity of our era. Imagine the awe that would have stirred in the hearts of our forebears, those devout believers who anchored their faith in God's design. They would have marveled at the unveiled truths manifesting before our eyes today. In the tableau of unfolding knowledge, God's word stands unswervingly faithful and true, its wisdom echoing through the ages.
Indeed, we stand on the cusp of an extraordinary epoch—a moment when the threads of discovery weave together intricate patterns of understanding. The convergence of science and faith, of empirical exploration and sacred revelation, paints a portrait of a universe and life that burst forth suddenly, in resplendent complexity. As witnesses to this convergence, we are privileged to see the harmony between the cosmic genesis and the divine Creator—an affirmation that resounds with resolute clarity in the tapestry of our existence.
In conclusion, first, science had the first surprise, with the Big Bang of the Universe, first proposed by Lemaitre in 1931. Then, in 2007, Eugene Koonin, proposed: The Biological Big Bang model for the major transitions biology. In 2013, Wilhelm Huck, Chemist stated: A working cell is more than the sum of its parts. "A functioning cell must be entirely correct at once, in all its complexity. And now, in 2023, the JWebb telescope makes another startling revelation: Early Universe Crackled With Bursts of Star Formation, Webb Shows, the title of a NASA news article, published on the 5th of June, states. Science and the Bible converge to the same outcome, and science confirms the Bible.
In scientific discovery, moments of profound revelation have punctuated our understanding of the cosmos and life's origins. The journey began with the proclamation of the Big Bang theory in 1931, conceived by Georges Lemaître. This monumental theory unveiled the birth of the universe itself, a cataclysmic event that set the stage for everything that followed. Then, in a parallel avenue of inquiry, 2007 brought forth another groundbreaking notion—the Biological Big Bang model. Eugene Koonin's audacious proposal illuminated the grand transitions in biology, suggesting that life's progression, much like the universe's, was marked by sudden transformative leaps. In 2013, the words of Wilhelm Huck, a chemist from the Netherlands, said that a functioning cell must possess intricate correctness from the very outset, corroborating the concept of irreducible complexity and interdependence underscoring the astonishing complexity of life's origin and development and that life had to emerge all at once, in all its complexity, and not in a gradualistic fashion, like proposed by chemical evolution.
And now, in 2023, the James Webb Space Telescope delivers a revelation that amplifies this theme of swift emergence. Its gaze deep into the past has uncovered the Universe's early beginning, a crescendo of star formation bursts. The NASA article published on June 5th succinctly captures this newfound insight into the cosmos. It's a revelation that harmonizes with the overarching narrative—one of sudden emergence and creation. NASAs article from Jun 5, 2023 states: Early Universe Crackled With Bursts of Star Formation, Webb Shows
Remarkably, this pace of discovery harmonizes with an ancient account—a narrative of creation that transcends centuries. The echo of Genesis resonates anew as the scientific narrative and the scriptural testimony align. Science, with its probing inquiry, and the words of the Bible, rooted in faith, converge on a shared revelation. The culmination of knowledge and spiritual understanding bears witness to the profundity of our era. Imagine the awe that would have stirred in the hearts of our forebears, those devout believers who anchored their faith in God's design. They would have marveled at the unveiled truths manifesting before our eyes today. In the tableau of unfolding knowledge, God's word stands unswervingly faithful and true, its wisdom echoing through the ages.
Indeed, we stand on the cusp of an extraordinary epoch—a moment when the threads of discovery weave together intricate patterns of understanding. The convergence of science and faith, of empirical exploration and sacred revelation, paints a portrait of a universe and life that burst forth suddenly, in resplendent complexity. As witnesses to this convergence, we are privileged to see the harmony between the cosmic genesis and the divine Creator—an affirmation that resounds with resolute clarity in the tapestry of our existence.
In conclusion, first, science had the first surprise, with the Big Bang of the Universe, first proposed by Lemaitre in 1931. Then, in 2007, Eugene Koonin, proposed: The Biological Big Bang model for the major transitions biology. In 2013, Wilhelm Huck, Chemist stated: A working cell is more than the sum of its parts. "A functioning cell must be entirely correct at once, in all its complexity. And now, in 2023, the JWebb telescope makes another startling revelation: Early Universe Crackled With Bursts of Star Formation, Webb Shows, the title of a NASA news article, published on the 5th of June, states. Science and the Bible converge to the same outcome, and science confirms the Bible.
286 Examining 47 crucial processes that influence the development, structure, and function of organisms reveals some astonishing interconnections Sat Sep 02, 2023 1:56 pm
Examining 47 crucial processes that influence the development, structure, and function of organisms reveals some astonishing interconnections Sat Sep 02, 2023 1:56 pm
Otangelo
Admin
Examining 47 crucial processes that influence the development, structure, and function of organisms reveals some astonishing interconnections
The sheer complexity and intricate interdependence observed in biological systems provide a strong foundation for arguments in favor of Intelligent Design (ID). Developmental biology encompasses a wide range of processes that dictate the growth, form, and function of organisms from conception to maturity. The following list encompasses processes ranging from the molecular to the organ level, each vital for the proper development, structure, and function of an organism. These processes, often interlinked, collectively orchestrate the intricate dance of development from a single cell to a multicellular organism.
1. Angiogenesis and Vasculogenesis: Formation of new blood vessels from pre-existing ones (angiogenesis) and de novo vessel formation (vasculogenesis).
2. Apoptosis: Programmed cell death essential for removing unwanted cells.
3. Cell-Cycle Regulation: Controls the progression of cells through the stages of growth and division.
4. Cell-cell adhesion and the ECM: Refers to how cells stick to each other and to the extracellular matrix, essential for tissue formation.
5. Cell-Cell Communication: Cells communicate to coordinate their actions.
6. Cell Fate Determination and Lineage Specification (Cell differentiation): Process by which cells become specialized in their function.
7. Cell Migration and Chemotaxis: Movement of cells, guided by certain chemical gradients.
8. Cell Polarity and Asymmetry: Defines distinct cellular 'sides' or 'ends', crucial for many cell functions.
9. Cellular Pluripotency: Cells can give rise to multiple cell types.
10. Cellular Senescence: State of stable cell cycle arrest.
11. Centrosomes: Organize microtubules and provide structure to cells.
12. Chromatin Dynamics: How DNA and proteins are organized in the nucleus.
13. Cytokinesis: Physical process of cell division.
14. Cytoskeletal Arrays: Framework of the cell, involved in cell shape, movement, and division.
15. DNA Methylation: Addition of methyl groups to DNA, often involved in gene silencing.
16. Egg-Polarity Genes: Determine the axes of the egg and subsequently the organism.
17. Epigenetic Codes: Changes in gene function without changing DNA sequence.
18. Gene Regulation Network: Interactions between genes, controlling when and where genes are expressed.
19. Germ Cell Formation and Migration: Development and movement of reproductive cells.
20. Germ Layer Formation (Gastrulation): Development of primary tissue layers in embryos.
21. Histone PTMs: Modifications to histone proteins affecting DNA accessibility.
22. Homeobox and Hox Genes: Control the body plan of an embryo along the head-tail axis.
23. Hormones: Chemical messengers coordinating bodily functions.
24. Immune System Development: Formation and maturation of immune cells.
25. Ion Channels and Electromagnetic Fields: Channels allowing ions to flow in/out of cells; electromagnetic fields can influence development.
26. Membrane Targets: Processes focusing on cell membrane components.
27. MicroRNA Regulation: Small RNAs regulating gene expression post-transcriptionally.
28. Morphogen Gradients: Concentration gradients of substances determining tissue development.
29. Neural Crest Cells Migration: Movement of cells contributing to diverse structures, including peripheral nerves.
30. Neural plate folding and convergence: Formation of the neural tube in early development.
31. Neuronal Pruning and Synaptogenesis: Refinement of neural connections and formation of synapses.
32. Neurulation and Neural Tube Formation: Development of the neural tube, precursor to the CNS.
33. Noncoding RNA from Junk DNA: RNA molecules not coding for protein but having various functions.
34. Oogenesis: Egg cell (oocyte) formation.
35. Oocyte Maturation and Fertilization: Development of mature egg and its fusion with sperm.
36. Pattern Formation: Processes determining organized spatial arrangement of cells/tissues.
37. Photoreceptor development: Formation of cells detecting light in the eye.
38. Regional specification: Defining distinct regions within developing tissues.
39. Segmentation and Somitogenesis: Division of body into segments and formation of somites in embryos.
40. Signaling Pathways: Series of molecular events relaying extracellular signals to intracellular targets.
41. Spatiotemporal gene expression: Time and place-specific gene expression.
42. Spermatogenesis: The process of sperm cell formation and maturation.
43. Stem Cell Regulation and Differentiation: Control of stem cell fate and their development into specialized cells.
44. Symbiotic Relationships and Microbiota Influence: Interactions with microbial partners and their influence on host development.
45. Syncytium formation: Multinucleated cell formation, especially important in muscle tissues.
46. Transposons and Retrotransposons: Mobile genetic elements, sometimes influencing gene regulation.
47. Tissue Induction and Organogenesis: Formation of tissues and organs from undifferentiated cells.
The processes and systems listed are essential to the complex orchestration of development and physiology in multicellular organisms. Many of these are interwoven and interdependent to ensure accurate and timely development, function, and maintenance. It's worth noting that this list only scratches the surface. The interconnectedness of these systems is immensely complex, with each one potentially influencing or being influenced by multiple others. They collectively underscore the intricacy and delicate choreography inherent to biology.
1. Harmony in Complexity
The vast array of processes, ranging from the microscopic level (like DNA methylation) to the macroscopic (like organogenesis), are so tightly interwoven that a disturbance in one can drastically impact another. This finely-tuned orchestration suggests a system that has been designed with precision and purpose, rather than one that arose from a series of unplanned, random events.
Chromatin Dynamics and Epigenetic Codes
Chromatin Dynamics (Point 12): At the microscopic level, chromatin dynamics describes how DNA and proteins are organized within the nucleus. DNA wraps around histone proteins, forming nucleosomes. The compactness of this structure dictates whether genes are accessible for transcription or not. Changes in chromatin structure play an essential role in controlling which genes are active at any given time.
Epigenetic Codes (Point 17): Epigenetics encompasses changes in gene function that don't involve alterations to the underlying DNA sequence. One of the primary mechanisms for this is DNA methylation (Point 15), where methyl groups are added to the DNA, usually leading to gene silencing.
Interdependencies and Implications
Gene Regulation Network (Point 18): Chromatin dynamics and epigenetic modifications directly influence the gene regulatory networks. These modifications decide which genes are turned on or off, ensuring that cells have the appropriate responses to environmental cues.
Cell Fate Determination and Lineage Specification (Point 6): Epigenetic codes and chromatin remodeling play crucial roles in determining cell fate. For instance, a stem cell's decision to become a muscle cell versus a nerve cell can be influenced by these modifications.
Tissue Induction and Organogenesis (Point 47): Proper tissue and organ formation requires specific sets of genes to be activated in a timely and spatial manner. Chromatin dynamics and epigenetic modifications help coordinate these gene expression patterns, ensuring organs form correctly and functionally.
Given the interplay between chromatin dynamics and epigenetic codes, one can see the harmony in complexity. If chromatin isn't organized correctly, or if the epigenetic codes go awry, the ripple effects can be vast, impacting everything from individual cell functions to the development of entire organs. Such a tightly coordinated system, where microscopic modifications can influence macroscopic outcomes, speaks to a design with intricate precision and purpose.
2. A House of Cards
Many proponents of ID describe the cellular processes and systems as a "house of cards." In this analogy, removing one card (or disrupting a single process) may cause the entire structure to collapse. Such intricate dependencies make it hard to envision a gradual, step-by-step evolutionary development. How would the system function if even one of its myriad processes was not yet in place?
Let's take a look into Cell-Cell Communication and its relevance to many of the processes listed previously.
Cell-Cell Communication and the Notch Signaling Pathway
In multicellular organisms, cells don't function in isolation. They constantly communicate with one another to maintain harmony and respond to changes in the environment. One of the most studied pathways in this realm is the Notch signaling pathway.
How Notch Signaling Works
Activation: Notch signaling is initiated when a ligand from a neighboring cell binds to the Notch receptor of another cell.
Cleavage and Migration: This binding event causes two proteolytic cleavages of the Notch receptor. The second cleavage releases the Notch intracellular domain (NICD), which then migrates to the cell's nucleus.
Gene Expression: Once inside the nucleus, the NICD associates with other proteins and acts as a transcriptional activator, turning on genes that will affect the cell's fate.
Interdependencies and Systems Biology Implications:
Cell Differentiation (Point 6): Notch signaling plays a critical role in determining cell fate and ensuring cells differentiate into the types needed for proper tissue and organ function.
Pattern Formation (Point 36): The pathway helps establish patterns of cells in tissues, ensuring the right cells are in the right places.
Gene Regulation Network (Point 18): Notch signaling interfaces with numerous other pathways, making it a node in the complex web of cellular communication. Disruptions here can have cascading effects on numerous processes.
Tissue Induction and Organogenesis (Point 47): Proper tissue formation often requires communication between cells, with Notch signaling being pivotal for many of these interactions.
Considering the Notch signaling pathway alone, it's evident that its perturbation can disrupt multiple processes. From a systems biology perspective, if this pathway wasn't functioning correctly or was only partially developed, it's challenging to see how many critical developmental processes would proceed effectively. Its intricate ties to various cellular and developmental processes underscore the vast interconnectedness in biological systems.
3. Irreducible Complexity
A cornerstone of the ID argument is that many biological systems are "irreducibly complex." This means that they need all their parts to be present and functioning simultaneously to work. In the vast web of interconnected processes, where one relies on another to operate, the absence or malfunctioning of even one process would render the whole system dysfunctional. This poses significant challenges to the idea of gradual evolution: if a system needs all its parts to function, how could it evolve piecemeal over time?
Epigenetic Regulation and Gene Expression
Taking a look at the list of 47 points, there is a profound interdependence between "DNA Methylation" (Point 15), "Epigenetic Codes" (Point 17), "Gene Regulation Network" (Point 18), and "MicroRNA Regulation" (Point 27).
DNA Methylation (Point 15): This involves the addition of a methyl group to a cytosine base in DNA. Methylation typically suppresses gene transcription, and thus, it's a mechanism by which genes can be "turned off."
Epigenetic Codes (Point 17): Epigenetics refers to changes in gene function without altering the DNA sequence itself. Methylation is an epigenetic modification, but there are others, such as histone modifications, which can impact how tightly DNA is wound around histone proteins, thereby regulating gene accessibility and expression.
Gene Regulation Network (Point 18): This is a complex network of interactions between genes, typically involving transcription factors, enhancers, silencers, and other regulatory elements that control when, where, and how genes are expressed.
MicroRNA Regulation (Point 27): MicroRNAs are small RNA molecules that do not code for proteins. Instead, they regulate gene expression post-transcriptionally. They can bind to messenger RNA (mRNA) molecules and prevent them from being translated into proteins, or even lead to their degradation.
The interdependedness between these processes ensures precise control over gene expression. For an organism to develop and function properly, genes need to be turned on and off at the right times and in the right places. But consider this: if DNA methylation patterns are awry, then certain genes might be wrongly activated or suppressed. The gene regulatory network relies on correct epigenetic codes to function properly, and aberrant microRNA expression can disrupt the entire balance. These systems' mutual dependencies make it challenging to envision how they could have evolved separately or in a stepwise fashion. For example, if a regulatory gene network evolved before the epigenetic controls were in place, how would it ensure precision in gene expression? If microRNAs emerged but the system to process them or the targets they bind to weren't present, would they confer any advantage?
This web of interdependence between epigenetic modifications, gene networks, and microRNA regulation exemplifies the intricacies and precision of cellular processes, underscoring the challenges faced by piecemeal evolutionary explanations.
4. The Language of Life
The cell operates with a myriad of 'codes' and 'languages.' From the genetic code in DNA to the intricate signaling pathways and feedback loops, cells communicate and operate in a way that is reminiscent of an intricately coded software program. The emergence of such a detailed and error-proof 'language system' from random events appears statistically implausible and points towards a designed system.
Neural Blueprint and Information Transfer
To showcase the interdependence within the 47 points, let's focus on the intricate processes associated with neural development and communication. Consider the following components:
Neural plate folding and convergence (Point 30): Early in development, the neural plate undergoes specific movements and foldings to form the neural tube, the precursor of the central nervous system. This requires accurate spatial organization.
Neurulation and Neural Tube Formation (Point 32): Once the neural plate has folded, it must properly close to form the neural tube. This structure eventually gives rise to the brain and spinal cord.
Cell-Cell Communication (Point 5): Cells must communicate effectively to coordinate these early developmental processes. Miscommunication or errors in signaling can lead to severe developmental defects.
Gene Regulation Network (Point 18): A precise network of gene interactions ensures that the right genes are activated (or suppressed) at the right times for neural tube formation.
Morphogen Gradients (Point 28): These are concentration gradients of substances that dictate tissue development. In the context of neural development, morphogens play critical roles in specifying which parts of the neural tube become the brain and which become the spinal cord.
Homeobox and Hox Genes (Point 22): These genes play a pivotal role in setting up the body plan of an embryo along its head-tail axis, including defining regions of the developing brain and spinal cord.
From a system's biology perspective, neural development is a marvel of coordination and communication. For the neural tube to form correctly, cells must communicate with each other, adhere to each other in specific ways, respond to morphogen gradients, and activate the right genes at the right times. All these processes are tightly interwoven, and a failure in one process can impact others. For instance, if the gene regulatory network doesn't activate the right set of genes due to some perturbation, it could potentially affect the morphogen gradients, which in turn might disturb the proper folding of the neural plate, leading to defects in neural tube formation. Given the intricate dance between these processes, it's hard to fathom how such a system could have evolved piecemeal. Without the precise coordination of these multiple factors, the entire process of neural development could be jeopardized. This mutual dependency paints a picture of an orchestrated design where all parts must work in concert for the successful creation of such a complex system.
5. Feedback Loops and Regulatory Mechanisms
The numerous feedback loops and regulatory mechanisms ensure that every cellular process is meticulously monitored and adjusted as necessary. The foresight required for such intricate regulation seems beyond the scope of random mutations and natural selection.
Tissue Development and Maintenance
Diving into the intricate world of cellular growth, differentiation, and communication, let's explore the interwoven dance of several processes from the 47 points:
Cell-Cycle Regulation (Point 3): Cells have inbuilt systems that control their growth and division. A cell must decide when to divide, based on numerous external and internal cues.
Apoptosis (Point 2): Paradoxically, while some cells are growing and dividing, others are programmed to die, ensuring that tissues are sculpted properly and potential rogue cells are eliminated.
Signaling Pathways (Point 40): These pathways relay extracellular signals to intracellular targets, determining whether a cell divides, differentiates, or dies.
Cell Fate Determination and Lineage Specification (Point 6): Within a developing tissue or organ, cells are assigned specific roles. This involves a complex interplay of signals that tell cells to differentiate into one type of cell versus another.
Epigenetic Codes (Point 17): These are modifications to the DNA or associated proteins that don't change the DNA sequence but control gene activity. Epigenetic changes can be induced by environmental factors and can influence cellular decisions like differentiation.
MicroRNA Regulation (Point 27): Small RNAs that don't code for protein but regulate other genes post-transcriptionally. These can fine-tune cellular responses by adjusting the levels of specific proteins in a cell.
Feedback Loops and Hormones (Point 23): Chemical messengers, like hormones, often function within feedback loops, where the output of a system acts as an input to control its behavior, ensuring homeostasis.
Tissue Induction and Organogenesis (Point 47): The formation of specific tissues and organs requires a concert of the above processes. Cells need to grow, communicate, decide their fate, differentiate, or even undergo programmed death, all under the watchful eyes of regulatory networks.
Morphogen Gradients (Point 28): Concentrations of specific molecules in an embryo provide cues to cells, guiding them in their development and spatial organization within tissues and organs.
In this intricate interdependence of cellular processes, each is indispensable. For tissue development and organogenesis to occur correctly, cells need the right mix of growth signals, differentiation cues, and spatial information. If signaling pathways go awry, it can lead to unchecked growth or improper differentiation. If apoptosis doesn't function correctly, it might lead to malformations or predispose tissues to cancers. If epigenetic codes aren't set right, genes essential for proper function might remain silent or get inappropriately activated. All these processes interlock in an elegant dance, each reliant on the other, ensuring that tissues and organs develop properly. Given the sheer complexity and the tight interdependence of these systems, one can argue the challenges it poses to a purely stepwise evolutionary process.
6. Information Storage and Retrieval
The cell's ability to store, retrieve, and implement vast amounts of information is unmatched. DNA, often likened to a data storage system, holds the blueprints for the entire organism. The intricate processes by which this information is accessed, read, and executed seem to be beyond the capacity of unguided evolutionary processes to produce.
Orchestrating Organism Development
Consider the awe-inspiring journey of a single fertilized egg (zygote) as it develops into a complex multicellular organism:
Oogenesis (Point 34) & Spermatogenesis (Point 42): The journey of life begins with the formation of gametes. These processes create the mature egg and sperm, each responsible for carrying half of the genetic information that will lead to a new organism. This initial formation of gametes is foundational to the progression of life.
Oocyte Maturation and Fertilization (Point 35): Following the formation of these gametes, the next step in the dance of life is their fusion. Once the oocyte and sperm unite, a zygote emerges, endowed with a complete set of DNA. This DNA is the architectural blueprint that directs the growth and development of the entire organism.
Gene Regulation Network (Point 18): As the zygote's journey begins, a need for orchestration arises. The gene regulation network offers this orchestration, a vast interconnected web of interactions, determining when, where, and how genes get expressed. This system can be visualized as a master conductor, deciding which sections of the orchestra play and at which moments.
Epigenetic Codes (Point 17): Complementing the conductor, there are specific markers, akin to bookmarks on our DNA, that dictate which musical notes (genes) are emphasized and which are muted. Epigenetic modifications ensure that certain genes are made accessible while others remain silent, all without altering the original score (DNA sequence).
MicroRNA Regulation (Point 27) & Noncoding RNA from Junk DNA (Point 33): Just as a symphony may require fine-tuning, these molecules offer a layer of adjustment to the genetic output after the primary transcript, enhancing or modulating the performance as necessary.
Cell-Cycle Regulation (Point 3): With the foundational notes set, the zygote embarks on a growth journey. This growth is meticulously orchestrated, ensuring that each cellular division is harmonious, with DNA replicated with precision.
Germ Layer Formation (Point 20): As this cellular symphony continues, differentiation begins, setting the stage for the future tissues and organs. Cells start aligning into three primary sections or layers: ectoderm, mesoderm, and endoderm, each layer contributing unique notes to the life song.
Cell Fate Determination and Lineage Specification (Point 6): Within these layers, the individual notes (cells) are further refined and specialized, ensuring that each plays its part in the evolving melody of life.
Signaling Pathways (Point 40) & Morphogen Gradients (Point 28): Communication becomes pivotal as cells continue to evolve and find their position in the overarching composition. These pathways and gradients act as messengers, ensuring each cell understands its role and positioning.
Tissue Induction and Organogenesis (Point 47): The crescendo approaches as cells, driven by unique cues, assemble to form the organs that are vital to life, such as the heart, lungs, and liver.
Cell-Cell Communication (Point 5) & Cell-cell adhesion and the ECM (Point 4): And as the composition reaches its zenith, for the entire system to function harmoniously, cells must communicate and connect, ensuring that every note is in place, creating a beautifully coordinated melody of life.
The life of an organism, as illustrated, is a complex interplay of various systems and processes, each building upon the other, forming a harmonious melody from inception to maturity. This journey, from a single cell to a fully formed organism, involves accessing, reading, and executing a vast amount of information stored within the DNA. At each step, multiple processes from the 47 points are at play, acting like meticulous architects interpreting and building a structure based on an intricate blueprint. Given the precision, coordination, and depth of information involved, it offers a profound reflection on the cell's unmatched information storage and retrieval system.
The intricate interdependence and sheer complexity observed in biological systems make it hard to reconcile with a purely evolutionary framework that relies on random mutations and natural selection. The precision, foresight, and harmony seen in these systems appear to be indicative of a design by an intelligent agent.
Interdependence and Intricacy in Biological Systems
When one looks at the formation and development of complex multicellular organisms, it's akin to witnessing a grand orchestra, where each musician (or process) plays an essential part in crafting a collective, harmonious sound. If even one musician is missing or plays out of tune, the entire performance can be compromised. Similarly, the 47 biological processes are so deeply interwoven that a disturbance or absence in even a single process can lead to systemic disruptions.
Foundational Importance: Just as an orchestra requires foundational instruments like percussion to set the rhythm, processes such as Oogenesis, Spermatogenesis, and Oocyte Maturation and Fertilization set the stage for life's beginning. Without these processes, the journey wouldn't even commence.
Regulation and Coordination: Once the foundational processes are in place, the need for regulation and coordination becomes paramount. The Gene Regulation Network, MicroRNA Regulation, and Epigenetic Codes serve as the conductors and coordinators, ensuring each 'musician' performs at the right time and in harmony with others.
Specialization and Differentiation: As the performance unfolds, specialized instruments like woodwinds or strings introduce unique melodies. Similarly, the Germ Layer Formation and Cell Fate Determination ensure cells differentiate and specialize, adding complexity to the organism's developmental 'symphony'.
Communication: In any orchestra, musicians must listen to and be in sync with each other. The biological equivalents are the Signaling Pathways and Cell-Cell Communication, which guarantee that cells 'listen' to each other and respond appropriately, maintaining the organism's intricate harmony.
Structural Integrity: Just as each section of an orchestra relies on the structure and positioning of its musicians, processes like Cell-cell adhesion and the ECM ensure the physical structure and integrity of tissues and organs.
Systemic Harmony: Finally, all these processes need to work in tandem. Tissue Induction, Organogenesis, and other processes ensure the organism's 'performance' is harmonized from start to finish.
Implications for Evolution and Complexity
The interconnectedness and dependency of these processes pose intriguing questions about the evolution of complex life. The traditional evolutionary model suggests a gradual accumulation of beneficial mutations over time. However, when considering irreducible complexity, a challenge arises: How can systems that rely so heavily on the simultaneous functioning of multiple components evolve incrementally? Such systems seem to defy a piecemeal evolutionary development, as the system wouldn't function (or would offer no evolutionary advantage) until all components are present and working together. The development of multicellular organisms is a marvel of complexity, coordination, and precision, revealing the awe-inspiring intricacies of life.
The Fine Balance of Life
Redundancy and Flexibility: While it's true that the intricacies of these systems point towards an irreducible complexity, nature has also ingeniously incorporated redundancy and flexibility. There are instances where multiple processes can achieve a similar outcome or where systems have backup mechanisms. This 'buffer' allows organisms to survive and adapt in fluctuating environments and under various stresses.
Fine-tuning: These systems are optimized for efficiency and effectiveness. Each process, while essential, has likely been the subject of countless iterations, shaped by environmental pressures and interactions with other processes. This ongoing 'tuning' has resulted in the beautifully orchestrated dance of cellular and molecular events we observe today.
The Starting Point of Complexity: If even the most primitive unicellular organisms required a subset of these 47 processes to survive, then how could such complexity arise spontaneously without guidance? The leap from non-life to even the simplest life form is monumental, given the intricate machinery required at the cellular level. Such complexity, right from the beginning, suggests a purposefully designed set up.
The Problem of Incremental Evolution: How can a partial system, that's non-functional until fully formed, provide a selective advantage? Without the advantage, the process won't be 'selected' and thus, won't evolve. If the machinery of the cell works like a finely tuned watch, missing one gear might render it non-functional. Evolutionary processes can't favor non-functional or less functional states.
The Interconnectedness Challenge: The interconnected nature of the 47 processes outlined implies that changes in one system could have ripple effects across others. A random mutation in one part might require synchronized changes in several others to maintain functionality. Such a level of concurrent and harmonized change seems beyond the capabilities of random mutation and natural selection.
Plasticity and Pre-programming: The capacity for organisms to adapt to their environment is often touted as evidence for evolution. However, this plasticity is evidence of pre-programmed adaptability—a foresight that allows organisms to respond to changing environments. Rather than being proof of random evolution, this built-in adaptability may suggest a designer who anticipated the varied and dynamic environments the organism would encounter.
Information Theory: One significant point is the infusion of information into the DNA. Information, as we understand it in other realms (like coding or linguistics), typically arises from intelligence. The intricate and specific information carried in the DNA, guiding the myriad of processes in the organism, is clear evidence of an intelligent input.[/size]
The sheer complexity and intricate interdependence observed in biological systems provide a strong foundation for arguments in favor of Intelligent Design (ID). Developmental biology encompasses a wide range of processes that dictate the growth, form, and function of organisms from conception to maturity. The following list encompasses processes ranging from the molecular to the organ level, each vital for the proper development, structure, and function of an organism. These processes, often interlinked, collectively orchestrate the intricate dance of development from a single cell to a multicellular organism.
1. Angiogenesis and Vasculogenesis: Formation of new blood vessels from pre-existing ones (angiogenesis) and de novo vessel formation (vasculogenesis).
2. Apoptosis: Programmed cell death essential for removing unwanted cells.
3. Cell-Cycle Regulation: Controls the progression of cells through the stages of growth and division.
4. Cell-cell adhesion and the ECM: Refers to how cells stick to each other and to the extracellular matrix, essential for tissue formation.
5. Cell-Cell Communication: Cells communicate to coordinate their actions.
6. Cell Fate Determination and Lineage Specification (Cell differentiation): Process by which cells become specialized in their function.
7. Cell Migration and Chemotaxis: Movement of cells, guided by certain chemical gradients.
8. Cell Polarity and Asymmetry: Defines distinct cellular 'sides' or 'ends', crucial for many cell functions.
9. Cellular Pluripotency: Cells can give rise to multiple cell types.
10. Cellular Senescence: State of stable cell cycle arrest.
11. Centrosomes: Organize microtubules and provide structure to cells.
12. Chromatin Dynamics: How DNA and proteins are organized in the nucleus.
13. Cytokinesis: Physical process of cell division.
14. Cytoskeletal Arrays: Framework of the cell, involved in cell shape, movement, and division.
15. DNA Methylation: Addition of methyl groups to DNA, often involved in gene silencing.
16. Egg-Polarity Genes: Determine the axes of the egg and subsequently the organism.
17. Epigenetic Codes: Changes in gene function without changing DNA sequence.
18. Gene Regulation Network: Interactions between genes, controlling when and where genes are expressed.
19. Germ Cell Formation and Migration: Development and movement of reproductive cells.
20. Germ Layer Formation (Gastrulation): Development of primary tissue layers in embryos.
21. Histone PTMs: Modifications to histone proteins affecting DNA accessibility.
22. Homeobox and Hox Genes: Control the body plan of an embryo along the head-tail axis.
23. Hormones: Chemical messengers coordinating bodily functions.
24. Immune System Development: Formation and maturation of immune cells.
25. Ion Channels and Electromagnetic Fields: Channels allowing ions to flow in/out of cells; electromagnetic fields can influence development.
26. Membrane Targets: Processes focusing on cell membrane components.
27. MicroRNA Regulation: Small RNAs regulating gene expression post-transcriptionally.
28. Morphogen Gradients: Concentration gradients of substances determining tissue development.
29. Neural Crest Cells Migration: Movement of cells contributing to diverse structures, including peripheral nerves.
30. Neural plate folding and convergence: Formation of the neural tube in early development.
31. Neuronal Pruning and Synaptogenesis: Refinement of neural connections and formation of synapses.
32. Neurulation and Neural Tube Formation: Development of the neural tube, precursor to the CNS.
33. Noncoding RNA from Junk DNA: RNA molecules not coding for protein but having various functions.
34. Oogenesis: Egg cell (oocyte) formation.
35. Oocyte Maturation and Fertilization: Development of mature egg and its fusion with sperm.
36. Pattern Formation: Processes determining organized spatial arrangement of cells/tissues.
37. Photoreceptor development: Formation of cells detecting light in the eye.
38. Regional specification: Defining distinct regions within developing tissues.
39. Segmentation and Somitogenesis: Division of body into segments and formation of somites in embryos.
40. Signaling Pathways: Series of molecular events relaying extracellular signals to intracellular targets.
41. Spatiotemporal gene expression: Time and place-specific gene expression.
42. Spermatogenesis: The process of sperm cell formation and maturation.
43. Stem Cell Regulation and Differentiation: Control of stem cell fate and their development into specialized cells.
44. Symbiotic Relationships and Microbiota Influence: Interactions with microbial partners and their influence on host development.
45. Syncytium formation: Multinucleated cell formation, especially important in muscle tissues.
46. Transposons and Retrotransposons: Mobile genetic elements, sometimes influencing gene regulation.
47. Tissue Induction and Organogenesis: Formation of tissues and organs from undifferentiated cells.
The processes and systems listed are essential to the complex orchestration of development and physiology in multicellular organisms. Many of these are interwoven and interdependent to ensure accurate and timely development, function, and maintenance. It's worth noting that this list only scratches the surface. The interconnectedness of these systems is immensely complex, with each one potentially influencing or being influenced by multiple others. They collectively underscore the intricacy and delicate choreography inherent to biology.
1. Harmony in Complexity
The vast array of processes, ranging from the microscopic level (like DNA methylation) to the macroscopic (like organogenesis), are so tightly interwoven that a disturbance in one can drastically impact another. This finely-tuned orchestration suggests a system that has been designed with precision and purpose, rather than one that arose from a series of unplanned, random events.
Chromatin Dynamics and Epigenetic Codes
Chromatin Dynamics (Point 12): At the microscopic level, chromatin dynamics describes how DNA and proteins are organized within the nucleus. DNA wraps around histone proteins, forming nucleosomes. The compactness of this structure dictates whether genes are accessible for transcription or not. Changes in chromatin structure play an essential role in controlling which genes are active at any given time.
Epigenetic Codes (Point 17): Epigenetics encompasses changes in gene function that don't involve alterations to the underlying DNA sequence. One of the primary mechanisms for this is DNA methylation (Point 15), where methyl groups are added to the DNA, usually leading to gene silencing.
Interdependencies and Implications
Gene Regulation Network (Point 18): Chromatin dynamics and epigenetic modifications directly influence the gene regulatory networks. These modifications decide which genes are turned on or off, ensuring that cells have the appropriate responses to environmental cues.
Cell Fate Determination and Lineage Specification (Point 6): Epigenetic codes and chromatin remodeling play crucial roles in determining cell fate. For instance, a stem cell's decision to become a muscle cell versus a nerve cell can be influenced by these modifications.
Tissue Induction and Organogenesis (Point 47): Proper tissue and organ formation requires specific sets of genes to be activated in a timely and spatial manner. Chromatin dynamics and epigenetic modifications help coordinate these gene expression patterns, ensuring organs form correctly and functionally.
Given the interplay between chromatin dynamics and epigenetic codes, one can see the harmony in complexity. If chromatin isn't organized correctly, or if the epigenetic codes go awry, the ripple effects can be vast, impacting everything from individual cell functions to the development of entire organs. Such a tightly coordinated system, where microscopic modifications can influence macroscopic outcomes, speaks to a design with intricate precision and purpose.
2. A House of Cards
Many proponents of ID describe the cellular processes and systems as a "house of cards." In this analogy, removing one card (or disrupting a single process) may cause the entire structure to collapse. Such intricate dependencies make it hard to envision a gradual, step-by-step evolutionary development. How would the system function if even one of its myriad processes was not yet in place?
Let's take a look into Cell-Cell Communication and its relevance to many of the processes listed previously.
Cell-Cell Communication and the Notch Signaling Pathway
In multicellular organisms, cells don't function in isolation. They constantly communicate with one another to maintain harmony and respond to changes in the environment. One of the most studied pathways in this realm is the Notch signaling pathway.
How Notch Signaling Works
Activation: Notch signaling is initiated when a ligand from a neighboring cell binds to the Notch receptor of another cell.
Cleavage and Migration: This binding event causes two proteolytic cleavages of the Notch receptor. The second cleavage releases the Notch intracellular domain (NICD), which then migrates to the cell's nucleus.
Gene Expression: Once inside the nucleus, the NICD associates with other proteins and acts as a transcriptional activator, turning on genes that will affect the cell's fate.
Interdependencies and Systems Biology Implications:
Cell Differentiation (Point 6): Notch signaling plays a critical role in determining cell fate and ensuring cells differentiate into the types needed for proper tissue and organ function.
Pattern Formation (Point 36): The pathway helps establish patterns of cells in tissues, ensuring the right cells are in the right places.
Gene Regulation Network (Point 18): Notch signaling interfaces with numerous other pathways, making it a node in the complex web of cellular communication. Disruptions here can have cascading effects on numerous processes.
Tissue Induction and Organogenesis (Point 47): Proper tissue formation often requires communication between cells, with Notch signaling being pivotal for many of these interactions.
Considering the Notch signaling pathway alone, it's evident that its perturbation can disrupt multiple processes. From a systems biology perspective, if this pathway wasn't functioning correctly or was only partially developed, it's challenging to see how many critical developmental processes would proceed effectively. Its intricate ties to various cellular and developmental processes underscore the vast interconnectedness in biological systems.
3. Irreducible Complexity
A cornerstone of the ID argument is that many biological systems are "irreducibly complex." This means that they need all their parts to be present and functioning simultaneously to work. In the vast web of interconnected processes, where one relies on another to operate, the absence or malfunctioning of even one process would render the whole system dysfunctional. This poses significant challenges to the idea of gradual evolution: if a system needs all its parts to function, how could it evolve piecemeal over time?
Epigenetic Regulation and Gene Expression
Taking a look at the list of 47 points, there is a profound interdependence between "DNA Methylation" (Point 15), "Epigenetic Codes" (Point 17), "Gene Regulation Network" (Point 18), and "MicroRNA Regulation" (Point 27).
DNA Methylation (Point 15): This involves the addition of a methyl group to a cytosine base in DNA. Methylation typically suppresses gene transcription, and thus, it's a mechanism by which genes can be "turned off."
Epigenetic Codes (Point 17): Epigenetics refers to changes in gene function without altering the DNA sequence itself. Methylation is an epigenetic modification, but there are others, such as histone modifications, which can impact how tightly DNA is wound around histone proteins, thereby regulating gene accessibility and expression.
Gene Regulation Network (Point 18): This is a complex network of interactions between genes, typically involving transcription factors, enhancers, silencers, and other regulatory elements that control when, where, and how genes are expressed.
MicroRNA Regulation (Point 27): MicroRNAs are small RNA molecules that do not code for proteins. Instead, they regulate gene expression post-transcriptionally. They can bind to messenger RNA (mRNA) molecules and prevent them from being translated into proteins, or even lead to their degradation.
The interdependedness between these processes ensures precise control over gene expression. For an organism to develop and function properly, genes need to be turned on and off at the right times and in the right places. But consider this: if DNA methylation patterns are awry, then certain genes might be wrongly activated or suppressed. The gene regulatory network relies on correct epigenetic codes to function properly, and aberrant microRNA expression can disrupt the entire balance. These systems' mutual dependencies make it challenging to envision how they could have evolved separately or in a stepwise fashion. For example, if a regulatory gene network evolved before the epigenetic controls were in place, how would it ensure precision in gene expression? If microRNAs emerged but the system to process them or the targets they bind to weren't present, would they confer any advantage?
This web of interdependence between epigenetic modifications, gene networks, and microRNA regulation exemplifies the intricacies and precision of cellular processes, underscoring the challenges faced by piecemeal evolutionary explanations.
4. The Language of Life
The cell operates with a myriad of 'codes' and 'languages.' From the genetic code in DNA to the intricate signaling pathways and feedback loops, cells communicate and operate in a way that is reminiscent of an intricately coded software program. The emergence of such a detailed and error-proof 'language system' from random events appears statistically implausible and points towards a designed system.
Neural Blueprint and Information Transfer
To showcase the interdependence within the 47 points, let's focus on the intricate processes associated with neural development and communication. Consider the following components:
Neural plate folding and convergence (Point 30): Early in development, the neural plate undergoes specific movements and foldings to form the neural tube, the precursor of the central nervous system. This requires accurate spatial organization.
Neurulation and Neural Tube Formation (Point 32): Once the neural plate has folded, it must properly close to form the neural tube. This structure eventually gives rise to the brain and spinal cord.
Cell-Cell Communication (Point 5): Cells must communicate effectively to coordinate these early developmental processes. Miscommunication or errors in signaling can lead to severe developmental defects.
Gene Regulation Network (Point 18): A precise network of gene interactions ensures that the right genes are activated (or suppressed) at the right times for neural tube formation.
Morphogen Gradients (Point 28): These are concentration gradients of substances that dictate tissue development. In the context of neural development, morphogens play critical roles in specifying which parts of the neural tube become the brain and which become the spinal cord.
Homeobox and Hox Genes (Point 22): These genes play a pivotal role in setting up the body plan of an embryo along its head-tail axis, including defining regions of the developing brain and spinal cord.
From a system's biology perspective, neural development is a marvel of coordination and communication. For the neural tube to form correctly, cells must communicate with each other, adhere to each other in specific ways, respond to morphogen gradients, and activate the right genes at the right times. All these processes are tightly interwoven, and a failure in one process can impact others. For instance, if the gene regulatory network doesn't activate the right set of genes due to some perturbation, it could potentially affect the morphogen gradients, which in turn might disturb the proper folding of the neural plate, leading to defects in neural tube formation. Given the intricate dance between these processes, it's hard to fathom how such a system could have evolved piecemeal. Without the precise coordination of these multiple factors, the entire process of neural development could be jeopardized. This mutual dependency paints a picture of an orchestrated design where all parts must work in concert for the successful creation of such a complex system.
5. Feedback Loops and Regulatory Mechanisms
The numerous feedback loops and regulatory mechanisms ensure that every cellular process is meticulously monitored and adjusted as necessary. The foresight required for such intricate regulation seems beyond the scope of random mutations and natural selection.
Tissue Development and Maintenance
Diving into the intricate world of cellular growth, differentiation, and communication, let's explore the interwoven dance of several processes from the 47 points:
Cell-Cycle Regulation (Point 3): Cells have inbuilt systems that control their growth and division. A cell must decide when to divide, based on numerous external and internal cues.
Apoptosis (Point 2): Paradoxically, while some cells are growing and dividing, others are programmed to die, ensuring that tissues are sculpted properly and potential rogue cells are eliminated.
Signaling Pathways (Point 40): These pathways relay extracellular signals to intracellular targets, determining whether a cell divides, differentiates, or dies.
Cell Fate Determination and Lineage Specification (Point 6): Within a developing tissue or organ, cells are assigned specific roles. This involves a complex interplay of signals that tell cells to differentiate into one type of cell versus another.
Epigenetic Codes (Point 17): These are modifications to the DNA or associated proteins that don't change the DNA sequence but control gene activity. Epigenetic changes can be induced by environmental factors and can influence cellular decisions like differentiation.
MicroRNA Regulation (Point 27): Small RNAs that don't code for protein but regulate other genes post-transcriptionally. These can fine-tune cellular responses by adjusting the levels of specific proteins in a cell.
Feedback Loops and Hormones (Point 23): Chemical messengers, like hormones, often function within feedback loops, where the output of a system acts as an input to control its behavior, ensuring homeostasis.
Tissue Induction and Organogenesis (Point 47): The formation of specific tissues and organs requires a concert of the above processes. Cells need to grow, communicate, decide their fate, differentiate, or even undergo programmed death, all under the watchful eyes of regulatory networks.
Morphogen Gradients (Point 28): Concentrations of specific molecules in an embryo provide cues to cells, guiding them in their development and spatial organization within tissues and organs.
In this intricate interdependence of cellular processes, each is indispensable. For tissue development and organogenesis to occur correctly, cells need the right mix of growth signals, differentiation cues, and spatial information. If signaling pathways go awry, it can lead to unchecked growth or improper differentiation. If apoptosis doesn't function correctly, it might lead to malformations or predispose tissues to cancers. If epigenetic codes aren't set right, genes essential for proper function might remain silent or get inappropriately activated. All these processes interlock in an elegant dance, each reliant on the other, ensuring that tissues and organs develop properly. Given the sheer complexity and the tight interdependence of these systems, one can argue the challenges it poses to a purely stepwise evolutionary process.
6. Information Storage and Retrieval
The cell's ability to store, retrieve, and implement vast amounts of information is unmatched. DNA, often likened to a data storage system, holds the blueprints for the entire organism. The intricate processes by which this information is accessed, read, and executed seem to be beyond the capacity of unguided evolutionary processes to produce.
Orchestrating Organism Development
Consider the awe-inspiring journey of a single fertilized egg (zygote) as it develops into a complex multicellular organism:
Oogenesis (Point 34) & Spermatogenesis (Point 42): The journey of life begins with the formation of gametes. These processes create the mature egg and sperm, each responsible for carrying half of the genetic information that will lead to a new organism. This initial formation of gametes is foundational to the progression of life.
Oocyte Maturation and Fertilization (Point 35): Following the formation of these gametes, the next step in the dance of life is their fusion. Once the oocyte and sperm unite, a zygote emerges, endowed with a complete set of DNA. This DNA is the architectural blueprint that directs the growth and development of the entire organism.
Gene Regulation Network (Point 18): As the zygote's journey begins, a need for orchestration arises. The gene regulation network offers this orchestration, a vast interconnected web of interactions, determining when, where, and how genes get expressed. This system can be visualized as a master conductor, deciding which sections of the orchestra play and at which moments.
Epigenetic Codes (Point 17): Complementing the conductor, there are specific markers, akin to bookmarks on our DNA, that dictate which musical notes (genes) are emphasized and which are muted. Epigenetic modifications ensure that certain genes are made accessible while others remain silent, all without altering the original score (DNA sequence).
MicroRNA Regulation (Point 27) & Noncoding RNA from Junk DNA (Point 33): Just as a symphony may require fine-tuning, these molecules offer a layer of adjustment to the genetic output after the primary transcript, enhancing or modulating the performance as necessary.
Cell-Cycle Regulation (Point 3): With the foundational notes set, the zygote embarks on a growth journey. This growth is meticulously orchestrated, ensuring that each cellular division is harmonious, with DNA replicated with precision.
Germ Layer Formation (Point 20): As this cellular symphony continues, differentiation begins, setting the stage for the future tissues and organs. Cells start aligning into three primary sections or layers: ectoderm, mesoderm, and endoderm, each layer contributing unique notes to the life song.
Cell Fate Determination and Lineage Specification (Point 6): Within these layers, the individual notes (cells) are further refined and specialized, ensuring that each plays its part in the evolving melody of life.
Signaling Pathways (Point 40) & Morphogen Gradients (Point 28): Communication becomes pivotal as cells continue to evolve and find their position in the overarching composition. These pathways and gradients act as messengers, ensuring each cell understands its role and positioning.
Tissue Induction and Organogenesis (Point 47): The crescendo approaches as cells, driven by unique cues, assemble to form the organs that are vital to life, such as the heart, lungs, and liver.
Cell-Cell Communication (Point 5) & Cell-cell adhesion and the ECM (Point 4): And as the composition reaches its zenith, for the entire system to function harmoniously, cells must communicate and connect, ensuring that every note is in place, creating a beautifully coordinated melody of life.
The life of an organism, as illustrated, is a complex interplay of various systems and processes, each building upon the other, forming a harmonious melody from inception to maturity. This journey, from a single cell to a fully formed organism, involves accessing, reading, and executing a vast amount of information stored within the DNA. At each step, multiple processes from the 47 points are at play, acting like meticulous architects interpreting and building a structure based on an intricate blueprint. Given the precision, coordination, and depth of information involved, it offers a profound reflection on the cell's unmatched information storage and retrieval system.
The intricate interdependence and sheer complexity observed in biological systems make it hard to reconcile with a purely evolutionary framework that relies on random mutations and natural selection. The precision, foresight, and harmony seen in these systems appear to be indicative of a design by an intelligent agent.
Interdependence and Intricacy in Biological Systems
When one looks at the formation and development of complex multicellular organisms, it's akin to witnessing a grand orchestra, where each musician (or process) plays an essential part in crafting a collective, harmonious sound. If even one musician is missing or plays out of tune, the entire performance can be compromised. Similarly, the 47 biological processes are so deeply interwoven that a disturbance or absence in even a single process can lead to systemic disruptions.
Foundational Importance: Just as an orchestra requires foundational instruments like percussion to set the rhythm, processes such as Oogenesis, Spermatogenesis, and Oocyte Maturation and Fertilization set the stage for life's beginning. Without these processes, the journey wouldn't even commence.
Regulation and Coordination: Once the foundational processes are in place, the need for regulation and coordination becomes paramount. The Gene Regulation Network, MicroRNA Regulation, and Epigenetic Codes serve as the conductors and coordinators, ensuring each 'musician' performs at the right time and in harmony with others.
Specialization and Differentiation: As the performance unfolds, specialized instruments like woodwinds or strings introduce unique melodies. Similarly, the Germ Layer Formation and Cell Fate Determination ensure cells differentiate and specialize, adding complexity to the organism's developmental 'symphony'.
Communication: In any orchestra, musicians must listen to and be in sync with each other. The biological equivalents are the Signaling Pathways and Cell-Cell Communication, which guarantee that cells 'listen' to each other and respond appropriately, maintaining the organism's intricate harmony.
Structural Integrity: Just as each section of an orchestra relies on the structure and positioning of its musicians, processes like Cell-cell adhesion and the ECM ensure the physical structure and integrity of tissues and organs.
Systemic Harmony: Finally, all these processes need to work in tandem. Tissue Induction, Organogenesis, and other processes ensure the organism's 'performance' is harmonized from start to finish.
Implications for Evolution and Complexity
The interconnectedness and dependency of these processes pose intriguing questions about the evolution of complex life. The traditional evolutionary model suggests a gradual accumulation of beneficial mutations over time. However, when considering irreducible complexity, a challenge arises: How can systems that rely so heavily on the simultaneous functioning of multiple components evolve incrementally? Such systems seem to defy a piecemeal evolutionary development, as the system wouldn't function (or would offer no evolutionary advantage) until all components are present and working together. The development of multicellular organisms is a marvel of complexity, coordination, and precision, revealing the awe-inspiring intricacies of life.
The Fine Balance of Life
Redundancy and Flexibility: While it's true that the intricacies of these systems point towards an irreducible complexity, nature has also ingeniously incorporated redundancy and flexibility. There are instances where multiple processes can achieve a similar outcome or where systems have backup mechanisms. This 'buffer' allows organisms to survive and adapt in fluctuating environments and under various stresses.
Fine-tuning: These systems are optimized for efficiency and effectiveness. Each process, while essential, has likely been the subject of countless iterations, shaped by environmental pressures and interactions with other processes. This ongoing 'tuning' has resulted in the beautifully orchestrated dance of cellular and molecular events we observe today.
The Starting Point of Complexity: If even the most primitive unicellular organisms required a subset of these 47 processes to survive, then how could such complexity arise spontaneously without guidance? The leap from non-life to even the simplest life form is monumental, given the intricate machinery required at the cellular level. Such complexity, right from the beginning, suggests a purposefully designed set up.
The Problem of Incremental Evolution: How can a partial system, that's non-functional until fully formed, provide a selective advantage? Without the advantage, the process won't be 'selected' and thus, won't evolve. If the machinery of the cell works like a finely tuned watch, missing one gear might render it non-functional. Evolutionary processes can't favor non-functional or less functional states.
The Interconnectedness Challenge: The interconnected nature of the 47 processes outlined implies that changes in one system could have ripple effects across others. A random mutation in one part might require synchronized changes in several others to maintain functionality. Such a level of concurrent and harmonized change seems beyond the capabilities of random mutation and natural selection.
Plasticity and Pre-programming: The capacity for organisms to adapt to their environment is often touted as evidence for evolution. However, this plasticity is evidence of pre-programmed adaptability—a foresight that allows organisms to respond to changing environments. Rather than being proof of random evolution, this built-in adaptability may suggest a designer who anticipated the varied and dynamic environments the organism would encounter.
Information Theory: One significant point is the infusion of information into the DNA. Information, as we understand it in other realms (like coding or linguistics), typically arises from intelligence. The intricate and specific information carried in the DNA, guiding the myriad of processes in the organism, is clear evidence of an intelligent input.[/size]
Last edited by Otangelo on Wed Sep 06, 2023 7:43 am; edited 2 times in total
287 Re: My articles Sun Sep 03, 2023 5:10 am
Re: My articles Sun Sep 03, 2023 5:10 am
Otangelo
Admin
Eukaryotes
As everywhere through evolutionary biology, the claim is that things went from less, to more complex, over long periods of time.
D. A. Peattie: The eukaryote has structural features that allow it to communicate better than prokaryotes, features that permit cellular aggregation and multicellular life. In contrast, the more primitive prokaryotes are less well-equipped for intercellular communication and cannot readily organize into multicellular organisms. Not only do eukaryotic cells allow larger and more complex organisms to be made, but they are themselves larger and more complex than prokaryotic cells. Whether eukaryotic cells live singly or as part of a multicellular organism, their activities can be much more complex and diversified than those of their prokaryotic counterparts. In prokaryotes, all internal cellular events take place within a single compartment, the cytoplasm. Eukaryotes contain many subcellular compartments, called organelles. Even single-celled eukaryotes can display remarkable complexity of function; some have features as specialized and diverse as sensory bristles, mouth parts, muscle-like contractile bundles, or stinging darts.
Structure of a typical animal cell
On a very fundamental level, eukaryotes and prokaryotes are similar. They share many aspects of their basic chemistry, physiology and metabolism. Both cell types are constructed of and use similar kinds of molecules and macromolecules to accomplish their cellular work. In both, for example, membranes are constructed mainly of fatty substances called lipids, and molecules that perform the cell's biological and mechanical work are called proteins.
Eukaryotes and prokaryotes both use the same chemical relay system to make protein. A permanent record of the code for all of the proteins the cell will require is stored in the form of DNA. Because DNA is the master copy of the cell's (or organism's) genetic make-up, the information it contains is absolutely crucial to the maintenance and perpetuation of the cell. As if to safeguard this archive, the cell does not use the DNA directly in protein synthesis but instead copies the information onto a temporary template of RNA, a chemical relative of DNA. Both the DNA and the RNA constitute a "recipe" for the cell's proteins. The recipe specifies the order in which amino acids, the chemical subunits of proteins, should be strung together to make the functional protein. Protein synthesis both in eukaryotes and prokaryotes takes place on structures called ribosomes, which are composed of RNA and protein. This illustrates one way in which prokaryotes and eukaryotes are similar and highlights the idea that differences between these organisms are often architectural. In other words, both cell types use the same bricks and mortar, but the structures they build with these materials vary dramatically.
The prokaryotic cell can be compared to a studio apartment: a one-room living space that has a kitchen area abutting the living room, which converts into a bedroom at night. All necessary items fit into their own locations in one room. There is an everyday; washable rug. Room temperature is comfortable-not too hot, not too cold. Conditions are adequate for everything that must occur in the apartment, but not optimal for any specific activity. In a similar way, all of the prokaryote's functions fit into a single compartment. The DNA is attached to the cell's membrane. Ribosomes float freely in the single compartment. Cellular respiration-the process by which nutrients are metabolized to release energy-is carried out at the cell membrane; there is no dedicated compartment for respiration. A eukaryotic cell can be compared to a mansion, where specific rooms are designed for particular activities. The mansion is more diverse in the activities it supports than the studio apartment. It can accommodate overnight guests comfortably and support social activities for adults in the living room or dining room, for children in the playroom. The baby's room is warm and furnished with bright colors and a soft, thick carpet. The kitchen has a stove, a refrigerator and a tile floor. Items are kept in the room that is most appropriate for them, under conditions ideal for the activities in that specific room. A eukaryotic cell resembles a mansion in that it is subdivided into many compartments. Each compartment is furnished with items and conditions suitable for a specific function, yet the compartments work together to allow the cell to maintain itself, to replicate and to perform more specialized activities.
Taking a closer look, we find three main structural aspects that differentiate prokaryotes from eukaryotes. The definitive difference is the presence of a true (eu) nucleus (karyon) in the eukaryotic cell. The nucleus, a double-membrane casing, sequesters the DNA in its own compartment and keeps it separate from the rest of the cell. In contrast, no such housing is provided for the DNA of a prokaryote. Instead the genetic material is tethered to the cell membrane and is otherwise allowed to float freely in the cell's interior. It is interesting to note that the DNA of eukaryotes is attached to the nuclear membrane, in a manner reminiscent of the attachment of prokaryotic DNA to the cell's outer membrane. 28
The greatest discontinuity in evolution: The gap from prokaryotes to eukaryotes

Ro Y. STANIER et. al., (1963) “The basic divergence in cellular structure, which separates the bacteria and blue-green algae from all other cellular organisms, represents the greatest single evolutionary discontinuity to be found in the presentday world” 31
E. V. Koonin (2002):The eukaryotic chromatin remodeling machinery, the cell cycle regulation systems, the nuclear envelope, the cytoskeleton, and the programmed cell death (PCD, or apoptosis) apparatus all are such major eukaryotic innovations, which do not appear to have direct prokaryotic predecessors.25
E. Derelle et.al.,(2006): The unicellular green marine alga Ostreococcus tauri is the world's smallest free-living eukaryote known to date, and encodes the fewest number of genes. It has been hypothesized, based on its small cellular and genome sizes, that it may reveal the “bare limits” of life as a free-living photosynthetic eukaryote, presumably having disposed of redundancies and presenting a simple organization and very little noncoding sequence. 27 It has a genome size of 12.560,000 base pairs, 8,166 genes and 7745 proteins. in comparison, the simplest free-living bacteria today is Pelagibacter ubique get by with about 1,300 genes and 1,308,759 base pairs and code for 1,354 proteins.
T. Cavalier-Smith (2010): This radical transformation of cell structure (eukaryogenesis) is the most complex and extensive case of quantum evolution in the history of life. Beforehand earth was a sexless, purely bacterial and viral world. Afterwards sexy, endoskeletal eukaryotes evolved morphological complexity: diatoms, butterflies, corals, whales, kelps, and trees 32
E. Szathmáry (2015): The divide between prokaryotes and eukaryotes is the biggest known evolutionary discontinuity. There is no space here to enter the whole maze of the recent debate about the origin of the eukaryotic cells; suffice it to say that the picture seems more obscure than 20 y ago. How did eukaryotic life evolve? This is one of the most controversial and puzzling questions in evolutionary history. Life began as single-celled, independent organisms that evolved into cells containing membrane-bound, specialized structures known as organelles. What’s clear is that this new type of cell, the eukaryote, is more complex than its predecessors. What’s unclear is how these changes took place. 24
A.Kauko (2018): The origin of eukaryotes is one of the central transitions in the history of life; without eukaryotes there would be no complex multicellular life.36
F.Rana (2019): The origin of eukaryotes is one of the hardest and most intriguing problems in the study of the evolution of life, and arguably, in the whole of biology. On average, the volume of eukaryotic cells is about 15,000 times larger than that of prokaryotic cells.30
Josip Skejo (2021): Eukaryotic cells are vastly more complex than prokaryotic cells as evident by their endomembrane system 26
A. Spang (2022): Archaea and Bacteria are often referred to as primary domains of life while eukaryotes form a secondary domain of life. The prevalence of horizontal gene transfer (HGT) via both mobile genetic elements (MGEs) and viruses but also directly between distinct organisms has to some extent questioned the concept of a Tree of Life (TOL), which may be more correctly represented as a network including both vertical and horizontal branches.
Arizona State University (2022): The transition from prokaryote to eukaryote has remained a central mystery biologists are still trying to untangle. How this crucial transition came to be remains a central mystery in biology.40
Origin of eukaryotes
M. A. O’Malley (2015): There are very roughly two main hypotheses for the evolution of eukaryotes: one sees the process as mutation-driven, with lateral acquisitions of genes and organisms also involved but in a causally secondary way; the other sees eukaryogenesis as driven causally by the acquisition of the mitochondrion. The acquisition of the mitochondrion is often portrayed as a one-off event that instigated a rapid transformation with major evolutionary outcomes 38
Eugene V. Koonin (2015): The origin of eukaryotes is one of the hardest and most intriguing problems in the study of the evolution of life, and arguably, in the whole of biology. Compared to archaea and bacteria (collectively, prokaryotes), eukaryotic cells display a qualitatively higher level of complexity of intracellular organization. Unlike the great majority of prokaryotes, eukaryotic cells possess an extended system of intracellular membranes that includes the eponymous eukaryotic organelle, the nucleus, and fully compartmentalizes the intracellular space. In eukaryotic cells, proteins, nucleic acids and small molecules are distributed by specific trafficking mechanisms rather than by free diffusion as is largely the case in bacteria and archaea. Thus, eukaryotic cells function on different physical principles compared to prokaryotic cells, which is directly due to their (comparatively) enormous size. The gulf between the cellular organizations of eukaryotes and prokaryotes is all the more striking because no intermediates have been found. The actin and tubulin cytoskeletons, the nuclear pore, the spliceosome, the proteasome, and the ubiquitin signalling system are only a few of the striking examples of the organizational complexity that seems to be a ‘birthright’ of eukaryotic cells. The formidable problem that these fundamental complex features present to evolutionary biologists makes Darwin’s famous account of the evolution of the eye look like a simple, straightforward case. Indeed, so intimidating is the challenge of eukaryogenesis that the infamous notion of ‘irreducible complexity’ has sneaked into serious scientific debate:
C. G. Kurland: Genomics and the Irreducible Nature of Eukaryote Cells (2006): Data from many sources give no direct evidence that eukaryotes evolved by genome fusion between archaea and bacteria. Because their cells appear simpler, prokaryotes have traditionally been considered ancestors of eukaryotes. Here, we review recent data from proteomics and genome sequences suggesting that eukaryotes are a unique primordial lineage. Mitochondria, mitosomes, and hydrogenosomes are a related family of organelles that distinguish eukaryotes from all prokaryotes. Recent analyses also suggest that early eukaryotes had many introns, and RNAs and proteins found in modern spliceosomes. Nuclei, nucleoli, Golgi apparatus, centrioles, and endoplasmic reticulum are examples of cellular signature structures (CSSs) that distinguish eukaryote cells from archaea and bacteria. Comparative genomics, aided by proteomics of CSSs such as the mitochondria, nucleoli, and spliceosomes, reveals hundreds of proteins with no orthologs (Orthologs are genes in different species that evolved from a common ancestral gene by speciation) evident in the genomes of prokaryotes; these are the eukaryotic signature proteins (ESPs). The many ESPs within the subcellular structures of eukaryote cells provide landmarks to track the trajectory of eukaryote genomes from their origins. In contrast, hypotheses that attribute eukaryote origins to genome fusion between archaea and bacteria are surprisingly uninformative about the emergence of the cellular and genomic signatures of eukaryotes (CSSs and ESPs). The failure of genome fusion to directly explain any characteristic feature of the eukaryote cell is a critical starting point for studying eukaryote origins. 34
As everywhere through evolutionary biology, the claim is that things went from less, to more complex, over long periods of time.
D. A. Peattie: The eukaryote has structural features that allow it to communicate better than prokaryotes, features that permit cellular aggregation and multicellular life. In contrast, the more primitive prokaryotes are less well-equipped for intercellular communication and cannot readily organize into multicellular organisms. Not only do eukaryotic cells allow larger and more complex organisms to be made, but they are themselves larger and more complex than prokaryotic cells. Whether eukaryotic cells live singly or as part of a multicellular organism, their activities can be much more complex and diversified than those of their prokaryotic counterparts. In prokaryotes, all internal cellular events take place within a single compartment, the cytoplasm. Eukaryotes contain many subcellular compartments, called organelles. Even single-celled eukaryotes can display remarkable complexity of function; some have features as specialized and diverse as sensory bristles, mouth parts, muscle-like contractile bundles, or stinging darts.

Structure of a typical animal cell
On a very fundamental level, eukaryotes and prokaryotes are similar. They share many aspects of their basic chemistry, physiology and metabolism. Both cell types are constructed of and use similar kinds of molecules and macromolecules to accomplish their cellular work. In both, for example, membranes are constructed mainly of fatty substances called lipids, and molecules that perform the cell's biological and mechanical work are called proteins.
Eukaryotes and prokaryotes both use the same chemical relay system to make protein. A permanent record of the code for all of the proteins the cell will require is stored in the form of DNA. Because DNA is the master copy of the cell's (or organism's) genetic make-up, the information it contains is absolutely crucial to the maintenance and perpetuation of the cell. As if to safeguard this archive, the cell does not use the DNA directly in protein synthesis but instead copies the information onto a temporary template of RNA, a chemical relative of DNA. Both the DNA and the RNA constitute a "recipe" for the cell's proteins. The recipe specifies the order in which amino acids, the chemical subunits of proteins, should be strung together to make the functional protein. Protein synthesis both in eukaryotes and prokaryotes takes place on structures called ribosomes, which are composed of RNA and protein. This illustrates one way in which prokaryotes and eukaryotes are similar and highlights the idea that differences between these organisms are often architectural. In other words, both cell types use the same bricks and mortar, but the structures they build with these materials vary dramatically.
The prokaryotic cell can be compared to a studio apartment: a one-room living space that has a kitchen area abutting the living room, which converts into a bedroom at night. All necessary items fit into their own locations in one room. There is an everyday; washable rug. Room temperature is comfortable-not too hot, not too cold. Conditions are adequate for everything that must occur in the apartment, but not optimal for any specific activity. In a similar way, all of the prokaryote's functions fit into a single compartment. The DNA is attached to the cell's membrane. Ribosomes float freely in the single compartment. Cellular respiration-the process by which nutrients are metabolized to release energy-is carried out at the cell membrane; there is no dedicated compartment for respiration. A eukaryotic cell can be compared to a mansion, where specific rooms are designed for particular activities. The mansion is more diverse in the activities it supports than the studio apartment. It can accommodate overnight guests comfortably and support social activities for adults in the living room or dining room, for children in the playroom. The baby's room is warm and furnished with bright colors and a soft, thick carpet. The kitchen has a stove, a refrigerator and a tile floor. Items are kept in the room that is most appropriate for them, under conditions ideal for the activities in that specific room. A eukaryotic cell resembles a mansion in that it is subdivided into many compartments. Each compartment is furnished with items and conditions suitable for a specific function, yet the compartments work together to allow the cell to maintain itself, to replicate and to perform more specialized activities.
Taking a closer look, we find three main structural aspects that differentiate prokaryotes from eukaryotes. The definitive difference is the presence of a true (eu) nucleus (karyon) in the eukaryotic cell. The nucleus, a double-membrane casing, sequesters the DNA in its own compartment and keeps it separate from the rest of the cell. In contrast, no such housing is provided for the DNA of a prokaryote. Instead the genetic material is tethered to the cell membrane and is otherwise allowed to float freely in the cell's interior. It is interesting to note that the DNA of eukaryotes is attached to the nuclear membrane, in a manner reminiscent of the attachment of prokaryotic DNA to the cell's outer membrane. 28
The greatest discontinuity in evolution: The gap from prokaryotes to eukaryotes

Ro Y. STANIER et. al., (1963) “The basic divergence in cellular structure, which separates the bacteria and blue-green algae from all other cellular organisms, represents the greatest single evolutionary discontinuity to be found in the presentday world” 31
E. V. Koonin (2002):The eukaryotic chromatin remodeling machinery, the cell cycle regulation systems, the nuclear envelope, the cytoskeleton, and the programmed cell death (PCD, or apoptosis) apparatus all are such major eukaryotic innovations, which do not appear to have direct prokaryotic predecessors.25
E. Derelle et.al.,(2006): The unicellular green marine alga Ostreococcus tauri is the world's smallest free-living eukaryote known to date, and encodes the fewest number of genes. It has been hypothesized, based on its small cellular and genome sizes, that it may reveal the “bare limits” of life as a free-living photosynthetic eukaryote, presumably having disposed of redundancies and presenting a simple organization and very little noncoding sequence. 27 It has a genome size of 12.560,000 base pairs, 8,166 genes and 7745 proteins. in comparison, the simplest free-living bacteria today is Pelagibacter ubique get by with about 1,300 genes and 1,308,759 base pairs and code for 1,354 proteins.
T. Cavalier-Smith (2010): This radical transformation of cell structure (eukaryogenesis) is the most complex and extensive case of quantum evolution in the history of life. Beforehand earth was a sexless, purely bacterial and viral world. Afterwards sexy, endoskeletal eukaryotes evolved morphological complexity: diatoms, butterflies, corals, whales, kelps, and trees 32
E. Szathmáry (2015): The divide between prokaryotes and eukaryotes is the biggest known evolutionary discontinuity. There is no space here to enter the whole maze of the recent debate about the origin of the eukaryotic cells; suffice it to say that the picture seems more obscure than 20 y ago. How did eukaryotic life evolve? This is one of the most controversial and puzzling questions in evolutionary history. Life began as single-celled, independent organisms that evolved into cells containing membrane-bound, specialized structures known as organelles. What’s clear is that this new type of cell, the eukaryote, is more complex than its predecessors. What’s unclear is how these changes took place. 24
A.Kauko (2018): The origin of eukaryotes is one of the central transitions in the history of life; without eukaryotes there would be no complex multicellular life.36
F.Rana (2019): The origin of eukaryotes is one of the hardest and most intriguing problems in the study of the evolution of life, and arguably, in the whole of biology. On average, the volume of eukaryotic cells is about 15,000 times larger than that of prokaryotic cells.30
Josip Skejo (2021): Eukaryotic cells are vastly more complex than prokaryotic cells as evident by their endomembrane system 26
A. Spang (2022): Archaea and Bacteria are often referred to as primary domains of life while eukaryotes form a secondary domain of life. The prevalence of horizontal gene transfer (HGT) via both mobile genetic elements (MGEs) and viruses but also directly between distinct organisms has to some extent questioned the concept of a Tree of Life (TOL), which may be more correctly represented as a network including both vertical and horizontal branches.
Arizona State University (2022): The transition from prokaryote to eukaryote has remained a central mystery biologists are still trying to untangle. How this crucial transition came to be remains a central mystery in biology.40
Origin of eukaryotes
M. A. O’Malley (2015): There are very roughly two main hypotheses for the evolution of eukaryotes: one sees the process as mutation-driven, with lateral acquisitions of genes and organisms also involved but in a causally secondary way; the other sees eukaryogenesis as driven causally by the acquisition of the mitochondrion. The acquisition of the mitochondrion is often portrayed as a one-off event that instigated a rapid transformation with major evolutionary outcomes 38
Eugene V. Koonin (2015): The origin of eukaryotes is one of the hardest and most intriguing problems in the study of the evolution of life, and arguably, in the whole of biology. Compared to archaea and bacteria (collectively, prokaryotes), eukaryotic cells display a qualitatively higher level of complexity of intracellular organization. Unlike the great majority of prokaryotes, eukaryotic cells possess an extended system of intracellular membranes that includes the eponymous eukaryotic organelle, the nucleus, and fully compartmentalizes the intracellular space. In eukaryotic cells, proteins, nucleic acids and small molecules are distributed by specific trafficking mechanisms rather than by free diffusion as is largely the case in bacteria and archaea. Thus, eukaryotic cells function on different physical principles compared to prokaryotic cells, which is directly due to their (comparatively) enormous size. The gulf between the cellular organizations of eukaryotes and prokaryotes is all the more striking because no intermediates have been found. The actin and tubulin cytoskeletons, the nuclear pore, the spliceosome, the proteasome, and the ubiquitin signalling system are only a few of the striking examples of the organizational complexity that seems to be a ‘birthright’ of eukaryotic cells. The formidable problem that these fundamental complex features present to evolutionary biologists makes Darwin’s famous account of the evolution of the eye look like a simple, straightforward case. Indeed, so intimidating is the challenge of eukaryogenesis that the infamous notion of ‘irreducible complexity’ has sneaked into serious scientific debate:
C. G. Kurland: Genomics and the Irreducible Nature of Eukaryote Cells (2006): Data from many sources give no direct evidence that eukaryotes evolved by genome fusion between archaea and bacteria. Because their cells appear simpler, prokaryotes have traditionally been considered ancestors of eukaryotes. Here, we review recent data from proteomics and genome sequences suggesting that eukaryotes are a unique primordial lineage. Mitochondria, mitosomes, and hydrogenosomes are a related family of organelles that distinguish eukaryotes from all prokaryotes. Recent analyses also suggest that early eukaryotes had many introns, and RNAs and proteins found in modern spliceosomes. Nuclei, nucleoli, Golgi apparatus, centrioles, and endoplasmic reticulum are examples of cellular signature structures (CSSs) that distinguish eukaryote cells from archaea and bacteria. Comparative genomics, aided by proteomics of CSSs such as the mitochondria, nucleoli, and spliceosomes, reveals hundreds of proteins with no orthologs (Orthologs are genes in different species that evolved from a common ancestral gene by speciation) evident in the genomes of prokaryotes; these are the eukaryotic signature proteins (ESPs). The many ESPs within the subcellular structures of eukaryote cells provide landmarks to track the trajectory of eukaryote genomes from their origins. In contrast, hypotheses that attribute eukaryote origins to genome fusion between archaea and bacteria are surprisingly uninformative about the emergence of the cellular and genomic signatures of eukaryotes (CSSs and ESPs). The failure of genome fusion to directly explain any characteristic feature of the eukaryote cell is a critical starting point for studying eukaryote origins. 34
288 Re: My articles Fri Dec 29, 2023 6:11 am
Re: My articles Fri Dec 29, 2023 6:11 am
Otangelo
Admin
The Rational Exploration of the Universe's Origins point to a Creator
When I hear atheists or skeptics respond with "I don't know" to the question of explaining the origin of our existence, it seems to me like a position of willful ignorance, especially in light of the evidence we have.
Building on the understanding that nothingness, defined as the absolute absence of anything, lacks causal powers, it's evident that something must have always existed to give rise to the reality we experience today. This fundamental insight leads us to two primary alternatives: either the universe has always existed in some form, or there is a higher, supernatural power that has always existed. The concept of an eternal universe, one that has always existed in some form, has been a subject of scientific and philosophical debate. However, we have solid evidence suggesting that the physical universe cannot be eternally past. The Big Bang theory, for instance, points to a specific origin in time, a moment of creation where the universe began to expand from an extremely hot and dense state. Additionally, concepts like the second law of thermodynamics, which talks about the increase of entropy, suggest that the universe is running down like a clock. If it had been eternal, it would have reached a state of heat death an infinite time ago, which it clearly has not.
Given these limitations to the concept of an eternal universe, the alternative perspective points to the existence of a higher, supernatural power that has always existed. This entity, often referred to as God, is posited as the ultimate cause of everything. If we agree that nothing can come from nothing, and something has always existed, then this something must have the power to bring the universe into existence. This line of reasoning leads to the conclusion that God, as a necessary being, exists outside of time and space and is the uncaused cause of the universe. This understanding of God as the ultimate reality, the prime mover, or the uncaused cause, is not just a religious or philosophical concept but is also aligned with the notion in cosmology that the universe had a beginning. If the universe had a beginning, it must have had a cause, and this cause, by necessity, must be outside the realm of the universe's physical laws and properties. It must be powerful, timeless, spaceless, immaterial, and personal, possessing the will to create. Considering the evidence against an eternal physical universe and the logical impossibility of something coming from absolute nothing, the existence of a supernatural, always-existing entity seems not just plausible but necessary. This line of thought leads us to recognize a higher power, an ultimate cause that itself is uncaused – a perspective that aligns with the concept of God in many religious and philosophical traditions.
The intricacy and specificity we observe in nature are best explained, in my opinion, by the action of an intelligent designer. Refusing to acknowledge this evidence, despite its abundance and the capacity we have for reason and understanding, feels unjustifiable to me. Recognizing the existence of God, given the evidence, seems far more rational and logically sound than denial or skepticism. The discoveries and knowledge we've accumulated about the natural world powerfully point towards the existence of God. To ignore or dismiss this evidence is not a position of informed skepticism, but rather one of willful ignorance. This isn't just about understanding the physical world; it's about acknowledging the profound implications of such knowledge on our understanding of life, the universe, and our own existence. Science's deepening grasp of worldly complexities uncovers evidence suggestive of a sophisticated and intentionally designed universe. This path of discovery yields profound insights, that are not based on lack of knowledge ( God of the gaps), but unraveling the evidence, and logically conclude that it points to a creator:
The complexity and intricacy observed in the universe and biological systems imply a deliberate design, hinting at an intelligent designer. The analogy of the universe as a winding-down clock suggests an initial fully wound state, indicating a definite beginning and thus a cause. The universe abides by specific laws and constants. The fine-tuning of these constants to support life prompts inquiries into who or what meticulously adjusted these parameters. The complexity and functionality of cells resemble advanced factories, indicating a design and intelligence level surpassing mere evolutionary processes. Cells' ability to encode detailed information in genes and translate it into physical forms showcases a marvel of biological engineering. DNA's role as a highly efficient information storage system, embodying life's blueprint, reflects sophistication and deliberate design. The presence of morality, consciousness, and human capacities for language and logic suggest a non-material dimension to our existence, pointing towards a higher intelligence or divine influence.
Scientific investigation is not only a tool for earthly understanding but also a means to comprehend God's designs. This pursuit serves to enhance our knowledge of the world, aiding our endeavors and deepening our understanding of the divine architect's intricate plans. Thus, the moral of this story is not merely the recognition of a profound intelligence behind the universe's creation but also an appreciation of this magnificently designed cosmos, and the mind behind all this.

When I hear atheists or skeptics respond with "I don't know" to the question of explaining the origin of our existence, it seems to me like a position of willful ignorance, especially in light of the evidence we have.
Building on the understanding that nothingness, defined as the absolute absence of anything, lacks causal powers, it's evident that something must have always existed to give rise to the reality we experience today. This fundamental insight leads us to two primary alternatives: either the universe has always existed in some form, or there is a higher, supernatural power that has always existed. The concept of an eternal universe, one that has always existed in some form, has been a subject of scientific and philosophical debate. However, we have solid evidence suggesting that the physical universe cannot be eternally past. The Big Bang theory, for instance, points to a specific origin in time, a moment of creation where the universe began to expand from an extremely hot and dense state. Additionally, concepts like the second law of thermodynamics, which talks about the increase of entropy, suggest that the universe is running down like a clock. If it had been eternal, it would have reached a state of heat death an infinite time ago, which it clearly has not.
Given these limitations to the concept of an eternal universe, the alternative perspective points to the existence of a higher, supernatural power that has always existed. This entity, often referred to as God, is posited as the ultimate cause of everything. If we agree that nothing can come from nothing, and something has always existed, then this something must have the power to bring the universe into existence. This line of reasoning leads to the conclusion that God, as a necessary being, exists outside of time and space and is the uncaused cause of the universe. This understanding of God as the ultimate reality, the prime mover, or the uncaused cause, is not just a religious or philosophical concept but is also aligned with the notion in cosmology that the universe had a beginning. If the universe had a beginning, it must have had a cause, and this cause, by necessity, must be outside the realm of the universe's physical laws and properties. It must be powerful, timeless, spaceless, immaterial, and personal, possessing the will to create. Considering the evidence against an eternal physical universe and the logical impossibility of something coming from absolute nothing, the existence of a supernatural, always-existing entity seems not just plausible but necessary. This line of thought leads us to recognize a higher power, an ultimate cause that itself is uncaused – a perspective that aligns with the concept of God in many religious and philosophical traditions.
The intricacy and specificity we observe in nature are best explained, in my opinion, by the action of an intelligent designer. Refusing to acknowledge this evidence, despite its abundance and the capacity we have for reason and understanding, feels unjustifiable to me. Recognizing the existence of God, given the evidence, seems far more rational and logically sound than denial or skepticism. The discoveries and knowledge we've accumulated about the natural world powerfully point towards the existence of God. To ignore or dismiss this evidence is not a position of informed skepticism, but rather one of willful ignorance. This isn't just about understanding the physical world; it's about acknowledging the profound implications of such knowledge on our understanding of life, the universe, and our own existence. Science's deepening grasp of worldly complexities uncovers evidence suggestive of a sophisticated and intentionally designed universe. This path of discovery yields profound insights, that are not based on lack of knowledge ( God of the gaps), but unraveling the evidence, and logically conclude that it points to a creator:
The complexity and intricacy observed in the universe and biological systems imply a deliberate design, hinting at an intelligent designer. The analogy of the universe as a winding-down clock suggests an initial fully wound state, indicating a definite beginning and thus a cause. The universe abides by specific laws and constants. The fine-tuning of these constants to support life prompts inquiries into who or what meticulously adjusted these parameters. The complexity and functionality of cells resemble advanced factories, indicating a design and intelligence level surpassing mere evolutionary processes. Cells' ability to encode detailed information in genes and translate it into physical forms showcases a marvel of biological engineering. DNA's role as a highly efficient information storage system, embodying life's blueprint, reflects sophistication and deliberate design. The presence of morality, consciousness, and human capacities for language and logic suggest a non-material dimension to our existence, pointing towards a higher intelligence or divine influence.
Scientific investigation is not only a tool for earthly understanding but also a means to comprehend God's designs. This pursuit serves to enhance our knowledge of the world, aiding our endeavors and deepening our understanding of the divine architect's intricate plans. Thus, the moral of this story is not merely the recognition of a profound intelligence behind the universe's creation but also an appreciation of this magnificently designed cosmos, and the mind behind all this.

289 Re: My articles Fri Dec 29, 2023 10:12 am
Re: My articles Fri Dec 29, 2023 10:12 am
Otangelo
Admin
https://www.dailymail.co.uk/sciencetech/article-12880829/Are-living-simulation-Scientist-claims-simply-characters-advanced-virtual-world-says-easy-way-prove-it.html
The fact there are limits to how fast light and sound can travel suggest they may be governed by the speed of a computer processor, according to the expert.
The laws of physics that govern the universe are also akin to computer code, he says, while elementary particles that make up matter are like pixels.
Commentary: Well, if we live in a computer simulation, the question, of course, is, who or what runs the simulation, and why?
I think our reality is not a simulation, but - actually - reality, but run by God who set it up. He did set up the laws of physics - arbitrarily- to create an intelligible, stable world, that, otherwise, would be chaotic, if it somehow would exist, by unguided means.
The suggestion that the limits on the speed of light and sound might be comparable to the limitations of a computer processor draws parallels between the fundamental laws of physics and the rules of a computer simulation. This analogy extends to viewing elementary particles as akin to pixels, the basic units of digital images. Such perspectives are often inspired by the remarkable regularities and mathematical precision found in the laws of nature, leading some to speculate about the nature of reality itself – whether it could be a simulation.
However, the idea that our reality is a simulation brings up profound philosophical and theological questions. If we were living in a simulated reality, it would naturally lead to the question of who or what is running this simulation. This line of thinking inevitably leads to the concept of a creator or a higher power – a programmer or designer of the simulation.
My belief is that our reality is not a simulation but is indeed actual reality, created and governed by God. God is the ultimate source and sustainer of the universe. God established the laws of physics not randomly, but with the specific intention of creating a coherent, intelligible world. The laws of physics, rather than being arbitrary, are finely tuned to allow for the existence of life and a stable universe. The complexity and order of the universe are evidence of divine design, rather than the outcome of a cosmic algorithm or happenstance simulation.
Expanding on this, the idea that the universe is orderly and governed by consistent laws is a cornerstone of both science and many religious beliefs. This orderliness allows us to understand the universe through scientific inquiry, the laws of nature are intelligible and accessible to human reasoning. The concept that these laws are not just random but are set up in a way that enables life to exist and thrive implies a purpose and intention behind their design.
Moreover, the notion that the universe is not a product of unguided processes but is instead the creation of a divine intelligence offers a framework for understanding the complexity and beauty of the world around us. It provides a foundation for values, morality, and a sense of meaning and purpose in life. The belief in a divine creator who set up the universe with intention and purpose stands in contrast to the idea of a random, unguided simulation, offers a richer and more profound understanding of our existence , that aligns better with our reality, and scientific evidence.
The fact there are limits to how fast light and sound can travel suggest they may be governed by the speed of a computer processor, according to the expert.
The laws of physics that govern the universe are also akin to computer code, he says, while elementary particles that make up matter are like pixels.
Commentary: Well, if we live in a computer simulation, the question, of course, is, who or what runs the simulation, and why?
I think our reality is not a simulation, but - actually - reality, but run by God who set it up. He did set up the laws of physics - arbitrarily- to create an intelligible, stable world, that, otherwise, would be chaotic, if it somehow would exist, by unguided means.
The suggestion that the limits on the speed of light and sound might be comparable to the limitations of a computer processor draws parallels between the fundamental laws of physics and the rules of a computer simulation. This analogy extends to viewing elementary particles as akin to pixels, the basic units of digital images. Such perspectives are often inspired by the remarkable regularities and mathematical precision found in the laws of nature, leading some to speculate about the nature of reality itself – whether it could be a simulation.
However, the idea that our reality is a simulation brings up profound philosophical and theological questions. If we were living in a simulated reality, it would naturally lead to the question of who or what is running this simulation. This line of thinking inevitably leads to the concept of a creator or a higher power – a programmer or designer of the simulation.
My belief is that our reality is not a simulation but is indeed actual reality, created and governed by God. God is the ultimate source and sustainer of the universe. God established the laws of physics not randomly, but with the specific intention of creating a coherent, intelligible world. The laws of physics, rather than being arbitrary, are finely tuned to allow for the existence of life and a stable universe. The complexity and order of the universe are evidence of divine design, rather than the outcome of a cosmic algorithm or happenstance simulation.
Expanding on this, the idea that the universe is orderly and governed by consistent laws is a cornerstone of both science and many religious beliefs. This orderliness allows us to understand the universe through scientific inquiry, the laws of nature are intelligible and accessible to human reasoning. The concept that these laws are not just random but are set up in a way that enables life to exist and thrive implies a purpose and intention behind their design.
Moreover, the notion that the universe is not a product of unguided processes but is instead the creation of a divine intelligence offers a framework for understanding the complexity and beauty of the world around us. It provides a foundation for values, morality, and a sense of meaning and purpose in life. The belief in a divine creator who set up the universe with intention and purpose stands in contrast to the idea of a random, unguided simulation, offers a richer and more profound understanding of our existence , that aligns better with our reality, and scientific evidence.
290 Re: My articles Mon Jan 01, 2024 5:09 am
Re: My articles Mon Jan 01, 2024 5:09 am
Otangelo
Admin
Ecclesiastes speaks to a profound mystery: the notion that the divine plan is vast and complex, beyond the full comprehension of human beings.
The Bible provides narratives about the creation, the unfolding of human history, and eschatological events like those described in Revelation. They offer a framework for understanding morality and ethics, positioning human beings within a larger story that gives meaning to our actions. They provide a source of hope and comfort; despite the apparent chaos and suffering in the world, there is a divine plan that will ultimately lead to a good and just conclusion.
They help to form a sense of community and identity among believers, who see themselves as part of a continuous tradition that stretches from creation through to the end times. The centrality of Jesus in the New Testament reflects the Christian belief that in the person of Jesus, the divine was made accessible and understandable to humanity in a unique way. Jesus is the culmination of God's revelation, the moment when the divine plan was most clearly manifested on Earth. His life, death, and resurrection are pivotal events in human history, providing the clearest insight into the nature of God and His intentions for humanity. God may have chosen to reveal Himself in this manner to invite humans into a relationship based on trust and faith rather than on comprehensive knowledge. This approach to revelation could be intended to develop human moral and spiritual qualities, such as humility, faith, and dependence on God, rather than encouraging a sense of self-sufficiency based on complete knowledge. The concept of progressive revelation suggests that God has revealed Himself and His purposes gradually over time in ways that are accessible and appropriate to different people and cultures.
In summary, the scriptures seem to provide enough detail to guide belief and practice, to offer comfort, and to create a sense of community and shared purpose, while also leaving much unrevealed, thus creating space for faith, for a diversity of interpretations, and for the ongoing, active engagement of believers in their quest to understand and relate to the divine. The core message of the New Testament revolves around the theme of redemption and the concept of Jesus as the savior who offers a path to salvation through his sacrificial death and resurrection. This message is encapsulated in the concept of the Gospel, which means "good news." The good news is that through Jesus, the barrier of sin that separates humanity from God is removed for those who believe.
Isaiah 53 is a pivotal Old Testament passage that foretells the coming of Jesus as a suffering servant who takes upon himself the iniquities of others.
"But he was pierced for our transgressions, he was crushed for our iniquities; the punishment that brought us peace was on him, and by his wounds we are healed." - **Isaiah 53:5 (NIV)**
This theme is further elaborated in the New Testament, with numerous passages underscoring the redemptive work of Christ:
"For God so loved the world that he gave his one and only Son, that whoever believes in him shall not perish but have eternal life." - **John 3:16 (NIV)**
This verse from the Gospel of John is probably the most well-known and succinctly captures the essence of the Gospel message: the offer of salvation through belief in Jesus Christ.
"In him, we have redemption through his blood, the forgiveness of sins, in accordance with the riches of God’s grace." - **Ephesians 1:7 (NIV)**
Paul's letter to the Ephesians emphasizes the role of Jesus' sacrifice as the means for redemption and forgiveness, highlighting the grace of God in this plan.
"But God demonstrates his own love for us in this: While we were still sinners, Christ died for us." - **Romans 5:8 (NIV)**
The Book of Romans offers a theological exposition of the Gospel, with Paul explaining how Christ's death serves as a demonstration of God's love and a mechanism for justifying sinners.
"Therefore, if anyone is in Christ, the new creation has come: The old has gone, the new is here!" - **2 Corinthians 5:17 (NIV)**
This passage describes the transformative power of accepting Christ: a believer is made a "new creation," indicating a fresh start and a restored relationship with God.
"For Christ also suffered once for sins, the righteous for the unrighteous, to bring you to God. He was put to death in the body but made alive in the Spirit." - **1 Peter 3:18 (NIV)**
Here, Peter speaks to the singular nature of Christ's sacrifice as a substitutionary atonement for humanity's sins, meant to reconcile the unrighteous with God.
The culmination of the Gospel message is not only in the atoning death of Jesus but also in his resurrection, which is seen as the assurance of eternal life for believers:
"And if Christ has not been raised, your faith is futile; you are still in your sins." - **1 Corinthians 15:17 (NIV)**
The resurrection is central to Christian faith, as it provides the evidence of Jesus' victory over sin and death and the hope of resurrection for believers.
These verses, among many others, form the foundation of what is the core message of the New Testament: that Jesus' life, death, and resurrection constitute the means by which we can have a restored relationship with God. This relationship is marked by repentance, surrender, belief, and a commitment to follow Jesus and his teachings, leading to a life transformed by love and grace.
Romans 3, particularly verses 23-26, is a central text in Christian theology that addresses the concept of redemption through Jesus Christ. The passage explains that all people have sinned and fall short of God's glory. Sin creates a separation between humanity and God, who is holy and righteous. The consequences of sin are so severe that they cannot be remedied by human efforts.
Romans 3:23 ("for all have sinned and fall short of the glory of God") every person has sinned. This sin creates a need for atonement or reconciliation with God. God is both just and merciful. His justice requires that sin be punished, as it's an affront to His holiness. However, His mercy desires to forgive and restore the relationship with humanity. The death of Christ is the perfect solution to this, satisfying both God's justice (as the penalty for sin is paid) and mercy (offering forgiveness to sinners). Jesus Christ died in place of sinners, taking upon Himself the punishment that was due for our sins. This is an act of grace, where Christ bears the consequences of sin on behalf of humanity. The benefits of Christ's sacrifice are available to all, but they must be personally accepted. Those who believe in Jesus Christ and accept His sacrifice are forgiven and reconciled with God. On the other hand, those who reject this offer must bear the consequences of their sins themselves, as they have not accepted the atonement provided through Christ. The seriousness of sin and the holiness of God necessitate a significant act for redemption. The death of Christ on the cross is the only sufficient means to bridge the gap between a holy God and sinful humanity. This is unnegotiable, untradable central core, traditional Christian theology, and there is no space for interpretations and varying beliefs.
The belief in the necessity of Christ's suffering and death on the cross, and the idea that there was no other way for humanity to be reconciled with God, is deeply rooted in Christian theology. God is inherently holy and just. His holiness means He is separate from sin and cannot tolerate it. His justice demands that sin, which is seen as rebellion against God, must be punished. The penalty for sin, according to the Bible, is death and separation from God (Romans 6:23). Humanity, being sinful and finite, cannot make adequate atonement for sin. No human being is perfect, and therefore, no one can pay the infinite price required to atone for sins committed. This is a key aspect of Christian soteriology (the study of salvation). It holds that Christ, being both fully divine and fully human, was the only one capable of bearing the full penalty for human sin. His divinity gives His sacrifice infinite value, and His humanity makes His sacrifice relevant for all humans. Jesus' death was also about obedience to the will of the Father. In the Garden of Gethsemane, Jesus prays, "not as I will, but as you will" (Matthew 26:39), demonstrating His submission to God's plan. This obedience is seen as part of what made His sacrifice acceptable. The death of Christ on the cross is also seen as the supreme demonstration of God's love and mercy. John 3:16 states, "For God so loved the world, that he gave his only Son, that whoever believes in him should not perish but have eternal life." The cross is a necessary demonstration of God's love, offering redemption to each one of us. Jesus' death is fulfilling the prophecies of the Old Testament and the requirements of the Law. He is often referred to as the "Lamb of God," an allusion to the sacrificial system of the Old Testament, where a lamb without blemish was sacrificed for the atonement of sin. The resurrection of Christ is crucial in this context. It is God's validation of Jesus' sacrifice, demonstrating His power over sin and death and confirming Jesus as the Son of God. There was no other way apart from the crucifixion for the reconciliation between God and humanity. This is founded on the notions of God's holiness and justice, human sinfulness, and the unique nature of Christ as fully God and fully man. The crucifixion is a necessary act of divine love and justice to atone for human sin and to offer salvation.
The claim that Jesus saves us because He overcame evil, and not because He saves us from God's wrath, presents an interpretation that focuses on the victory over evil and sin. However, traditional Christian theology, as reflected in the Bible, does indeed emphasize that Jesus' sacrifice on the cross was also to save humanity from God's wrath against sin.
Romans 5:9: "Since we have now been justified by His blood, how much more shall we be saved from God's wrath through Him!" This verse clearly states that through Jesus’ sacrifice (His blood), believers are not only justified but also saved from the wrath of God.
1 Thessalonians 1:10: "And to wait for His Son from heaven, whom He raised from the dead—Jesus, who rescues us from the coming wrath." Here, Paul speaks about Jesus as the one who rescues believers from the wrath that is to come, referring to God's future judgment.
John 3:36: "Whoever believes in the Son has eternal life, but whoever rejects the Son will not see life, for God's wrath remains on them." This verse contrasts the eternal life received by believing in Jesus with the continued state of being under God's wrath for those who reject Him.
Romans 1:18: "The wrath of God is being revealed from heaven against all the godlessness and wickedness of people, who suppress the truth by their wickedness." This verse sets the context for why salvation from God’s wrath is necessary—it is revealed against all ungodliness and unrighteousness.
Ephesians 2:3: "All of us also lived among them at one time, gratifying the cravings of our flesh and following its desires and thoughts. Like the rest, we were by nature deserving of wrath." Paul indicates that all people were once subject to God's wrath due to their sinful nature, but implies that this changed for those in Christ.
Romans 2:5: "But because of your stubbornness and your unrepentant heart, you are storing up wrath against yourself for the day of God’s wrath, when his righteous judgment will be revealed." This speaks to the concept of God’s wrath being stored up because of sin.
These verses collectively demonstrate that a significant aspect of Jesus’ mission, was to save humanity from the wrath of God, which is directed against sin. The concept of God's wrath in the Bible is tied to His holiness and justice, and the salvation offered through Jesus Christ as the means by which believers are rescued from this wrath. This aspect of salvation does not negate the victory over evil and sin but rather complements it, presenting a holistic view of salvation.
There are several passages in the Bible that substantiate that Jesus saved humanity from God's wrath, aligning with the PSA perspective:
Romans 5:9: "Since we have now been justified by his blood, how much more shall we be saved from God’s wrath through him!" This verse directly links Jesus’ death (symbolized by His blood) to salvation from God’s wrath.
1 Thessalonians 1:10: "And to wait for his Son from heaven, whom he raised from the dead—Jesus, who rescues us from the coming wrath." This passage speaks of Jesus as the rescuer from the future wrath of God.
John 3:36: "Whoever believes in the Son has eternal life, but whoever rejects the Son will not see life, for God’s wrath remains on them." This contrasts the state of those who believe in Jesus (and thereby escape God's wrath) with those who don't.
Ephesians 2:3: "All of us also lived among them at one time, gratifying the cravings of our flesh and following its desires and thoughts. Like the rest, we were by nature deserving of wrath." This implies that before faith in Christ, all were under the wrath of God due to sin.
Romans 2:5: "But because of your stubbornness and your unrepentant heart, you are storing up wrath against yourself for the day of God’s wrath, when his righteous judgment will be revealed." This suggests a future outpouring of God’s wrath from which believers in Christ are spared.
Colossians 3:5-6: "Put to death, therefore, whatever belongs to your earthly nature... Because of these, the wrath of God is coming." This passage warns of God's wrath against sin, implying that salvation through Christ offers a way to avoid this wrath.
Hebrews 2:17: "For this reason he had to be made like them, fully human in every way, in order that he might become a merciful and faithful high priest in service to God, and that he might make atonement for the sins of the people." While not explicitly mentioning wrath, this speaks to the idea of Jesus making atonement for sin, which in PSA is understood as absorbing God's wrath against sin.
Revelation 6:16-17: "They called to the mountains and the rocks, ‘Fall on us and hide us from the face of him who sits on the throne and from the wrath of the Lamb! For the great day of their wrath has come, and who can withstand it?’" This apocalyptic vision describes the wrath of God and the Lamb (Christ) in the context of final judgment.
These passages support the notion that Jesus' death was a means of saving humanity from the wrath of God that is due because of sin. Jesus as beared the penalty for sin on humanity's behalf, thus satisfying God's justice and allowing for the extension of mercy to sinners.
There are statements and parables in the Gospels, that imply or indirectly reference Jesus role in saving humanity from God's wrath. These include:
John 3:16-17: "For God so loved the world, that he gave his only Son, that whoever believes in him should not perish but have eternal life. For God did not send his Son into the world to condemn the world, but in order that the world might be saved through him." This passage is often interpreted as Jesus stating His mission to save humanity, with "not perish" implying rescue from God's judgment or wrath.
Mark 10:45: "For even the Son of Man did not come to be served, but to serve, and to give his life as a ransom for many." The concept of giving His life as a ransom is interpreted by some as Jesus taking upon Himself the punishment or wrath deserved by humanity.
Matthew 26:28: "This is my blood of the covenant, which is poured out for many for the forgiveness of sins." In this statement during the Last Supper, Jesus refers to His impending death as a sacrificial act leading to the forgiveness of sins, which can be interpreted as a saving act from the consequences of sin, including God's wrath.
John 12:47: "If anyone hears my words and does not keep them, I do not judge him; for I did not come to judge the world but to save the world." Here Jesus speaks of His mission as one of salvation, not immediate judgment, which can be understood as offering salvation from the ultimate judgment of God.
Luke 19:10: "For the Son of Man came to seek and to save the lost." This statement, while not explicitly mentioning wrath, speaks to Jesus' mission of salvation, which is often interpreted in the context of saving from sin and its consequences, including divine wrath.
The Christus Victor view detracts from the core message of the gospel.
The Christus Victor view sees Christ's death and resurrection as a victory over the powers of sin, death, and the devil. It emphasizes Jesus as a conqueror who liberates humanity from bondage. The Penal Substitutionary Atonement is the core message. It posits that Jesus died as a substitute for sinners, bearing the punishment that was due to us, thus satisfying the wrath of God against sin. If one excludes the penal substitution, it misses a crucial aspect of the Christian understanding of salvation. The New Testament frequently presents Jesus' death as a substitutionary sacrifice for sins (e.g., 1 Peter 2:24, Isaiah 53:5-6, Romans 3:25). This sacrifice was necessary to reconcile sinful humanity with a holy God. Penal substitution stresses God's justice. It suggests that God's love and mercy don't negate His justice. Instead, Jesus satisfies divine justice by bearing the penalty for sin, thereby offering mercy to sinners (Romans 3:26). This emphasizes a personal aspect of salvation – that Christ died for each individual's sins. This personal dimension is a key component of the gospel message, reinforcing a personal relationship with God through Christ. While Christus Victor is a significant aspect of atonement theology, focusing solely on it leads to a less comprehensive understanding of the multifaceted nature of Jesus' work. The New Testament presents a multi-dimensional view of atonement, including ransom, victory, substitution, and moral influence. A balanced theological perspective acknowledges the validity of multiple atonement theories found in Scripture. For instance, Christ's victory over evil powers (Colossians 2:15) and His role as a substitutionary sacrifice (1 John 2:2) are both biblically grounded. While the Christus Victor model is a valuable aspect of understanding Christ's work, it is important to also recognize and maintain the centrality of the penal substitutionary view in the overall Christian doctrine of salvation. This balanced approach allows for a fuller appreciation of the complexity and depth of what the New Testament teaches about the meaning and significance of Jesus' death and resurrection.
Various Bible verses emphasize how sin affects God and the necessity of justice and atonement in the Christian understanding:
Romans 3:23-25: "For all have sinned and fall short of the glory of God, and are justified freely by his grace through the redemption that came by Christ Jesus. God presented Christ as a sacrifice of atonement, through the shedding of his blood—to be received by faith. He did this to demonstrate his justice, because in his forbearance he had left the sins committed beforehand unpunished."
Isaiah 53:5-6: "But he was pierced for our transgressions, he was crushed for our iniquities; the punishment that brought us peace was upon him, and by his wounds we are healed. We all, like sheep, have gone astray, each of us has turned to his own way; and the LORD has laid on him the iniquity of us all."
Psalm 51:4: "Against you, you only, have I sinned and done what is evil in your sight; so you are right in your verdict and justified when you judge."
Hebrews 9:22: "In fact, the law requires that nearly everything be cleansed with blood, and without the shedding of blood there is no forgiveness."
1 John 2:2: "He is the atoning sacrifice for our sins, and not only for ours but also for the sins of the whole world."
2 Corinthians 5:21: "God made him who had no sin to be sin for us, so that in him we might become the righteousness of God."
Romans 6:23: "For the wages of sin is death, but the gift of God is eternal life in Christ Jesus our Lord."
These verses collectively highlight that sin is a serious offense against God, requiring a response that addresses both the nature of sin and the holiness and justice of God. The concept of atonement, particularly through the sacrifice of Jesus, is central in these passages, underscoring the belief that God’s justice required a payment or penalty for sin, which was satisfied through Christ’s death. This reflects the Christian doctrine of penal substitution, where Christ takes upon himself the penalty for human sin, reconciling humanity with God.
The Bible provides narratives about the creation, the unfolding of human history, and eschatological events like those described in Revelation. They offer a framework for understanding morality and ethics, positioning human beings within a larger story that gives meaning to our actions. They provide a source of hope and comfort; despite the apparent chaos and suffering in the world, there is a divine plan that will ultimately lead to a good and just conclusion.
They help to form a sense of community and identity among believers, who see themselves as part of a continuous tradition that stretches from creation through to the end times. The centrality of Jesus in the New Testament reflects the Christian belief that in the person of Jesus, the divine was made accessible and understandable to humanity in a unique way. Jesus is the culmination of God's revelation, the moment when the divine plan was most clearly manifested on Earth. His life, death, and resurrection are pivotal events in human history, providing the clearest insight into the nature of God and His intentions for humanity. God may have chosen to reveal Himself in this manner to invite humans into a relationship based on trust and faith rather than on comprehensive knowledge. This approach to revelation could be intended to develop human moral and spiritual qualities, such as humility, faith, and dependence on God, rather than encouraging a sense of self-sufficiency based on complete knowledge. The concept of progressive revelation suggests that God has revealed Himself and His purposes gradually over time in ways that are accessible and appropriate to different people and cultures.
In summary, the scriptures seem to provide enough detail to guide belief and practice, to offer comfort, and to create a sense of community and shared purpose, while also leaving much unrevealed, thus creating space for faith, for a diversity of interpretations, and for the ongoing, active engagement of believers in their quest to understand and relate to the divine. The core message of the New Testament revolves around the theme of redemption and the concept of Jesus as the savior who offers a path to salvation through his sacrificial death and resurrection. This message is encapsulated in the concept of the Gospel, which means "good news." The good news is that through Jesus, the barrier of sin that separates humanity from God is removed for those who believe.
Isaiah 53 is a pivotal Old Testament passage that foretells the coming of Jesus as a suffering servant who takes upon himself the iniquities of others.
"But he was pierced for our transgressions, he was crushed for our iniquities; the punishment that brought us peace was on him, and by his wounds we are healed." - **Isaiah 53:5 (NIV)**
This theme is further elaborated in the New Testament, with numerous passages underscoring the redemptive work of Christ:
"For God so loved the world that he gave his one and only Son, that whoever believes in him shall not perish but have eternal life." - **John 3:16 (NIV)**
This verse from the Gospel of John is probably the most well-known and succinctly captures the essence of the Gospel message: the offer of salvation through belief in Jesus Christ.
"In him, we have redemption through his blood, the forgiveness of sins, in accordance with the riches of God’s grace." - **Ephesians 1:7 (NIV)**
Paul's letter to the Ephesians emphasizes the role of Jesus' sacrifice as the means for redemption and forgiveness, highlighting the grace of God in this plan.
"But God demonstrates his own love for us in this: While we were still sinners, Christ died for us." - **Romans 5:8 (NIV)**
The Book of Romans offers a theological exposition of the Gospel, with Paul explaining how Christ's death serves as a demonstration of God's love and a mechanism for justifying sinners.
"Therefore, if anyone is in Christ, the new creation has come: The old has gone, the new is here!" - **2 Corinthians 5:17 (NIV)**
This passage describes the transformative power of accepting Christ: a believer is made a "new creation," indicating a fresh start and a restored relationship with God.
"For Christ also suffered once for sins, the righteous for the unrighteous, to bring you to God. He was put to death in the body but made alive in the Spirit." - **1 Peter 3:18 (NIV)**
Here, Peter speaks to the singular nature of Christ's sacrifice as a substitutionary atonement for humanity's sins, meant to reconcile the unrighteous with God.
The culmination of the Gospel message is not only in the atoning death of Jesus but also in his resurrection, which is seen as the assurance of eternal life for believers:
"And if Christ has not been raised, your faith is futile; you are still in your sins." - **1 Corinthians 15:17 (NIV)**
The resurrection is central to Christian faith, as it provides the evidence of Jesus' victory over sin and death and the hope of resurrection for believers.
These verses, among many others, form the foundation of what is the core message of the New Testament: that Jesus' life, death, and resurrection constitute the means by which we can have a restored relationship with God. This relationship is marked by repentance, surrender, belief, and a commitment to follow Jesus and his teachings, leading to a life transformed by love and grace.
Romans 3, particularly verses 23-26, is a central text in Christian theology that addresses the concept of redemption through Jesus Christ. The passage explains that all people have sinned and fall short of God's glory. Sin creates a separation between humanity and God, who is holy and righteous. The consequences of sin are so severe that they cannot be remedied by human efforts.
Romans 3:23 ("for all have sinned and fall short of the glory of God") every person has sinned. This sin creates a need for atonement or reconciliation with God. God is both just and merciful. His justice requires that sin be punished, as it's an affront to His holiness. However, His mercy desires to forgive and restore the relationship with humanity. The death of Christ is the perfect solution to this, satisfying both God's justice (as the penalty for sin is paid) and mercy (offering forgiveness to sinners). Jesus Christ died in place of sinners, taking upon Himself the punishment that was due for our sins. This is an act of grace, where Christ bears the consequences of sin on behalf of humanity. The benefits of Christ's sacrifice are available to all, but they must be personally accepted. Those who believe in Jesus Christ and accept His sacrifice are forgiven and reconciled with God. On the other hand, those who reject this offer must bear the consequences of their sins themselves, as they have not accepted the atonement provided through Christ. The seriousness of sin and the holiness of God necessitate a significant act for redemption. The death of Christ on the cross is the only sufficient means to bridge the gap between a holy God and sinful humanity. This is unnegotiable, untradable central core, traditional Christian theology, and there is no space for interpretations and varying beliefs.
The belief in the necessity of Christ's suffering and death on the cross, and the idea that there was no other way for humanity to be reconciled with God, is deeply rooted in Christian theology. God is inherently holy and just. His holiness means He is separate from sin and cannot tolerate it. His justice demands that sin, which is seen as rebellion against God, must be punished. The penalty for sin, according to the Bible, is death and separation from God (Romans 6:23). Humanity, being sinful and finite, cannot make adequate atonement for sin. No human being is perfect, and therefore, no one can pay the infinite price required to atone for sins committed. This is a key aspect of Christian soteriology (the study of salvation). It holds that Christ, being both fully divine and fully human, was the only one capable of bearing the full penalty for human sin. His divinity gives His sacrifice infinite value, and His humanity makes His sacrifice relevant for all humans. Jesus' death was also about obedience to the will of the Father. In the Garden of Gethsemane, Jesus prays, "not as I will, but as you will" (Matthew 26:39), demonstrating His submission to God's plan. This obedience is seen as part of what made His sacrifice acceptable. The death of Christ on the cross is also seen as the supreme demonstration of God's love and mercy. John 3:16 states, "For God so loved the world, that he gave his only Son, that whoever believes in him should not perish but have eternal life." The cross is a necessary demonstration of God's love, offering redemption to each one of us. Jesus' death is fulfilling the prophecies of the Old Testament and the requirements of the Law. He is often referred to as the "Lamb of God," an allusion to the sacrificial system of the Old Testament, where a lamb without blemish was sacrificed for the atonement of sin. The resurrection of Christ is crucial in this context. It is God's validation of Jesus' sacrifice, demonstrating His power over sin and death and confirming Jesus as the Son of God. There was no other way apart from the crucifixion for the reconciliation between God and humanity. This is founded on the notions of God's holiness and justice, human sinfulness, and the unique nature of Christ as fully God and fully man. The crucifixion is a necessary act of divine love and justice to atone for human sin and to offer salvation.
The claim that Jesus saves us because He overcame evil, and not because He saves us from God's wrath, presents an interpretation that focuses on the victory over evil and sin. However, traditional Christian theology, as reflected in the Bible, does indeed emphasize that Jesus' sacrifice on the cross was also to save humanity from God's wrath against sin.
Romans 5:9: "Since we have now been justified by His blood, how much more shall we be saved from God's wrath through Him!" This verse clearly states that through Jesus’ sacrifice (His blood), believers are not only justified but also saved from the wrath of God.
1 Thessalonians 1:10: "And to wait for His Son from heaven, whom He raised from the dead—Jesus, who rescues us from the coming wrath." Here, Paul speaks about Jesus as the one who rescues believers from the wrath that is to come, referring to God's future judgment.
John 3:36: "Whoever believes in the Son has eternal life, but whoever rejects the Son will not see life, for God's wrath remains on them." This verse contrasts the eternal life received by believing in Jesus with the continued state of being under God's wrath for those who reject Him.
Romans 1:18: "The wrath of God is being revealed from heaven against all the godlessness and wickedness of people, who suppress the truth by their wickedness." This verse sets the context for why salvation from God’s wrath is necessary—it is revealed against all ungodliness and unrighteousness.
Ephesians 2:3: "All of us also lived among them at one time, gratifying the cravings of our flesh and following its desires and thoughts. Like the rest, we were by nature deserving of wrath." Paul indicates that all people were once subject to God's wrath due to their sinful nature, but implies that this changed for those in Christ.
Romans 2:5: "But because of your stubbornness and your unrepentant heart, you are storing up wrath against yourself for the day of God’s wrath, when his righteous judgment will be revealed." This speaks to the concept of God’s wrath being stored up because of sin.
These verses collectively demonstrate that a significant aspect of Jesus’ mission, was to save humanity from the wrath of God, which is directed against sin. The concept of God's wrath in the Bible is tied to His holiness and justice, and the salvation offered through Jesus Christ as the means by which believers are rescued from this wrath. This aspect of salvation does not negate the victory over evil and sin but rather complements it, presenting a holistic view of salvation.
There are several passages in the Bible that substantiate that Jesus saved humanity from God's wrath, aligning with the PSA perspective:
Romans 5:9: "Since we have now been justified by his blood, how much more shall we be saved from God’s wrath through him!" This verse directly links Jesus’ death (symbolized by His blood) to salvation from God’s wrath.
1 Thessalonians 1:10: "And to wait for his Son from heaven, whom he raised from the dead—Jesus, who rescues us from the coming wrath." This passage speaks of Jesus as the rescuer from the future wrath of God.
John 3:36: "Whoever believes in the Son has eternal life, but whoever rejects the Son will not see life, for God’s wrath remains on them." This contrasts the state of those who believe in Jesus (and thereby escape God's wrath) with those who don't.
Ephesians 2:3: "All of us also lived among them at one time, gratifying the cravings of our flesh and following its desires and thoughts. Like the rest, we were by nature deserving of wrath." This implies that before faith in Christ, all were under the wrath of God due to sin.
Romans 2:5: "But because of your stubbornness and your unrepentant heart, you are storing up wrath against yourself for the day of God’s wrath, when his righteous judgment will be revealed." This suggests a future outpouring of God’s wrath from which believers in Christ are spared.
Colossians 3:5-6: "Put to death, therefore, whatever belongs to your earthly nature... Because of these, the wrath of God is coming." This passage warns of God's wrath against sin, implying that salvation through Christ offers a way to avoid this wrath.
Hebrews 2:17: "For this reason he had to be made like them, fully human in every way, in order that he might become a merciful and faithful high priest in service to God, and that he might make atonement for the sins of the people." While not explicitly mentioning wrath, this speaks to the idea of Jesus making atonement for sin, which in PSA is understood as absorbing God's wrath against sin.
Revelation 6:16-17: "They called to the mountains and the rocks, ‘Fall on us and hide us from the face of him who sits on the throne and from the wrath of the Lamb! For the great day of their wrath has come, and who can withstand it?’" This apocalyptic vision describes the wrath of God and the Lamb (Christ) in the context of final judgment.
These passages support the notion that Jesus' death was a means of saving humanity from the wrath of God that is due because of sin. Jesus as beared the penalty for sin on humanity's behalf, thus satisfying God's justice and allowing for the extension of mercy to sinners.
There are statements and parables in the Gospels, that imply or indirectly reference Jesus role in saving humanity from God's wrath. These include:
John 3:16-17: "For God so loved the world, that he gave his only Son, that whoever believes in him should not perish but have eternal life. For God did not send his Son into the world to condemn the world, but in order that the world might be saved through him." This passage is often interpreted as Jesus stating His mission to save humanity, with "not perish" implying rescue from God's judgment or wrath.
Mark 10:45: "For even the Son of Man did not come to be served, but to serve, and to give his life as a ransom for many." The concept of giving His life as a ransom is interpreted by some as Jesus taking upon Himself the punishment or wrath deserved by humanity.
Matthew 26:28: "This is my blood of the covenant, which is poured out for many for the forgiveness of sins." In this statement during the Last Supper, Jesus refers to His impending death as a sacrificial act leading to the forgiveness of sins, which can be interpreted as a saving act from the consequences of sin, including God's wrath.
John 12:47: "If anyone hears my words and does not keep them, I do not judge him; for I did not come to judge the world but to save the world." Here Jesus speaks of His mission as one of salvation, not immediate judgment, which can be understood as offering salvation from the ultimate judgment of God.
Luke 19:10: "For the Son of Man came to seek and to save the lost." This statement, while not explicitly mentioning wrath, speaks to Jesus' mission of salvation, which is often interpreted in the context of saving from sin and its consequences, including divine wrath.
The Christus Victor view detracts from the core message of the gospel.
The Christus Victor view sees Christ's death and resurrection as a victory over the powers of sin, death, and the devil. It emphasizes Jesus as a conqueror who liberates humanity from bondage. The Penal Substitutionary Atonement is the core message. It posits that Jesus died as a substitute for sinners, bearing the punishment that was due to us, thus satisfying the wrath of God against sin. If one excludes the penal substitution, it misses a crucial aspect of the Christian understanding of salvation. The New Testament frequently presents Jesus' death as a substitutionary sacrifice for sins (e.g., 1 Peter 2:24, Isaiah 53:5-6, Romans 3:25). This sacrifice was necessary to reconcile sinful humanity with a holy God. Penal substitution stresses God's justice. It suggests that God's love and mercy don't negate His justice. Instead, Jesus satisfies divine justice by bearing the penalty for sin, thereby offering mercy to sinners (Romans 3:26). This emphasizes a personal aspect of salvation – that Christ died for each individual's sins. This personal dimension is a key component of the gospel message, reinforcing a personal relationship with God through Christ. While Christus Victor is a significant aspect of atonement theology, focusing solely on it leads to a less comprehensive understanding of the multifaceted nature of Jesus' work. The New Testament presents a multi-dimensional view of atonement, including ransom, victory, substitution, and moral influence. A balanced theological perspective acknowledges the validity of multiple atonement theories found in Scripture. For instance, Christ's victory over evil powers (Colossians 2:15) and His role as a substitutionary sacrifice (1 John 2:2) are both biblically grounded. While the Christus Victor model is a valuable aspect of understanding Christ's work, it is important to also recognize and maintain the centrality of the penal substitutionary view in the overall Christian doctrine of salvation. This balanced approach allows for a fuller appreciation of the complexity and depth of what the New Testament teaches about the meaning and significance of Jesus' death and resurrection.
Various Bible verses emphasize how sin affects God and the necessity of justice and atonement in the Christian understanding:
Romans 3:23-25: "For all have sinned and fall short of the glory of God, and are justified freely by his grace through the redemption that came by Christ Jesus. God presented Christ as a sacrifice of atonement, through the shedding of his blood—to be received by faith. He did this to demonstrate his justice, because in his forbearance he had left the sins committed beforehand unpunished."
Isaiah 53:5-6: "But he was pierced for our transgressions, he was crushed for our iniquities; the punishment that brought us peace was upon him, and by his wounds we are healed. We all, like sheep, have gone astray, each of us has turned to his own way; and the LORD has laid on him the iniquity of us all."
Psalm 51:4: "Against you, you only, have I sinned and done what is evil in your sight; so you are right in your verdict and justified when you judge."
Hebrews 9:22: "In fact, the law requires that nearly everything be cleansed with blood, and without the shedding of blood there is no forgiveness."
1 John 2:2: "He is the atoning sacrifice for our sins, and not only for ours but also for the sins of the whole world."
2 Corinthians 5:21: "God made him who had no sin to be sin for us, so that in him we might become the righteousness of God."
Romans 6:23: "For the wages of sin is death, but the gift of God is eternal life in Christ Jesus our Lord."
These verses collectively highlight that sin is a serious offense against God, requiring a response that addresses both the nature of sin and the holiness and justice of God. The concept of atonement, particularly through the sacrifice of Jesus, is central in these passages, underscoring the belief that God’s justice required a payment or penalty for sin, which was satisfied through Christ’s death. This reflects the Christian doctrine of penal substitution, where Christ takes upon himself the penalty for human sin, reconciling humanity with God.
Last edited by Otangelo on Tue Jan 02, 2024 6:20 am; edited 2 times in total
291 Re: My articles Mon Jan 01, 2024 5:31 pm
Re: My articles Mon Jan 01, 2024 5:31 pm
Otangelo
Admin
The Resurrection as Validation
The resurrection of Jesus Christ symbolizes the victory over sin and death. The resurrection is God the Father's confirmation that Christ's sacrifice was sufficient to atone for the sins of humanity.
The Sinlessness of Christ
Hebrews 4:15: "For we do not have a high priest who is unable to empathize with our weaknesses, but we have one who has been tempted in every way, just as we are—yet he did not sin."
1 Peter 2:22: "He committed no sin, and no deceit was found in his mouth."
Exegesis: These verses affirm the sinless nature of Jesus. His sinlessness is crucial because it qualifies Him as the perfect sacrifice, capable of atoning for the sins of humanity. Unlike humans, who are born with a sinful nature, Jesus, being sinless, was not subject to the law of sin and death.
The Price of Sin and the Sacrifice of Christ
Romans 6:23: "For the wages of sin is death, but the gift of God is eternal life in Christ Jesus our Lord."
1 John 2:2: "He is the atoning sacrifice for our sins, and not only for ours but also for the sins of the whole world."
Exegesis: Sin, as stated in Romans, earns the wage of death. This death is not just physical but also spiritual—eternal separation from God. Jesus' death on the cross, being a sinless sacrifice, paid this price in full, satisfying the justice of God. It is through His sacrifice that humanity can be reconciled with God.
The Resurrection as Validation
Romans 1:4: "And who through the Spirit of holiness was declared with power to be the Son of God by his resurrection from the dead: Jesus Christ our Lord."
1 Corinthians 15:17: "And if Christ has not been raised, your faith is futile; you are still in your sins."
Exegesis: The resurrection is God's seal of approval on the work of Christ. If Christ had remained dead, it would imply that His sacrifice was insufficient. However, the resurrection confirms that not only was Jesus' sacrifice sufficient, but it also demonstrates His power over death, affirming His divinity and the truth of His teachings.
The Implications for Believers
Romans 8:11: "And if the Spirit of him who raised Jesus from the dead is living in you, he who raised Christ from the dead will also give life to your mortal bodies because of his Spirit who lives in you."
1 Peter 1:3: "Praise be to the God and Father of our Lord Jesus Christ! In his great mercy he has given us new birth into a living hope through the resurrection of Jesus Christ from the dead."
Exegesis: For believers, the resurrection is not just a historical event but a source of hope and assurance. It promises them new life and a future resurrection, similar to Christ's. The same power that raised Jesus from the dead is at work in those who believe, ensuring their ultimate victory over sin and death.
The resurrection of Jesus Christ is a pivotal event that signifies the acceptance of Christ's sacrifice by the Father. It underscores the sinless nature of Christ, His victory over sin and death, and the hope of resurrection for believers. This event is foundational to Christian faith, offering assurance of eternal life and reconciliation with God.
The resurrection of Jesus Christ symbolizes the victory over sin and death. The resurrection is God the Father's confirmation that Christ's sacrifice was sufficient to atone for the sins of humanity.
The Sinlessness of Christ
Hebrews 4:15: "For we do not have a high priest who is unable to empathize with our weaknesses, but we have one who has been tempted in every way, just as we are—yet he did not sin."
1 Peter 2:22: "He committed no sin, and no deceit was found in his mouth."
Exegesis: These verses affirm the sinless nature of Jesus. His sinlessness is crucial because it qualifies Him as the perfect sacrifice, capable of atoning for the sins of humanity. Unlike humans, who are born with a sinful nature, Jesus, being sinless, was not subject to the law of sin and death.
The Price of Sin and the Sacrifice of Christ
Romans 6:23: "For the wages of sin is death, but the gift of God is eternal life in Christ Jesus our Lord."
1 John 2:2: "He is the atoning sacrifice for our sins, and not only for ours but also for the sins of the whole world."
Exegesis: Sin, as stated in Romans, earns the wage of death. This death is not just physical but also spiritual—eternal separation from God. Jesus' death on the cross, being a sinless sacrifice, paid this price in full, satisfying the justice of God. It is through His sacrifice that humanity can be reconciled with God.
The Resurrection as Validation
Romans 1:4: "And who through the Spirit of holiness was declared with power to be the Son of God by his resurrection from the dead: Jesus Christ our Lord."
1 Corinthians 15:17: "And if Christ has not been raised, your faith is futile; you are still in your sins."
Exegesis: The resurrection is God's seal of approval on the work of Christ. If Christ had remained dead, it would imply that His sacrifice was insufficient. However, the resurrection confirms that not only was Jesus' sacrifice sufficient, but it also demonstrates His power over death, affirming His divinity and the truth of His teachings.
The Implications for Believers
Romans 8:11: "And if the Spirit of him who raised Jesus from the dead is living in you, he who raised Christ from the dead will also give life to your mortal bodies because of his Spirit who lives in you."
1 Peter 1:3: "Praise be to the God and Father of our Lord Jesus Christ! In his great mercy he has given us new birth into a living hope through the resurrection of Jesus Christ from the dead."
Exegesis: For believers, the resurrection is not just a historical event but a source of hope and assurance. It promises them new life and a future resurrection, similar to Christ's. The same power that raised Jesus from the dead is at work in those who believe, ensuring their ultimate victory over sin and death.
The resurrection of Jesus Christ is a pivotal event that signifies the acceptance of Christ's sacrifice by the Father. It underscores the sinless nature of Christ, His victory over sin and death, and the hope of resurrection for believers. This event is foundational to Christian faith, offering assurance of eternal life and reconciliation with God.
292 Re: My articles Thu Jan 04, 2024 4:35 am
Re: My articles Thu Jan 04, 2024 4:35 am
Otangelo
Admin
Contrasting Worldviews: Earthly Investments vs. Eternal Promises
It's intriguing to consider how some of the world's wealthiest individuals, like Mark Zuckerberg and Jeff Bezos, invest heavily in projects to either extend or protect their lives. Zuckerberg is reportedly building an expensive bunker in Hawaii to safeguard against potential nuclear threats, while Bezos is funding biotechnology research aimed at prolonging human life. These actions underscore the high value they place on life on Earth.
From a Christian perspective, this approach contrasts sharply with the teachings of Jesus, who emphasized the futility of worrying excessively about earthly concerns, as it does not extend one's lifespan. Faith in Christ offers a path to eternal life, without the need for such earthly investments. God promises a new heaven, and a new earth for those who believe and follow Christ. God plans to create a new heaven and a new earth, free from evil and invites everyone to be a part of this eternal kingdom.
Christ also encourages us to invest already here and now in the heavens bank, and the heavenly kingdom. How? Serving each other. The greatest in heaven will be those who served here on earth. Jesus came to serve us. Now we are called to serve each other in love. That means taking care, being graceful, loving, gentle, helping, etc.
This view contrasts with a naturalistic worldview, which typically focuses on maximizing the present life, as it does not anticipate an existence beyond death. The hope in this perspective is to experience all the good in the now, as the is no life, no future beyond this life. Faith in Christ is priceless and offers an invaluable new perspective, peace, and purpose that transcends material wealth and earthly concerns.

It's intriguing to consider how some of the world's wealthiest individuals, like Mark Zuckerberg and Jeff Bezos, invest heavily in projects to either extend or protect their lives. Zuckerberg is reportedly building an expensive bunker in Hawaii to safeguard against potential nuclear threats, while Bezos is funding biotechnology research aimed at prolonging human life. These actions underscore the high value they place on life on Earth.
From a Christian perspective, this approach contrasts sharply with the teachings of Jesus, who emphasized the futility of worrying excessively about earthly concerns, as it does not extend one's lifespan. Faith in Christ offers a path to eternal life, without the need for such earthly investments. God promises a new heaven, and a new earth for those who believe and follow Christ. God plans to create a new heaven and a new earth, free from evil and invites everyone to be a part of this eternal kingdom.
Christ also encourages us to invest already here and now in the heavens bank, and the heavenly kingdom. How? Serving each other. The greatest in heaven will be those who served here on earth. Jesus came to serve us. Now we are called to serve each other in love. That means taking care, being graceful, loving, gentle, helping, etc.
This view contrasts with a naturalistic worldview, which typically focuses on maximizing the present life, as it does not anticipate an existence beyond death. The hope in this perspective is to experience all the good in the now, as the is no life, no future beyond this life. Faith in Christ is priceless and offers an invaluable new perspective, peace, and purpose that transcends material wealth and earthly concerns.

293 Re: My articles Sat Jan 06, 2024 5:18 am
Re: My articles Sat Jan 06, 2024 5:18 am
Otangelo
Admin
The Narrow Path: Discerning True Faith today
"Enter by the narrow gate. For the gate is wide and the way is easy that leads to destruction, and those who enter by it are many. For the gate is narrow and the way is hard that leads to life, and those who find it are few." - Matthew 7:13-14 (ESV)
This passage, being part of the Sermon on the Mount, presents a metaphorical framework for understanding the choices individuals make in life, particularly about faith and morality. Jesus describes two paths: one wide and easy, leading to destruction, and the other narrow and difficult, leading to life. Jesus makes a distinction between living a life that is self-serving and one that aligns with being born again spiritually, repenting from a sinful, godless life, surrendering to His love and grace, receiving Him as Savior, and following Him, having Him as center, and not one's self.
In all ages, this metaphor has reflected the diversity of beliefs and lifestyles. The rise of secularism and atheism in recent times is evidence of the wide road, where individuals choose paths that do not acknowledge Christ as who He said to be: Jesus said, "I am the way, and the truth, and the life. No one comes to the Father except through me." (John 14:6). Similarly, the presence of multiple religious sects promoting false teachings, that do not align with the truth of Jesus' message, are also are part of the broader road leading to destruction. Moreover, some identify as Christians yet live outside His will. This discrepancy between confession and practice is another aspect of the broad path, where the label of faith is present but the commitment is shallow. In this context, the prophecy that Jesus speaks of doesn't merely become a statement about the afterlife, but also a commentary on the human condition and the challenges of living a life of genuine faith. The fact that the majority of people do not choose the path of strict adherence to Jesus' teachings is a confirmation of the prophecy, so unbelievers are unwillingly through their condition confirming Jesus' prophecy.
The narrow path is characterized by those who not only confess their faith in Jesus but also prove that their faith is genuine, by producing good fruits coming from the spirit that dwells in them, that experienced a change of their hearts through the rebirth in spirit, those who strive to live in a way that is consistent with the Lord's teachings, despite societal pressures or the popularity of alternative lifestyles. This path is marked by a commitment to spiritual growth, ethical behavior, and often, service to others, which can be more demanding and less traveled.
This dichotomy of paths serves as a call to self-examination for us who confess Christ, encouraging believers to reflect on the authenticity of their practice and the depth of their commitment to the principles taught by Jesus.
"Not everyone who says to me, 'Lord, Lord,' will enter the kingdom of heaven, but the one who does the will of my Father who is in heaven. On that day many will say to me, 'Lord, Lord, did we not prophesy in your name, and cast out demons in your name, and do many mighty works in your name?' And then will I declare to them, 'I never knew you; depart from me, you workers of lawlessness.'" - Matthew 7:21-23 (ESV)
The key distinction Jesus makes is not between believers and non-believers, but between true and nominal disciples. The former genuinely adhere to God's will, while the latter perform acts of piety or power in Jesus' name but lack a true relationship with Him. The passage begins with "Not everyone who says to me, 'Lord, Lord,'...". This repetition of "Lord" signifies a claim to acknowledge Jesus' authority. However, Jesus emphasizes that calling Him "Lord" is not enough; it must be accompanied by obedience to God's will. This implies that verbal confession alone does not equate to salvation. The individuals Jesus refers to claim to have performed miracles, including exorcism and prophecy. These deeds of power might be seen as evidence of divine favor in many religious contexts. However, Jesus suggests that miraculous works in His name are not the ultimate test of one's relationship with Him. Jesus' declaration, "I never knew you; depart from me, you workers of lawlessness," indicates a relational and moral disconnect. The term "knew" goes beyond intellectual knowledge to intimate recognition and approval. Those He condemns are labeled "workers of lawlessness," suggesting that despite their prophetic claims, their lives are marked by disobedience to God's moral law. The passage also challenges the listener to reflect on the authenticity of their faith. Are they merely using Jesus' name for authority or personal gain, or is their profession of faith accompanied by a life transformed by His teachings and characterized by the fruit of the Spirit? The scenario described is eschatological, pertaining to the final judgment. It is a warning that in the last assessment, Jesus will distinguish between those who had a true relationship with Him and those who did not, regardless of their religious activities.
Jesus' teachings in the Sermon on the Mount (Matthew 6:5-6) caution against practicing righteousness before others to be seen by them. This includes public prayers that are performed for the sake of attention rather than genuine communion with God. The principle is that true faith is lived out authentically, not for show. Within some Christian traditions, such as Catholicism and Orthodoxy, veneration of Mary and the saints plays a significant role. However, this can cross into idolatry when veneration supersedes or obscures the centrality of Christ. The Bible is clear that prayer should be directed to God through Christ alone, (1 Timothy 2:5), which speaks of Jesus as the sole mediator between God and humanity. Spiritism and similar beliefs often incorporate elements of Christianity but diverge fundamentally on the person and role of Jesus Christ. The New Testament is clear in passages like John 14:6 and Acts 4:12 that salvation is found in no one else but Jesus. The notion that moral improvement can lead to God apart from faith in Christ is counter to the message of the gospel, which states that it is by grace through faith that one is saved, not by works (Ephesians 2:8-9). The metaphor of the narrow path that Jesus presents (Matthew 7:13-14) is a call to a discipleship that is not just nominal but transformative. This discipleship is marked by a personal relationship with Jesus, a commitment to His teachings, and a lifestyle that reflects His lordship. The Bible indicates that there will be a judgment based on the truth of one's faith and the authenticity of one's relationship with Christ. The New Testament stresses the importance of belief in Jesus' atoning sacrifice as the basis for salvation, a gift of grace rather than a result of personal merit.
In this context, Jesus' teachings serve as a mirror for self-reflection, urging us to examine our beliefs and practices. Are they aligned with the teachings of the Lord, and do they reflect a genuine faith in Jesus Christ as Lord and Savior? This call for discernment is central to Christian faith and life.

Claim: Love me or be tortured forever. Sounds about right lol
Response: The response is a common critique that frames the Christian doctrine of salvation and judgment in misunderstood terms. The quip "Love me or be tortured forever" misrepresents the Christian message as a coercive ultimatum, which it is not. Christianity posits that God's love is freely given and that He desires a relationship with each person (1 Timothy 2:4). The choice to accept or reject this relationship is the free will of the individual.
God's love is sacrificial and redemptive, as demonstrated through Jesus' life, death, and resurrection. The call to love God is not a demand but an invitation to reciprocate the love already shown to humanity (1 John 4:19).
The concept of eternal separation from God, often described as hell, is a state resulting from one's choice to live independently of God's will and reject His grace, not a punitive measure imposed by God. It's an eternal continuation of the rejection of God already chosen by individuals in this life. The Christian God is perfectly just and holy. Sin, which separates humanity from God, must be addressed. The consequence of sin is not so much an active infliction from God but a natural outcome of being apart from the source of life and goodness. Far from being about coercion or torture, the gospel message is that Christ's sacrifice offers a way to overcome the separation caused by sin. It is an act of mercy, providing an avenue for reconciliation (2 Corinthians 5:18-19). Christian doctrine holds that God respects human freedom so profoundly that He allows people to reject Him, even if that means they choose separation from Him. This respect for free will underscores the authenticity of the relationship God seeks with each person. The Christian narrative is one of love intertwined with judgment. Judgment is not about God's desire to punish but about His commitment to truth and righteousness. It's about the reality of sin and its consequences being addressed through Jesus, with the offer of forgiveness for those who accept it (Romans 3:23-26). The "narrow path" that Jesus speaks of is a metaphor for the journey of faith that leads to eternal life with God. This life is marked by transformation and the fruit of a relationship with God, not a grim, fear-based compliance.
"Enter by the narrow gate. For the gate is wide and the way is easy that leads to destruction, and those who enter by it are many. For the gate is narrow and the way is hard that leads to life, and those who find it are few." - Matthew 7:13-14 (ESV)
This passage, being part of the Sermon on the Mount, presents a metaphorical framework for understanding the choices individuals make in life, particularly about faith and morality. Jesus describes two paths: one wide and easy, leading to destruction, and the other narrow and difficult, leading to life. Jesus makes a distinction between living a life that is self-serving and one that aligns with being born again spiritually, repenting from a sinful, godless life, surrendering to His love and grace, receiving Him as Savior, and following Him, having Him as center, and not one's self.
In all ages, this metaphor has reflected the diversity of beliefs and lifestyles. The rise of secularism and atheism in recent times is evidence of the wide road, where individuals choose paths that do not acknowledge Christ as who He said to be: Jesus said, "I am the way, and the truth, and the life. No one comes to the Father except through me." (John 14:6). Similarly, the presence of multiple religious sects promoting false teachings, that do not align with the truth of Jesus' message, are also are part of the broader road leading to destruction. Moreover, some identify as Christians yet live outside His will. This discrepancy between confession and practice is another aspect of the broad path, where the label of faith is present but the commitment is shallow. In this context, the prophecy that Jesus speaks of doesn't merely become a statement about the afterlife, but also a commentary on the human condition and the challenges of living a life of genuine faith. The fact that the majority of people do not choose the path of strict adherence to Jesus' teachings is a confirmation of the prophecy, so unbelievers are unwillingly through their condition confirming Jesus' prophecy.
The narrow path is characterized by those who not only confess their faith in Jesus but also prove that their faith is genuine, by producing good fruits coming from the spirit that dwells in them, that experienced a change of their hearts through the rebirth in spirit, those who strive to live in a way that is consistent with the Lord's teachings, despite societal pressures or the popularity of alternative lifestyles. This path is marked by a commitment to spiritual growth, ethical behavior, and often, service to others, which can be more demanding and less traveled.
This dichotomy of paths serves as a call to self-examination for us who confess Christ, encouraging believers to reflect on the authenticity of their practice and the depth of their commitment to the principles taught by Jesus.
"Not everyone who says to me, 'Lord, Lord,' will enter the kingdom of heaven, but the one who does the will of my Father who is in heaven. On that day many will say to me, 'Lord, Lord, did we not prophesy in your name, and cast out demons in your name, and do many mighty works in your name?' And then will I declare to them, 'I never knew you; depart from me, you workers of lawlessness.'" - Matthew 7:21-23 (ESV)
The key distinction Jesus makes is not between believers and non-believers, but between true and nominal disciples. The former genuinely adhere to God's will, while the latter perform acts of piety or power in Jesus' name but lack a true relationship with Him. The passage begins with "Not everyone who says to me, 'Lord, Lord,'...". This repetition of "Lord" signifies a claim to acknowledge Jesus' authority. However, Jesus emphasizes that calling Him "Lord" is not enough; it must be accompanied by obedience to God's will. This implies that verbal confession alone does not equate to salvation. The individuals Jesus refers to claim to have performed miracles, including exorcism and prophecy. These deeds of power might be seen as evidence of divine favor in many religious contexts. However, Jesus suggests that miraculous works in His name are not the ultimate test of one's relationship with Him. Jesus' declaration, "I never knew you; depart from me, you workers of lawlessness," indicates a relational and moral disconnect. The term "knew" goes beyond intellectual knowledge to intimate recognition and approval. Those He condemns are labeled "workers of lawlessness," suggesting that despite their prophetic claims, their lives are marked by disobedience to God's moral law. The passage also challenges the listener to reflect on the authenticity of their faith. Are they merely using Jesus' name for authority or personal gain, or is their profession of faith accompanied by a life transformed by His teachings and characterized by the fruit of the Spirit? The scenario described is eschatological, pertaining to the final judgment. It is a warning that in the last assessment, Jesus will distinguish between those who had a true relationship with Him and those who did not, regardless of their religious activities.
Jesus' teachings in the Sermon on the Mount (Matthew 6:5-6) caution against practicing righteousness before others to be seen by them. This includes public prayers that are performed for the sake of attention rather than genuine communion with God. The principle is that true faith is lived out authentically, not for show. Within some Christian traditions, such as Catholicism and Orthodoxy, veneration of Mary and the saints plays a significant role. However, this can cross into idolatry when veneration supersedes or obscures the centrality of Christ. The Bible is clear that prayer should be directed to God through Christ alone, (1 Timothy 2:5), which speaks of Jesus as the sole mediator between God and humanity. Spiritism and similar beliefs often incorporate elements of Christianity but diverge fundamentally on the person and role of Jesus Christ. The New Testament is clear in passages like John 14:6 and Acts 4:12 that salvation is found in no one else but Jesus. The notion that moral improvement can lead to God apart from faith in Christ is counter to the message of the gospel, which states that it is by grace through faith that one is saved, not by works (Ephesians 2:8-9). The metaphor of the narrow path that Jesus presents (Matthew 7:13-14) is a call to a discipleship that is not just nominal but transformative. This discipleship is marked by a personal relationship with Jesus, a commitment to His teachings, and a lifestyle that reflects His lordship. The Bible indicates that there will be a judgment based on the truth of one's faith and the authenticity of one's relationship with Christ. The New Testament stresses the importance of belief in Jesus' atoning sacrifice as the basis for salvation, a gift of grace rather than a result of personal merit.
In this context, Jesus' teachings serve as a mirror for self-reflection, urging us to examine our beliefs and practices. Are they aligned with the teachings of the Lord, and do they reflect a genuine faith in Jesus Christ as Lord and Savior? This call for discernment is central to Christian faith and life.

Claim: Love me or be tortured forever. Sounds about right lol
Response: The response is a common critique that frames the Christian doctrine of salvation and judgment in misunderstood terms. The quip "Love me or be tortured forever" misrepresents the Christian message as a coercive ultimatum, which it is not. Christianity posits that God's love is freely given and that He desires a relationship with each person (1 Timothy 2:4). The choice to accept or reject this relationship is the free will of the individual.
God's love is sacrificial and redemptive, as demonstrated through Jesus' life, death, and resurrection. The call to love God is not a demand but an invitation to reciprocate the love already shown to humanity (1 John 4:19).
The concept of eternal separation from God, often described as hell, is a state resulting from one's choice to live independently of God's will and reject His grace, not a punitive measure imposed by God. It's an eternal continuation of the rejection of God already chosen by individuals in this life. The Christian God is perfectly just and holy. Sin, which separates humanity from God, must be addressed. The consequence of sin is not so much an active infliction from God but a natural outcome of being apart from the source of life and goodness. Far from being about coercion or torture, the gospel message is that Christ's sacrifice offers a way to overcome the separation caused by sin. It is an act of mercy, providing an avenue for reconciliation (2 Corinthians 5:18-19). Christian doctrine holds that God respects human freedom so profoundly that He allows people to reject Him, even if that means they choose separation from Him. This respect for free will underscores the authenticity of the relationship God seeks with each person. The Christian narrative is one of love intertwined with judgment. Judgment is not about God's desire to punish but about His commitment to truth and righteousness. It's about the reality of sin and its consequences being addressed through Jesus, with the offer of forgiveness for those who accept it (Romans 3:23-26). The "narrow path" that Jesus speaks of is a metaphor for the journey of faith that leads to eternal life with God. This life is marked by transformation and the fruit of a relationship with God, not a grim, fear-based compliance.
294 Re: My articles Thu Jan 11, 2024 6:44 am
Re: My articles Thu Jan 11, 2024 6:44 am
Otangelo
Admin
The Heart Transformed: Divine Intervention and the Global Witness of Faith
The conception of God as the necessary, foundational being is a profound recognition of a higher power that is both the origin and sustainer of all that exists. God is not merely a first cause but an ongoing force that upholds the universe. The Bible presents God as the ultimate reality, whose existence provides the foundation for our own. The incarnation of God in Jesus Christ is central. It embodies the intersection of the divine with the human experience, demonstrating God's willingness to enter into the very fabric of human life and suffering. The narrative of Christ's life, death, and resurrection as recorded in the Gospels is the ultimate act of divine love and grace—a self-sacrifice for the redemption and restoration of a world estranged from its Creator. God's holiness and justice are essential attributes that complement His love and grace. God is perfectly just and will judge all people according to their deeds and the intentions of their hearts. This judgment is based on His revealed word, which serves as a moral compass and a guide for living a life aligned with His will. The reality of God encompasses every aspect of existence. From the vast expanse of the cosmos to the innermost parts of the human heart, nothing is beyond His reach or knowledge. To live in recognition of God's reality is to see oneself as part of a greater purpose and narrative. It is to understand that our existence is not an accident but a deliberate act of creation by a God who is intimately involved with His creation and desires a relationship with it. The omnipresence of God implies that we are constantly within His care and under His governance. This belief provides comfort, purpose, and a sense of direction. It calls for a response of faith, trust, and obedience to the divine will as the path to true fulfillment and peace. In a world where the tangible often overshadows the transcendent, the acknowledgment of God's reality invites us to look beyond the immediate and the material, to the eternal and the spiritual. God's knowledge of everything, His omniscience, is a reassurance that our lives are seen and valued by Him. Every moment is known, every struggle witnessed, and every joy shared by a God who is both beyond us and with us. This profound truth has the power to transform how we view ourselves, our lives, and our place in the universe.
There is nothing harder than man's heart. Another proof is Christs church, the body of Christ. Today, one can encounter true believers all over the globe. This is a fulfilled prophecy in
Ezekiel 36:26, which states:
"I will give you a new heart and put a new spirit in you; I will remove from you your heart of stone and give you a heart of flesh."
This passage speaks about spiritual renewal and transformation. The existence of Christianity is proof of fullfilled prophecy in the Bible.
The passage from Ezekiel 36:26 is a profound promise of transformation, one that speaks directly to the capacity for change within the human condition. It suggests a metamorphosis from a state of spiritual insensitivity, symbolized by the "heart of stone," to one of receptivity and responsiveness, indicated by the "heart of flesh." This prophecy underscores the idea of an inner renaissance that is enacted by God’s intervention, a pivotal theme in both the Hebrew Scriptures and the New Testament. The global existence of Christianity, with its diverse expressions of faith and practice, indeed, is a manifestation of this promise. Across cultures and societies, individuals testify to personal experiences of profound change that they attribute to their faith in Jesus Christ. Such accounts often include elements of repentance, a newfound sense of purpose, and an orientation towards love, compassion, and justice, aligning with the characteristics of the "new heart" mentioned in Ezekiel. Moreover, the Christian concept of the Church as the Body of Christ extends this individual transformation to a corporate reality. It envisions a community that transcends ethnic, linguistic, and cultural boundaries, united in a common identity and purpose. The church, in its ideal form, is meant to be a demonstration of God's transformative power at work in the world – a community where the values of the Kingdom of God are lived out and where the "new spirit" leads to actions that reflect God's love and righteousness.
This prophecy from Ezekiel also has eschatological implications, anticipating a time when God's plan for humanity reaches its fulfillment. The outpouring of the Holy Spirit at Pentecost is the initial fulfillment of this promise, with the ultimate fulfillment still to come in the full realization of God's Kingdom. The transformation of the human heart is, therefore, both an already accomplished fact and a future hope, a dynamic tension that animates Christian life and eschatology. In a broader sense, the transformation of the human heart is a theme that resonates beyond Christianity. Many religious and philosophical systems grapple with the problem of the human condition and offer various solutions for inner change. Yet, Christianity's claim is unique because such change is not merely the result of human effort but is a work of divine grace through the presence of the Holy Spirit. The enduring and widespread presence of Christianity, despite historical and cultural challenges, is a testament to the power of this divine transformation. It is a vivid illustration of the "new heart" at work across time and space, a fulfillment of ancient prophecy that continues to unfold in the lives of believers today.

The conception of God as the necessary, foundational being is a profound recognition of a higher power that is both the origin and sustainer of all that exists. God is not merely a first cause but an ongoing force that upholds the universe. The Bible presents God as the ultimate reality, whose existence provides the foundation for our own. The incarnation of God in Jesus Christ is central. It embodies the intersection of the divine with the human experience, demonstrating God's willingness to enter into the very fabric of human life and suffering. The narrative of Christ's life, death, and resurrection as recorded in the Gospels is the ultimate act of divine love and grace—a self-sacrifice for the redemption and restoration of a world estranged from its Creator. God's holiness and justice are essential attributes that complement His love and grace. God is perfectly just and will judge all people according to their deeds and the intentions of their hearts. This judgment is based on His revealed word, which serves as a moral compass and a guide for living a life aligned with His will. The reality of God encompasses every aspect of existence. From the vast expanse of the cosmos to the innermost parts of the human heart, nothing is beyond His reach or knowledge. To live in recognition of God's reality is to see oneself as part of a greater purpose and narrative. It is to understand that our existence is not an accident but a deliberate act of creation by a God who is intimately involved with His creation and desires a relationship with it. The omnipresence of God implies that we are constantly within His care and under His governance. This belief provides comfort, purpose, and a sense of direction. It calls for a response of faith, trust, and obedience to the divine will as the path to true fulfillment and peace. In a world where the tangible often overshadows the transcendent, the acknowledgment of God's reality invites us to look beyond the immediate and the material, to the eternal and the spiritual. God's knowledge of everything, His omniscience, is a reassurance that our lives are seen and valued by Him. Every moment is known, every struggle witnessed, and every joy shared by a God who is both beyond us and with us. This profound truth has the power to transform how we view ourselves, our lives, and our place in the universe.
There is nothing harder than man's heart. Another proof is Christs church, the body of Christ. Today, one can encounter true believers all over the globe. This is a fulfilled prophecy in
Ezekiel 36:26, which states:
"I will give you a new heart and put a new spirit in you; I will remove from you your heart of stone and give you a heart of flesh."
This passage speaks about spiritual renewal and transformation. The existence of Christianity is proof of fullfilled prophecy in the Bible.
The passage from Ezekiel 36:26 is a profound promise of transformation, one that speaks directly to the capacity for change within the human condition. It suggests a metamorphosis from a state of spiritual insensitivity, symbolized by the "heart of stone," to one of receptivity and responsiveness, indicated by the "heart of flesh." This prophecy underscores the idea of an inner renaissance that is enacted by God’s intervention, a pivotal theme in both the Hebrew Scriptures and the New Testament. The global existence of Christianity, with its diverse expressions of faith and practice, indeed, is a manifestation of this promise. Across cultures and societies, individuals testify to personal experiences of profound change that they attribute to their faith in Jesus Christ. Such accounts often include elements of repentance, a newfound sense of purpose, and an orientation towards love, compassion, and justice, aligning with the characteristics of the "new heart" mentioned in Ezekiel. Moreover, the Christian concept of the Church as the Body of Christ extends this individual transformation to a corporate reality. It envisions a community that transcends ethnic, linguistic, and cultural boundaries, united in a common identity and purpose. The church, in its ideal form, is meant to be a demonstration of God's transformative power at work in the world – a community where the values of the Kingdom of God are lived out and where the "new spirit" leads to actions that reflect God's love and righteousness.
This prophecy from Ezekiel also has eschatological implications, anticipating a time when God's plan for humanity reaches its fulfillment. The outpouring of the Holy Spirit at Pentecost is the initial fulfillment of this promise, with the ultimate fulfillment still to come in the full realization of God's Kingdom. The transformation of the human heart is, therefore, both an already accomplished fact and a future hope, a dynamic tension that animates Christian life and eschatology. In a broader sense, the transformation of the human heart is a theme that resonates beyond Christianity. Many religious and philosophical systems grapple with the problem of the human condition and offer various solutions for inner change. Yet, Christianity's claim is unique because such change is not merely the result of human effort but is a work of divine grace through the presence of the Holy Spirit. The enduring and widespread presence of Christianity, despite historical and cultural challenges, is a testament to the power of this divine transformation. It is a vivid illustration of the "new heart" at work across time and space, a fulfillment of ancient prophecy that continues to unfold in the lives of believers today.

295 Re: My articles Sat Jan 20, 2024 3:53 am
Re: My articles Sat Jan 20, 2024 3:53 am
Otangelo
Admin
Exploring the Mysteries of the Divine: A Journey Through Theological and Philosophical Questions
God revealed Himself to mankind. But up to a certain point. If we knew everything that God knows, we would maybe be equal to God. But the thing is, if we investigate things up to where it's possible, we will always, at a certain point, arrive at an impenetrable barrier, where we cannot investigate further, and then, all we can do, is either speculate or confess ignorance. Why is there something rather than nothing? Why does God exist, rather than not? Why did he choose to create this world, rather than another? Why did he create just one tiny planet amid an unfathomably large universe? How is God 3 persons in one? How are these 3 persons one substance? How will He be able to judge the whole world, every word that each one of us spoke, and not get bored by it? How and why did He decide to create this world, despite of its brokenness, and go forward with a plan to save humanity, despite knowing how much pain it would cause Him? How was love stronger than fear in this case?
**Why is there something rather than nothing?** This question, often considered one of the most profound in philosophy, touches on the fundamental nature of existence. It asks why the universe exists at all, rather than not existing. Some theological perspectives suggest that the universe is a manifestation of divine will or a necessary expression of a creator's nature.
**Why does God exist, rather than not?** This question delves into the nature of God's existence. God is a necessary being. God's existence is the foundational truth upon which all other things depend.
**Why did He choose to create this world, rather than another?** This is a question about divine intentionality and purpose. The world reflects a divine plan or purpose, though the specifics of that plan are subject to various interpretations and beliefs.
**Why did He create just one tiny planet amid an unfathomably large universe?** This question highlights the vastness of the universe and the seemingly small place humanity occupies within it. Some see this as a reflection of the majesty and mystery of the divine creation, while others find in it a call to humility and a sense of wonder.
**How is God 3 persons in one? How are these 3 persons one substance?** The Trinity is a complex and nuanced theological concept. It's about the unity of Father, Son, and Holy Spirit as three distinct persons who are simultaneously one being.
**How will He be able to judge the whole world?** This question involves the omniscience and omnipotence attributed to God in many religious traditions. It implies a divine capacity to know and judge all actions and thoughts, which is beyond human comprehension.
**Why create a world with brokenness, and go forward with a plan for salvation?** This touches on the problem of evil and suffering in the world. Theological perspectives often interpret this as part of a larger divine plan, where free will, moral growth, and the possibility of redemption play crucial roles.
**How was love stronger than fear in the decision to create and save the world?** This question reflects on the motivations attributed to the divine in creation and salvation narratives. Love is the ultimate driving force behind divine actions, surpassing fear or any other considerations.
**What is the nature of consciousness?** Understanding how consciousness arises, what it truly is, and why it exists remains one of the most profound mysteries. The 'hard problem of consciousness' refers to the difficulty of explaining why and how we have qualia or subjective experiences.
**What is the ultimate nature of reality?** Questions about whether our perception of reality truly reflects the universe, or if there are aspects beyond our understanding, have long intrigued philosophers and scientists alike. Concepts like simulation theory add modern twists to this age-old question.
**What is the nature of time?** Understanding time – whether it is fundamental to the universe, an emergent property, or an illusion – is a deeply complex question that challenges our perceptions and understanding of the universe.
**What is the nature of dark matter and dark energy?** These components make up most of the universe's mass-energy content, but their true nature remains one of the biggest mysteries in cosmology.
**What is the nature of God?** This question delves into understanding the attributes and essence of God. Is God immanent or transcendent, and how these attributes reconcile with human experience.
**Why does evil and suffering exist if God is benevolent and omnipotent?** Known as the problem of evil, this question challenges how to reconcile the existence of evil and suffering in the world with the concept of a loving and all-powerful God.
**Is there a predestined plan or do we have free will?** This question examines the nature of human freedom and destiny, especially about divine omniscience and omnipotence.
**What is the nature of the soul or spirit, and its connection to God?** This explores the concept of the soul or spirit in humans, its origin, nature, and how it relates to the divine.
**How should humans respond to the divine?** This question looks at what constitutes appropriate recognition, worship, or response to God, varying greatly among different religious traditions.
**How do God's foreknowledge and human action interact?** This explores the compatibility of divine omniscience, particularly God's knowledge of future events, with the concept of human free will.
**Why does God seem hidden or silent to some people?** Often referred to as the 'hiddenness of God,' this question grapples with why God isn't more overtly manifest in the world, especially to those who seek evidence of the divine.
**What is the relationship between God and the laws of nature?** This explores how God interacts with or relates to the physical universe, including the creation and sustaining of natural laws.
These questions have been, and continue to be, the subject of extensive theological and philosophical exploration. They reflect the human quest to understand the divine and its relation to our existence, morality, purpose, and the universe at large. They serve as a reminder of the depth and complexity inherent in contemplating the divine and our relationship with it. Humans, despite our remarkable capacity for reasoning, discovery, and understanding, are bound by the limitations of our senses, cognitive abilities, and lifespan. Our ability to comprehend concepts that are infinite, eternal, or transcendent is inherently limited. This limitation is not just scientific or intellectual; it also encompasses the spiritual and existential dimensions of our existence.
The contrast between the all-knowing nature of the divine and the limited understanding of humanity is a central theme in many theological discussions. God, in these frameworks, is often described as omniscient, omnipotent, and omnipresent - attributes that transcend human capabilities. This distinction serves to emphasize the special status of the divine as fundamentally different from and greater than human beings. The acceptance of mysteries that cannot be fully understood or explained is an essential aspect of faith. The concept of mystery in theology does not imply a puzzle to be solved but rather an acknowledgment of the majesty and incomprehensibility of the divine nature. It invites a sense of awe, wonder, and humility in the face of the sacred.
It's a humbling reminder of our finite nature and an invitation to explore the depths of faith, spirituality, and wonder. The journey of seeking understanding, especially in matters concerning the divine, is as much about the questions we ask as it is about the answers we find or the mysteries we embrace.

God revealed Himself to mankind. But up to a certain point. If we knew everything that God knows, we would maybe be equal to God. But the thing is, if we investigate things up to where it's possible, we will always, at a certain point, arrive at an impenetrable barrier, where we cannot investigate further, and then, all we can do, is either speculate or confess ignorance. Why is there something rather than nothing? Why does God exist, rather than not? Why did he choose to create this world, rather than another? Why did he create just one tiny planet amid an unfathomably large universe? How is God 3 persons in one? How are these 3 persons one substance? How will He be able to judge the whole world, every word that each one of us spoke, and not get bored by it? How and why did He decide to create this world, despite of its brokenness, and go forward with a plan to save humanity, despite knowing how much pain it would cause Him? How was love stronger than fear in this case?
**Why is there something rather than nothing?** This question, often considered one of the most profound in philosophy, touches on the fundamental nature of existence. It asks why the universe exists at all, rather than not existing. Some theological perspectives suggest that the universe is a manifestation of divine will or a necessary expression of a creator's nature.
**Why does God exist, rather than not?** This question delves into the nature of God's existence. God is a necessary being. God's existence is the foundational truth upon which all other things depend.
**Why did He choose to create this world, rather than another?** This is a question about divine intentionality and purpose. The world reflects a divine plan or purpose, though the specifics of that plan are subject to various interpretations and beliefs.
**Why did He create just one tiny planet amid an unfathomably large universe?** This question highlights the vastness of the universe and the seemingly small place humanity occupies within it. Some see this as a reflection of the majesty and mystery of the divine creation, while others find in it a call to humility and a sense of wonder.
**How is God 3 persons in one? How are these 3 persons one substance?** The Trinity is a complex and nuanced theological concept. It's about the unity of Father, Son, and Holy Spirit as three distinct persons who are simultaneously one being.
**How will He be able to judge the whole world?** This question involves the omniscience and omnipotence attributed to God in many religious traditions. It implies a divine capacity to know and judge all actions and thoughts, which is beyond human comprehension.
**Why create a world with brokenness, and go forward with a plan for salvation?** This touches on the problem of evil and suffering in the world. Theological perspectives often interpret this as part of a larger divine plan, where free will, moral growth, and the possibility of redemption play crucial roles.
**How was love stronger than fear in the decision to create and save the world?** This question reflects on the motivations attributed to the divine in creation and salvation narratives. Love is the ultimate driving force behind divine actions, surpassing fear or any other considerations.
**What is the nature of consciousness?** Understanding how consciousness arises, what it truly is, and why it exists remains one of the most profound mysteries. The 'hard problem of consciousness' refers to the difficulty of explaining why and how we have qualia or subjective experiences.
**What is the ultimate nature of reality?** Questions about whether our perception of reality truly reflects the universe, or if there are aspects beyond our understanding, have long intrigued philosophers and scientists alike. Concepts like simulation theory add modern twists to this age-old question.
**What is the nature of time?** Understanding time – whether it is fundamental to the universe, an emergent property, or an illusion – is a deeply complex question that challenges our perceptions and understanding of the universe.
**What is the nature of dark matter and dark energy?** These components make up most of the universe's mass-energy content, but their true nature remains one of the biggest mysteries in cosmology.
**What is the nature of God?** This question delves into understanding the attributes and essence of God. Is God immanent or transcendent, and how these attributes reconcile with human experience.
**Why does evil and suffering exist if God is benevolent and omnipotent?** Known as the problem of evil, this question challenges how to reconcile the existence of evil and suffering in the world with the concept of a loving and all-powerful God.
**Is there a predestined plan or do we have free will?** This question examines the nature of human freedom and destiny, especially about divine omniscience and omnipotence.
**What is the nature of the soul or spirit, and its connection to God?** This explores the concept of the soul or spirit in humans, its origin, nature, and how it relates to the divine.
**How should humans respond to the divine?** This question looks at what constitutes appropriate recognition, worship, or response to God, varying greatly among different religious traditions.
**How do God's foreknowledge and human action interact?** This explores the compatibility of divine omniscience, particularly God's knowledge of future events, with the concept of human free will.
**Why does God seem hidden or silent to some people?** Often referred to as the 'hiddenness of God,' this question grapples with why God isn't more overtly manifest in the world, especially to those who seek evidence of the divine.
**What is the relationship between God and the laws of nature?** This explores how God interacts with or relates to the physical universe, including the creation and sustaining of natural laws.
These questions have been, and continue to be, the subject of extensive theological and philosophical exploration. They reflect the human quest to understand the divine and its relation to our existence, morality, purpose, and the universe at large. They serve as a reminder of the depth and complexity inherent in contemplating the divine and our relationship with it. Humans, despite our remarkable capacity for reasoning, discovery, and understanding, are bound by the limitations of our senses, cognitive abilities, and lifespan. Our ability to comprehend concepts that are infinite, eternal, or transcendent is inherently limited. This limitation is not just scientific or intellectual; it also encompasses the spiritual and existential dimensions of our existence.
The contrast between the all-knowing nature of the divine and the limited understanding of humanity is a central theme in many theological discussions. God, in these frameworks, is often described as omniscient, omnipotent, and omnipresent - attributes that transcend human capabilities. This distinction serves to emphasize the special status of the divine as fundamentally different from and greater than human beings. The acceptance of mysteries that cannot be fully understood or explained is an essential aspect of faith. The concept of mystery in theology does not imply a puzzle to be solved but rather an acknowledgment of the majesty and incomprehensibility of the divine nature. It invites a sense of awe, wonder, and humility in the face of the sacred.
It's a humbling reminder of our finite nature and an invitation to explore the depths of faith, spirituality, and wonder. The journey of seeking understanding, especially in matters concerning the divine, is as much about the questions we ask as it is about the answers we find or the mysteries we embrace.

296 Re: My articles Sat Jan 20, 2024 8:38 am
Re: My articles Sat Jan 20, 2024 8:38 am
Otangelo
Admin
Breaking Free to New Life: The Resurrection of Christ and the Birth of the Church
The metaphor of an eggshell being damaged takes on profound significance when viewed in the light of Jesus Christ's life, death, and resurrection, and its impact on the birth and growth of the Church. The damaging of the eggshell from the outside represents the external forces— the Sanhedrin, the Jews, and the Romans—that led to Jesus' crucifixion. These forces, driven by a multitude of societal, political, and spiritual factors, converged to bring an end to His earthly life, culminating in His burial in a tomb.
However, the story does not end there. Just as a vital force within a chick cannot be contained by the shell, Jesus' essence, His very being as the Son of God and the source of life, could not be held by the tomb. His resurrection is the ultimate testament to His divine nature and the power of God over death. This miraculous event did not just signify the triumph of life over death, but it initiated something entirely new and transformative: the birth of the Church of God. This birth of the Church can be likened to a chick emerging from within an egg. The resurrection of Christ symbolizes a powerful internal force, a divine intervention that breaks the barriers of death and sin. It marks the beginning of a new era where believers, reborn in the Spirit, become part of the body of Christ. This internal transformation within each believer reflects the same power that raised Jesus from the dead.
The implications of this metaphor extend deeply into the life of the Church and its believers: Just as a chick emerges into a new life, believers are spiritually reborn in Christ. This rebirth is a transformation from within, a heart changed by the Holy Spirit, leading to a new way of living that reflects the teachings and love of Christ. The resurrection of Jesus demonstrates that no external force can ultimately thwart God’s will. It serves as a reminder that God's power is supreme, able to bring life and hope even in situations that seem hopeless. The Church, birthed from the resurrection, grows and thrives not by external human effort alone but through the internal working of the Holy Spirit in the hearts of believers. This growth is organic and dynamic, mirroring the natural and powerful emergence of life from within an eggshell. In this new life, believers are called to spread the Gospel, serving as witnesses to the resurrection of Christ.
Just as the chick breaking free from the eggshell is a natural and necessary process, so too is the believer's call to share their faith and serve others, an integral expression of their new life in Christ. The metaphor of the eggshell, when viewed through the lens of Jesus’ death and resurrection, beautifully encapsulates the essence of Christian faith. It speaks of a transformation that is both deeply personal and expansively communal. In the resurrection of Christ, believers find not only the promise of new life but also the power to live out that life in service to God and others. This is the heart of the Church, a community continually reborn and renewed by the vital, life-giving force of Christ’s resurrection.
The metaphor of an eggshell being damaged takes on profound significance when viewed in the light of Jesus Christ's life, death, and resurrection, and its impact on the birth and growth of the Church. The damaging of the eggshell from the outside represents the external forces— the Sanhedrin, the Jews, and the Romans—that led to Jesus' crucifixion. These forces, driven by a multitude of societal, political, and spiritual factors, converged to bring an end to His earthly life, culminating in His burial in a tomb.
However, the story does not end there. Just as a vital force within a chick cannot be contained by the shell, Jesus' essence, His very being as the Son of God and the source of life, could not be held by the tomb. His resurrection is the ultimate testament to His divine nature and the power of God over death. This miraculous event did not just signify the triumph of life over death, but it initiated something entirely new and transformative: the birth of the Church of God. This birth of the Church can be likened to a chick emerging from within an egg. The resurrection of Christ symbolizes a powerful internal force, a divine intervention that breaks the barriers of death and sin. It marks the beginning of a new era where believers, reborn in the Spirit, become part of the body of Christ. This internal transformation within each believer reflects the same power that raised Jesus from the dead.
The implications of this metaphor extend deeply into the life of the Church and its believers: Just as a chick emerges into a new life, believers are spiritually reborn in Christ. This rebirth is a transformation from within, a heart changed by the Holy Spirit, leading to a new way of living that reflects the teachings and love of Christ. The resurrection of Jesus demonstrates that no external force can ultimately thwart God’s will. It serves as a reminder that God's power is supreme, able to bring life and hope even in situations that seem hopeless. The Church, birthed from the resurrection, grows and thrives not by external human effort alone but through the internal working of the Holy Spirit in the hearts of believers. This growth is organic and dynamic, mirroring the natural and powerful emergence of life from within an eggshell. In this new life, believers are called to spread the Gospel, serving as witnesses to the resurrection of Christ.
Just as the chick breaking free from the eggshell is a natural and necessary process, so too is the believer's call to share their faith and serve others, an integral expression of their new life in Christ. The metaphor of the eggshell, when viewed through the lens of Jesus’ death and resurrection, beautifully encapsulates the essence of Christian faith. It speaks of a transformation that is both deeply personal and expansively communal. In the resurrection of Christ, believers find not only the promise of new life but also the power to live out that life in service to God and others. This is the heart of the Church, a community continually reborn and renewed by the vital, life-giving force of Christ’s resurrection.
297 Re: My articles Fri Jan 26, 2024 12:58 pm
Re: My articles Fri Jan 26, 2024 12:58 pm
Otangelo
Admin
Gods foreknowledge, and human free will
Romans 8:29: "For those God foreknew he also predestined to be conformed to the image of his Son, that he might be the firstborn among many brothers and sisters."
1 Peter 1:2: "who have been chosen according to the foreknowledge of God the Father, through the sanctifying work of the Spirit, to be obedient to Jesus Christ and sprinkled with his blood: Grace and peace be yours in abundance."
Acts 2:23: "This man was handed over to you by God's deliberate plan and foreknowledge; and you, with the help of wicked men, put him to death by nailing him to the cross."
1 Peter 1:20: "He was chosen before the creation of the world, but was revealed in these last times for your sake."
Ephesians 1:4-5: "For he chose us in him before the creation of the world to be holy and blameless in his sight. In love he predestined us for adoption to sonship through Jesus Christ, in accordance with his pleasure and will."
Molinism is a theological position that suggests that God possesses not only simple foreknowledge (knowing what will happen in the future) but also middle knowledge. Middle knowledge is the idea that God knows what individuals would freely choose in any given situation, even if those situations never actually occur. In this view, God's foreknowledge is harmonized with human free will because He knows what individuals would choose freely in various circumstances without necessarily determining their choices.
Open Theism argues that God's knowledge of the future is not exhaustive or predetermined. Instead, God has chosen to limit His knowledge of future human choices to respect human free will. In this perspective, God knows all possibilities but does not know the actual choices individuals will make until they make them. This view maintains a strong emphasis on human free will. This raises questions about how God can have a coherent and certain plan for the redemption of humanity if He doesn't know with certainty what will happen. This introduces an element of unpredictability and risk into God's plan. Many prophecies in the Bible, including those related to the coming of the Messiah, are detailed and specific. Open Theism's view that God doesn't know the exact choices individuals will make could create uncertainty about whether these prophecies would be fulfilled. It seems unlikely that God would provide such detailed prophecies if He didn't have a clear plan for their fulfillment. Open Theism is inconsistent with certain theological doctrines, particularly those emphasizing God's sovereignty, omnipotence, and omniscience. These attributes of God, as traditionally understood, are undermined by the limitations on God's knowledge proposed by Open Theism. Open Theism represents a departure from the long-standing theological tradition that has affirmed God's exhaustive foreknowledge. This tradition has been central to Christian thought for centuries and Open Theism's rejection of it raises questions about the continuity of theological beliefs. This also raises questions of how God can lead to questions about how God responds to and interacts with evil in the world. If God doesn't know what will happen, it may raise concerns about God's ability to intervene and bring about justice.
Romans 8:29: "For those God foreknew he also predestined to be conformed to the image of his Son, that he might be the firstborn among many brothers and sisters."
1 Peter 1:2: "who have been chosen according to the foreknowledge of God the Father, through the sanctifying work of the Spirit, to be obedient to Jesus Christ and sprinkled with his blood: Grace and peace be yours in abundance."
Acts 2:23: "This man was handed over to you by God's deliberate plan and foreknowledge; and you, with the help of wicked men, put him to death by nailing him to the cross."
1 Peter 1:20: "He was chosen before the creation of the world, but was revealed in these last times for your sake."
Ephesians 1:4-5: "For he chose us in him before the creation of the world to be holy and blameless in his sight. In love he predestined us for adoption to sonship through Jesus Christ, in accordance with his pleasure and will."
Molinism is a theological position that suggests that God possesses not only simple foreknowledge (knowing what will happen in the future) but also middle knowledge. Middle knowledge is the idea that God knows what individuals would freely choose in any given situation, even if those situations never actually occur. In this view, God's foreknowledge is harmonized with human free will because He knows what individuals would choose freely in various circumstances without necessarily determining their choices.
Open Theism argues that God's knowledge of the future is not exhaustive or predetermined. Instead, God has chosen to limit His knowledge of future human choices to respect human free will. In this perspective, God knows all possibilities but does not know the actual choices individuals will make until they make them. This view maintains a strong emphasis on human free will. This raises questions about how God can have a coherent and certain plan for the redemption of humanity if He doesn't know with certainty what will happen. This introduces an element of unpredictability and risk into God's plan. Many prophecies in the Bible, including those related to the coming of the Messiah, are detailed and specific. Open Theism's view that God doesn't know the exact choices individuals will make could create uncertainty about whether these prophecies would be fulfilled. It seems unlikely that God would provide such detailed prophecies if He didn't have a clear plan for their fulfillment. Open Theism is inconsistent with certain theological doctrines, particularly those emphasizing God's sovereignty, omnipotence, and omniscience. These attributes of God, as traditionally understood, are undermined by the limitations on God's knowledge proposed by Open Theism. Open Theism represents a departure from the long-standing theological tradition that has affirmed God's exhaustive foreknowledge. This tradition has been central to Christian thought for centuries and Open Theism's rejection of it raises questions about the continuity of theological beliefs. This also raises questions of how God can lead to questions about how God responds to and interacts with evil in the world. If God doesn't know what will happen, it may raise concerns about God's ability to intervene and bring about justice.
298 Re: My articles Sat Jan 27, 2024 10:20 am
Re: My articles Sat Jan 27, 2024 10:20 am
Otangelo
Admin
From Cosmic Precision to Divine Design: The Scientific Journey Towards a Grand Designer
The principle that "something cannot come from nothing" underpins the argument that the material universe, with its complexity and order, must have been derived from an existing entity. This foundational premise posits that because the universe exists as a highly structured and law-governed system, it cannot be the product of chance or emerge spontaneously from nothingness. The intricate laws and constants governing the universe's functioning suggest that it operates under a deliberate design, which, by nature, implies the involvement of a conscious mind. Given that designs and purposes are inherently the products of intelligent deliberation, it follows that the universe's existence and its governed structure point towards an eternal mind. This mind, characterized by neither beginning nor end, must have conceived and crafted the material universe. The argument strengthens with the understanding that such a mind, transcending time and space, is the ultimate source from which the universe, with all its laws and order, originated.
Imagine you're holding a very complex, intricately designed watch. You see the gears moving in harmony, keeping time perfectly. Now, think of the universe as that watch, but on an incomprehensibly larger scale. Just like the watch has fundamental components that allow it to function (like the gears, hands, and battery), the universe has its own basic building blocks. These are things like time, measured in seconds; distance, measured in meters; mass, in kilograms; electric current, in amperes; temperature, in kelvin; the amount of a substance, in moles; and the intensity of light, in candelas.
These building blocks are like the universe's DNA, setting the stage for everything that exists and happens within it. They're like the rules of the game, but with a twist: we can't trace these rules back to anything simpler. They're the bedrock, the foundation that everything else builds upon. And here's where it gets really interesting: these rules seem to be set up just right for life to exist. It's as if the universe has been finely tuned, like adjusting the strings of a guitar to get the perfect pitch, so that life can emerge and thrive.
The big question is, why are these rules set up this way? Why do these fundamental building blocks have the values they do? Science, as of now, doesn't have an answer. It's like finding the watch and marveling at its precision and design, yet not knowing who made it or why it was made that way. This situation leads some to ponder the possibility of a designer, a master watchmaker if you will. This isn't about proving something with a formula or an experiment; it's about looking at the universe and asking, "Could this all be a result of chance, or is there something more, some intention behind it?" Just like Isaac Newton, a great scientist, or Paley, who also saw the universe as a creation of a divine watchmaker, many find in the precision and complexity of the universe hints of a grander design, suggesting perhaps an intelligent force or being behind it all. This doesn't replace scientific inquiry but invites us to marvel at the mysteries still beyond our understanding.
Scientific evidence points towards a grand designer. The observation that the universe if finely tuned doesn't merely fill gaps in our current understanding with divine intervention, but rather suggests that the precision and values of the fundamental constants in our universe are so exact that they imply a deliberate design. The universe operates under a set of fundamental constants, such as the gravitational constant, the speed of light, and the charge of an electron, to name a few. These constants dictate the behavior of the cosmos, from the formation of atoms to the birth of stars. The remarkable aspect of these constants is their precision; they are finely tuned to values that allow the universe to be habitable. For instance, if the gravitational constant were slightly stronger or weaker, stars (including our sun) might not form or function in a way that supports life. The same precision applies to the other constants, each contributing to a delicate balance that makes the universe conducive to life.
The statistical improbability of these constants randomly aligning to create a life-sustaining universe is astronomically high. The odds are so extreme that simply attributing this fine-tuning to chance is irrational to the extreme, to say the least. This precision suggests purposeful design, as the likelihood of such a perfect balance occurring by sheer coincidence is vanishingly small.
Considering a grand designer is a legitimate plausible inference based on scientific evidence. The fine-tuning of the universe points to a purposeful arrangement of its fundamental constants, which, in turn, implies an intelligent designer. Acknowledging a grand designer does not halt scientific inquiry; rather, it expands the philosophical framework within which science operates. It encourages scientists and thinkers to explore not just how the universe functions, but why it exists in such a precisely calibrated form. This perspective fosters a holistic view of the cosmos, one that embraces both the rigor of scientific investigation and the profound implications of a universe seemingly designed with intention.
Louis Pasteur's observation that "a little science estranges men from God, but much science leads them back to Him" beautifully encapsulates the journey of scientific discovery and its relationship with the concept of a grand designer or God. Pasteur, a pioneering scientist known for his groundbreaking work in microbiology and chemistry, recognized the profound interplay between scientific inquiry and spiritual understanding. Extending this idea to the discussion of the universe's fine-tuning and the evidence pointing towards a grand designer, we can see how deeper scientific exploration brings us closer to contemplating the possibility of purposeful creation. At the surface level, scientific discoveries might seem to challenge traditional religious views, creating an apparent rift between science and creationism. The initial stages of scientific inquiry often focus on understanding the natural laws that govern the universe, leading some to perceive the world as a product of random processes and natural selection. This perspective can create a sense of estrangement from the notion of a grand designer, as it appears that the universe and life itself can be explained purely through physical laws and chance. However, as scientific exploration progresses and delves deeper into the fundamental aspects of the universe, such as the precise tuning of physical constants, the complexity and order inherent in the cosmos become increasingly apparent.
This complexity is not just superficial; it's foundational to the very existence and habitability of the universe. The precise conditions that allow for life are so statistically improbable that they prompt a reevaluation of the role of chance in the cosmos's creation. This deeper scientific inquiry reveals a universe so intricately balanced and finely adjusted with extraordinary precision that it seems to point beyond mere chance or necessity. The more we learn about the universe's fundamental properties, the more we are drawn to consider the possibility of a purposeful designer behind our existence. This realization aligns with Pasteur's insight, where the pursuit of more profound scientific understanding brings us closer to contemplating a grand designer's role in the cosmos. Pasteur's statement underscores the complementary nature of science and spirituality. Far from being mutually exclusive, they offer different lenses through which to explore and appreciate the marvels of the universe. Science equips us with the tools to understand the "how" of the universe's workings, while philosophy without keeping the framework of philosophical naturalism leads us to an intelligent powerful designer.

The principle that "something cannot come from nothing" underpins the argument that the material universe, with its complexity and order, must have been derived from an existing entity. This foundational premise posits that because the universe exists as a highly structured and law-governed system, it cannot be the product of chance or emerge spontaneously from nothingness. The intricate laws and constants governing the universe's functioning suggest that it operates under a deliberate design, which, by nature, implies the involvement of a conscious mind. Given that designs and purposes are inherently the products of intelligent deliberation, it follows that the universe's existence and its governed structure point towards an eternal mind. This mind, characterized by neither beginning nor end, must have conceived and crafted the material universe. The argument strengthens with the understanding that such a mind, transcending time and space, is the ultimate source from which the universe, with all its laws and order, originated.
Imagine you're holding a very complex, intricately designed watch. You see the gears moving in harmony, keeping time perfectly. Now, think of the universe as that watch, but on an incomprehensibly larger scale. Just like the watch has fundamental components that allow it to function (like the gears, hands, and battery), the universe has its own basic building blocks. These are things like time, measured in seconds; distance, measured in meters; mass, in kilograms; electric current, in amperes; temperature, in kelvin; the amount of a substance, in moles; and the intensity of light, in candelas.
These building blocks are like the universe's DNA, setting the stage for everything that exists and happens within it. They're like the rules of the game, but with a twist: we can't trace these rules back to anything simpler. They're the bedrock, the foundation that everything else builds upon. And here's where it gets really interesting: these rules seem to be set up just right for life to exist. It's as if the universe has been finely tuned, like adjusting the strings of a guitar to get the perfect pitch, so that life can emerge and thrive.
The big question is, why are these rules set up this way? Why do these fundamental building blocks have the values they do? Science, as of now, doesn't have an answer. It's like finding the watch and marveling at its precision and design, yet not knowing who made it or why it was made that way. This situation leads some to ponder the possibility of a designer, a master watchmaker if you will. This isn't about proving something with a formula or an experiment; it's about looking at the universe and asking, "Could this all be a result of chance, or is there something more, some intention behind it?" Just like Isaac Newton, a great scientist, or Paley, who also saw the universe as a creation of a divine watchmaker, many find in the precision and complexity of the universe hints of a grander design, suggesting perhaps an intelligent force or being behind it all. This doesn't replace scientific inquiry but invites us to marvel at the mysteries still beyond our understanding.
Scientific evidence points towards a grand designer. The observation that the universe if finely tuned doesn't merely fill gaps in our current understanding with divine intervention, but rather suggests that the precision and values of the fundamental constants in our universe are so exact that they imply a deliberate design. The universe operates under a set of fundamental constants, such as the gravitational constant, the speed of light, and the charge of an electron, to name a few. These constants dictate the behavior of the cosmos, from the formation of atoms to the birth of stars. The remarkable aspect of these constants is their precision; they are finely tuned to values that allow the universe to be habitable. For instance, if the gravitational constant were slightly stronger or weaker, stars (including our sun) might not form or function in a way that supports life. The same precision applies to the other constants, each contributing to a delicate balance that makes the universe conducive to life.
The statistical improbability of these constants randomly aligning to create a life-sustaining universe is astronomically high. The odds are so extreme that simply attributing this fine-tuning to chance is irrational to the extreme, to say the least. This precision suggests purposeful design, as the likelihood of such a perfect balance occurring by sheer coincidence is vanishingly small.
Considering a grand designer is a legitimate plausible inference based on scientific evidence. The fine-tuning of the universe points to a purposeful arrangement of its fundamental constants, which, in turn, implies an intelligent designer. Acknowledging a grand designer does not halt scientific inquiry; rather, it expands the philosophical framework within which science operates. It encourages scientists and thinkers to explore not just how the universe functions, but why it exists in such a precisely calibrated form. This perspective fosters a holistic view of the cosmos, one that embraces both the rigor of scientific investigation and the profound implications of a universe seemingly designed with intention.
Louis Pasteur's observation that "a little science estranges men from God, but much science leads them back to Him" beautifully encapsulates the journey of scientific discovery and its relationship with the concept of a grand designer or God. Pasteur, a pioneering scientist known for his groundbreaking work in microbiology and chemistry, recognized the profound interplay between scientific inquiry and spiritual understanding. Extending this idea to the discussion of the universe's fine-tuning and the evidence pointing towards a grand designer, we can see how deeper scientific exploration brings us closer to contemplating the possibility of purposeful creation. At the surface level, scientific discoveries might seem to challenge traditional religious views, creating an apparent rift between science and creationism. The initial stages of scientific inquiry often focus on understanding the natural laws that govern the universe, leading some to perceive the world as a product of random processes and natural selection. This perspective can create a sense of estrangement from the notion of a grand designer, as it appears that the universe and life itself can be explained purely through physical laws and chance. However, as scientific exploration progresses and delves deeper into the fundamental aspects of the universe, such as the precise tuning of physical constants, the complexity and order inherent in the cosmos become increasingly apparent.
This complexity is not just superficial; it's foundational to the very existence and habitability of the universe. The precise conditions that allow for life are so statistically improbable that they prompt a reevaluation of the role of chance in the cosmos's creation. This deeper scientific inquiry reveals a universe so intricately balanced and finely adjusted with extraordinary precision that it seems to point beyond mere chance or necessity. The more we learn about the universe's fundamental properties, the more we are drawn to consider the possibility of a purposeful designer behind our existence. This realization aligns with Pasteur's insight, where the pursuit of more profound scientific understanding brings us closer to contemplating a grand designer's role in the cosmos. Pasteur's statement underscores the complementary nature of science and spirituality. Far from being mutually exclusive, they offer different lenses through which to explore and appreciate the marvels of the universe. Science equips us with the tools to understand the "how" of the universe's workings, while philosophy without keeping the framework of philosophical naturalism leads us to an intelligent powerful designer.

299 Re: My articles Sun Feb 11, 2024 6:42 am
Re: My articles Sun Feb 11, 2024 6:42 am
Otangelo
Admin
The Pearl of Greatest Price: The Mutual Belonging of Christ and the Believer
The kingdom of heaven is like a merchant looking for fine pearls. When he found one of great value, he went away and sold everything he had and bought it." - Matthew 13:45-46 (NIV)
In this parable, the pearl of great price represents the immense value of the Kingdom of Heaven, suggesting that it is worth surrendering all earthly possessions to obtain it.
The pursuit of happiness, peace, joy, and love is a universal human experience, akin to the merchant's search for fine pearls in the parable. Just as the merchant in the story traverses far and wide, investing time and resources to find that one pearl of great price, so do we all explore various paths in life, seeking fulfillment and meaning. The pearl in the parable is a metaphor for something of ultimate value, and the merchant's discovery symbolizes the realization that finding this treasure is worth more than all other possessions combined. For many, the search for happiness and contentment can involve the pursuit of wealth, success, relationships, and experiences. Each person looks for these "pearls" in different places — in careers, in academic achievements, in social status, or in personal relationships. However, the parable suggests that once the merchant finds the pearl, he sells everything to possess it. This dramatic action implies that true joy and contentment, once discovered, demand a reevaluation of priorities. It suggests that the ephemeral pleasures of life may not offer lasting satisfaction and that true peace and happiness might come from something more profound. The Kingdom of Heaven is something of intrinsic value, worth all the sacrifices. Translated into the quest for happiness and fulfillment, it means that what we ultimately seek is not found in external circumstances but in the internal state of being — in spiritual peace, moral integrity, loving relationships, and a sense of purpose that aligns with one's deepest values.
In a broader sense, the merchant's search is a journey toward self-discovery and enlightenment. It is about identifying what is truly valuable in life. The parable emphasizes that the greatest treasure is Christ, and is worth even the greatest sacrifices. It is an invitation to look beyond the surface, to find joy in the intangible, and to consider what we ultimately seek is Christ. The ultimate treasure is not found in the accumulation of wealth, the pursuit of power, or the indulgence in hedonistic pleasures, but rather in the discovery and relationship with Christ. Christ embodies the quintessence of all that is good, loving, gracious, and just. This comprehensive goodness is not only inherent but is also reflected in the world He created, which was deemed "very good" upon its creation. The brokenness and imperfection we witness in the world today are consequences of the fall of man, an event that introduced sin and its resulting decay into the pristine creation. Despite this, the inherent goodness of creation and its Creator can still be discerned. The beauty in a sunset, the complexity of a living organism, the joy in human relationships—these experiences, though marred by the fall, still bear witness to the original goodness of God's creation. They invite us to look beyond the immediate physical reality and consider that God will create a new heaven and a new earth, with no evil, no pain, where everything will be perfect, and exist in harmony.
In recognizing Christ as the greatest treasure, we acknowledge that true richness lies in qualities that are eternal and transcendent, such as goodness, love, grace, and justice. These are attributes that bring lasting fulfillment and purpose, far surpassing the temporary satisfaction derived from worldly pursuits. The pursuit of Christ and His righteousness becomes the path to uncovering the deepest treasures of life. Moreover, this understanding calls for a transformation in how we live and perceive the world around us. It challenges us to live not for ourselves but in a manner that reflects the character of Christ—serving others, loving unconditionally, and seeking justice. As we align our lives with these values, we not only draw closer to the treasure that is Christ but also become conduits of His goodness in a broken world. In this light, every aspect of creation, every moment of existence, becomes an opportunity to witness the divine, to give glory to God, and to participate in the restoration of all things to their intended goodness. This perspective instills hope, for it assures us that despite the present brokenness, there is a promise of redemption and restoration, a future where the fullness of God's goodness will be once again fully manifest.
The assertion that God is 'hidden' and therefore not evident is a common theme in discussions about faith and belief. It revolves around the idea of divine hiddenness as a barrier to belief. This notion posits that if God were more evident or accessible to sensory or empirical observation, there would be more widespread belief. However, the doctrine of the Incarnation, holds that God made the divine nature accessible and visible in the person of Jesus Christ. God is not hidden but has revealed Himself in history. The life and teachings of Jesus, as recorded in the Gospels, are direct evidence of God’s interaction with the world. Jesus is the “image of the invisible God” (Colossians 1:15) and "the Word made flesh" (John 1:14), indicating that in Him the nature and character of God are made manifest. The eyewitness accounts of Jesus’ life, death, and resurrection as recorded in the Gospels are central to the knowability of God. These texts are not merely historical records but are also sacred scriptures that convey theological truths. The Gospels are a testament to the life of Jesus and are a means through which we can encounter the person of Christ. In addition to the textual evidence of the Gospels, the Shroud of Turin is a tangible connection to the historical Jesus. The Shroud is a physical artifact that bears witness to Christ’s existence and the events of the crucifixion, serving as a reminder of the incarnation and suffering of God in Christ.
Then, the evidence of God's existence and nature is not only philosophical or metaphysical but historical and tangible. The incarnation of Christ is the ultimate act of divine revelation, making searching for God not a quest into the unknown but a journey into the well-documented life of Jesus. The message of the Gospels, the transformation of the lives of the apostles, the historical impact of Jesus’ teachings, and the material evidence imprinted in the Shroud are all signs pointing towards the visibility of God. Our response to the idea of divine hiddenness is an invitation to engage with the person of Jesus Christ as the definitive revelation of God. This is not merely as an intellectual assent to facts but as a relational knowledge that involves trust, faith, and personal encounter.
The face of Christ depicted on the Shroud of Turin expresses serenity, which is a reflection of Christ's acceptance and foreknowledge of His fate. This serenity amidst unfathomable, extreme suffering is an emblem of His divine grace and forgiveness. The closed eyes and the peaceful demeanor signify the completion of His earthly mission and the fulfillment of prophecies concerning His death. The sublime nobility of His visage as the great King of Kings on the Shroud is consistent with His dual nature, being both fully human and fully divine. The human aspects of suffering are evident in the battered state of the countenance. Yet, the tranquility of the expression suggests a transcendence of mere human experience — an assurance of hope and resurrection. For us believers, the face on the Shroud communicates the depth of Christ's compassion and His willing sacrifice. It is a silent testimony to the narrative of redemption, reflecting a character that is at once both strong and gentle. The image is a bridge between the temporal and the eternal, a visual sermon that speaks of love, redemption, and the possibility of communion with the divine. The Shroud is more than a relic; it is a visual narrative that encapsulates the core of Christian belief — the death and resurrection of Jesus. Christ is the pearl not just of great, but the greatest value of all. Blessed are those who have found this pearl.
Christ belongs to us, believers. This is not in the sense of property, but rather in the relational and covenantal sense. We believers are described as the 'body of Christ' (1 Corinthians 12:27), indicating a profound union with Him. There is a shared identity and a shared inheritance, as outlined in Romans 8:17, which says, "Now if we are children, then we are heirs—heirs of God and co-heirs with Christ." In this union, the blessings that Christ has prepared for us believers are peace, grace, and ultimately eternal life. The blessings are not merely future promises but present realities that shape our experience of life. The spiritual blessings include the presence of the Holy Spirit, the assurance of God's love, and the transformation of character to bear the fruit of the Spirit: love, joy, peace, patience, kindness, goodness, faithfulness, gentleness, and self-control (Galatians 5:22-23).
Just as in a loving human relationship where the lives and fortunes of the individuals are intertwined, so too in the believer's relationship with Christ, there is a mutual belonging. Christ's sacrifice establishes the relationship, and we can respond with faith and love, which further cements this spiritual bond. We are called to be like Christ, to share in His sufferings, and to live out the values of the Kingdom of Heaven. Moreover, this belonging carries with it the idea of responsibility. We, belonging to Christ, are called to represent Him in the world, to live lives that reflect His love and grace. At the same time, the assurance that Christ belongs to us provides comfort and strength, knowing that we are not left to navigate the challenges of life alone.

The kingdom of heaven is like a merchant looking for fine pearls. When he found one of great value, he went away and sold everything he had and bought it." - Matthew 13:45-46 (NIV)
In this parable, the pearl of great price represents the immense value of the Kingdom of Heaven, suggesting that it is worth surrendering all earthly possessions to obtain it.
The pursuit of happiness, peace, joy, and love is a universal human experience, akin to the merchant's search for fine pearls in the parable. Just as the merchant in the story traverses far and wide, investing time and resources to find that one pearl of great price, so do we all explore various paths in life, seeking fulfillment and meaning. The pearl in the parable is a metaphor for something of ultimate value, and the merchant's discovery symbolizes the realization that finding this treasure is worth more than all other possessions combined. For many, the search for happiness and contentment can involve the pursuit of wealth, success, relationships, and experiences. Each person looks for these "pearls" in different places — in careers, in academic achievements, in social status, or in personal relationships. However, the parable suggests that once the merchant finds the pearl, he sells everything to possess it. This dramatic action implies that true joy and contentment, once discovered, demand a reevaluation of priorities. It suggests that the ephemeral pleasures of life may not offer lasting satisfaction and that true peace and happiness might come from something more profound. The Kingdom of Heaven is something of intrinsic value, worth all the sacrifices. Translated into the quest for happiness and fulfillment, it means that what we ultimately seek is not found in external circumstances but in the internal state of being — in spiritual peace, moral integrity, loving relationships, and a sense of purpose that aligns with one's deepest values.
In a broader sense, the merchant's search is a journey toward self-discovery and enlightenment. It is about identifying what is truly valuable in life. The parable emphasizes that the greatest treasure is Christ, and is worth even the greatest sacrifices. It is an invitation to look beyond the surface, to find joy in the intangible, and to consider what we ultimately seek is Christ. The ultimate treasure is not found in the accumulation of wealth, the pursuit of power, or the indulgence in hedonistic pleasures, but rather in the discovery and relationship with Christ. Christ embodies the quintessence of all that is good, loving, gracious, and just. This comprehensive goodness is not only inherent but is also reflected in the world He created, which was deemed "very good" upon its creation. The brokenness and imperfection we witness in the world today are consequences of the fall of man, an event that introduced sin and its resulting decay into the pristine creation. Despite this, the inherent goodness of creation and its Creator can still be discerned. The beauty in a sunset, the complexity of a living organism, the joy in human relationships—these experiences, though marred by the fall, still bear witness to the original goodness of God's creation. They invite us to look beyond the immediate physical reality and consider that God will create a new heaven and a new earth, with no evil, no pain, where everything will be perfect, and exist in harmony.
In recognizing Christ as the greatest treasure, we acknowledge that true richness lies in qualities that are eternal and transcendent, such as goodness, love, grace, and justice. These are attributes that bring lasting fulfillment and purpose, far surpassing the temporary satisfaction derived from worldly pursuits. The pursuit of Christ and His righteousness becomes the path to uncovering the deepest treasures of life. Moreover, this understanding calls for a transformation in how we live and perceive the world around us. It challenges us to live not for ourselves but in a manner that reflects the character of Christ—serving others, loving unconditionally, and seeking justice. As we align our lives with these values, we not only draw closer to the treasure that is Christ but also become conduits of His goodness in a broken world. In this light, every aspect of creation, every moment of existence, becomes an opportunity to witness the divine, to give glory to God, and to participate in the restoration of all things to their intended goodness. This perspective instills hope, for it assures us that despite the present brokenness, there is a promise of redemption and restoration, a future where the fullness of God's goodness will be once again fully manifest.
The assertion that God is 'hidden' and therefore not evident is a common theme in discussions about faith and belief. It revolves around the idea of divine hiddenness as a barrier to belief. This notion posits that if God were more evident or accessible to sensory or empirical observation, there would be more widespread belief. However, the doctrine of the Incarnation, holds that God made the divine nature accessible and visible in the person of Jesus Christ. God is not hidden but has revealed Himself in history. The life and teachings of Jesus, as recorded in the Gospels, are direct evidence of God’s interaction with the world. Jesus is the “image of the invisible God” (Colossians 1:15) and "the Word made flesh" (John 1:14), indicating that in Him the nature and character of God are made manifest. The eyewitness accounts of Jesus’ life, death, and resurrection as recorded in the Gospels are central to the knowability of God. These texts are not merely historical records but are also sacred scriptures that convey theological truths. The Gospels are a testament to the life of Jesus and are a means through which we can encounter the person of Christ. In addition to the textual evidence of the Gospels, the Shroud of Turin is a tangible connection to the historical Jesus. The Shroud is a physical artifact that bears witness to Christ’s existence and the events of the crucifixion, serving as a reminder of the incarnation and suffering of God in Christ.
Then, the evidence of God's existence and nature is not only philosophical or metaphysical but historical and tangible. The incarnation of Christ is the ultimate act of divine revelation, making searching for God not a quest into the unknown but a journey into the well-documented life of Jesus. The message of the Gospels, the transformation of the lives of the apostles, the historical impact of Jesus’ teachings, and the material evidence imprinted in the Shroud are all signs pointing towards the visibility of God. Our response to the idea of divine hiddenness is an invitation to engage with the person of Jesus Christ as the definitive revelation of God. This is not merely as an intellectual assent to facts but as a relational knowledge that involves trust, faith, and personal encounter.
The face of Christ depicted on the Shroud of Turin expresses serenity, which is a reflection of Christ's acceptance and foreknowledge of His fate. This serenity amidst unfathomable, extreme suffering is an emblem of His divine grace and forgiveness. The closed eyes and the peaceful demeanor signify the completion of His earthly mission and the fulfillment of prophecies concerning His death. The sublime nobility of His visage as the great King of Kings on the Shroud is consistent with His dual nature, being both fully human and fully divine. The human aspects of suffering are evident in the battered state of the countenance. Yet, the tranquility of the expression suggests a transcendence of mere human experience — an assurance of hope and resurrection. For us believers, the face on the Shroud communicates the depth of Christ's compassion and His willing sacrifice. It is a silent testimony to the narrative of redemption, reflecting a character that is at once both strong and gentle. The image is a bridge between the temporal and the eternal, a visual sermon that speaks of love, redemption, and the possibility of communion with the divine. The Shroud is more than a relic; it is a visual narrative that encapsulates the core of Christian belief — the death and resurrection of Jesus. Christ is the pearl not just of great, but the greatest value of all. Blessed are those who have found this pearl.
Christ belongs to us, believers. This is not in the sense of property, but rather in the relational and covenantal sense. We believers are described as the 'body of Christ' (1 Corinthians 12:27), indicating a profound union with Him. There is a shared identity and a shared inheritance, as outlined in Romans 8:17, which says, "Now if we are children, then we are heirs—heirs of God and co-heirs with Christ." In this union, the blessings that Christ has prepared for us believers are peace, grace, and ultimately eternal life. The blessings are not merely future promises but present realities that shape our experience of life. The spiritual blessings include the presence of the Holy Spirit, the assurance of God's love, and the transformation of character to bear the fruit of the Spirit: love, joy, peace, patience, kindness, goodness, faithfulness, gentleness, and self-control (Galatians 5:22-23).
Just as in a loving human relationship where the lives and fortunes of the individuals are intertwined, so too in the believer's relationship with Christ, there is a mutual belonging. Christ's sacrifice establishes the relationship, and we can respond with faith and love, which further cements this spiritual bond. We are called to be like Christ, to share in His sufferings, and to live out the values of the Kingdom of Heaven. Moreover, this belonging carries with it the idea of responsibility. We, belonging to Christ, are called to represent Him in the world, to live lives that reflect His love and grace. At the same time, the assurance that Christ belongs to us provides comfort and strength, knowing that we are not left to navigate the challenges of life alone.

300 Re: My articles Mon Feb 12, 2024 6:30 pm
Re: My articles Mon Feb 12, 2024 6:30 pm
Otangelo
Admin
The Dawn of Hope: Mary Magdalene's Encounter with the Risen Christ
The passage John 20:1-18 describes a deeply moving and significant moment in the Christian narrative. Mary Magdalene, one of Jesus' most devoted followers, visits Jesus' tomb early in the morning and finds it empty.
Distraught, she encounters Jesus but does not recognize him at first. When she does, she exclaims "Rabbouni!" (which means "Teacher" in Aramaic).
This passage captures in my view the most emotional moment in the entire Bible. This scene begins in the early morning. Mary Magdalene's visit to the tomb, driven by her devotion and love for Jesus, sets the stage for this emotional encounter.
The emptiness of the tomb initially signifies loss and despair for Mary. In her grief, the empty tomb represents the finality of death and the apparent end of the journey she shared with Jesus.
This moment of despair reflects a universal human experience of grief, where the absence of a loved one evokes sadness a feel of loss, and emptiness.
However, the narrative quickly shifts from despair to confusion and then to profound revelation. Mary's encounter with Jesus, whom she initially fails to recognize, speaks to the mysterious and transformative nature of the resurrection.
It is only through the calling of her name that the familiar relationship is re-established, highlighting her recognition of the voice.
When Mary recognizes Jesus and exclaims "Rabbouni," the emotional intensity of the scene reaches its apex. This exclamation is not merely a recognition but an expression of profound joy, relief, and reconnection.
The use of "Rabbouni" underscores a personal and intimate relationship with Jesus, reflecting both respect and deep affection.
This encounter between Mary Magdalene and the resurrected Jesus symbolizes the passage from grief and loss to hope and renewal.
Mary Magdalene's role in this narrative is significant. As a woman and a devoted follower, her witness to the resurrection and her commission to share the news with the disciples challenge contemporary cultural norms and elevate her role within the Christian tradition.
This moment marks the beginning of her transformation from mourner to apostle to the apostles, underscoring the theme of empowerment and the spread of the new faith.
In this narrative, the personal encounter with the divine, the transformation of understanding, and the commissioning to share the good news encapsulates the essence of the Christian message.
The emotional depth of this passage lies not only in the joy of the resurrection but in the profound changes it heralds for Mary Magdalene, the disciples, and ultimately, for us all.
Mary Magdalene was dramatically healed by Jesus and followed him closely thereafter, making her recognition of him deeply personal and emotionally charged. The resurrection was a pivotal event.
For Mary and other followers of Jesus, the crucifixion was a moment of despair and perceived defeat. Seeing Jesus alive again turned that despair into hope and joy, validating his teachings and promises.
Mary Magdalene's prominent role in this event emphasizes the significant place of women in the early Christian community. Despite societal norms of the time, this narrative places a woman at the center of one of Christianity's most pivotal moments, illustrating the inclusive nature of Jesus' ministry.
In a period where women were often regarded as second-class citizens and their testimonies were given little credence, the inclusion and elevation of women in key biblical narratives is significant.
Throughout the Gospels, Jesus consistently breaks societal norms by valuing and respecting women. He interacts with women publicly, teaches them alongside men, and includes them in his ministry.
This was revolutionary in a context where women were often relegated to the background and excluded from religious and scholarly discussions. In several pivotal biblical events, women are central figures.
The resurrection narrative, where women (specifically Mary Magdalene) are the first to witness the empty tomb and the risen Christ, is a prime example. This is a powerful statement, especially considering that women's testimonies were often disregarded in legal and societal contexts at the time.
Jesus' interactions with women often served to empower them. Examples include his conversation with the Samaritan woman at the well (John 4:1-26)
On that occasion, Jesus breaks cultural taboos to engage with her and reveal his identity as the Messiah, and his defense of Mary of Bethany's choice to sit and listen to his teachings (Luke 10:39-42).
The resurrection is central to Christian theology, symbolizing not just the defeat of death but also the potential for spiritual transformation. This event offers hope and a promise of new life beyond physical death, which is a cornerstone of our belief.
Jesus addressing Mary by name and her subsequent recognition of him underscores the personal nature of faith and the individual relationship that we believers can have with the Lord.
Our faith in the Lord is deeply personal. Following her encounter, Mary Magdalene goes to the disciples to announce that she has seen the Lord. This act highlights the importance of proclamation in Christianity - the responsibility of believers to share their experiences and the message of Jesus Christ.
The resurrection is God's seal of approval on the work of Christ. If Christ had remained dead, it would imply that His sacrifice was insufficient. However, the resurrection confirms that not only was Jesus' sacrifice sufficient, but it also demonstrates His power over death, affirming His divinity and the truth of His teachings.
For us believers, the resurrection is a source of hope and assurance. It promises us new life and a future resurrection, similar to Christ's. The same power that raised Jesus from the dead is at work in us who believe, in ensuring our ultimate victory over sin and death.
How wonderful will our encounter be, when we see the first time the Lord, in which we put our hope for our entire life? Hallelujah.
==========================================================================================================================================
The passage John 20:1-18 describes a deeply moving and significant moment in the Christian narrative. Mary Magdalene, one of Jesus' most devoted followers, visits Jesus' tomb early in the morning and finds it empty.
یوحنا ۲۰:۱-۱۸ کی آیتیں ایک گہری اور اہم لمحے کو بیان کرتی ہیں جو عیسائیت کی کہانی میں بہت اہم ہے۔ مریم مگدلینی، جو یسوع کی سب سے مخلص پیروکاروں میں سے ایک تھی، صبح سویرے یسوع کی قبر پر جاتی ہے اور اسے خالی پاتی ہے۔
Distraught, she encounters Jesus but does not recognize him at first. When she does, she exclaims "Rabbouni!" (which means "Teacher" in Aramaic).
پریشان حال، وہ یسوع سے ملاقات کرتی ہے لیکن شروع میں اسے پہچان نہیں پاتی۔ جب وہ پہچانتی ہے، تو وہ "ربونی!" کہتی ہے (جس کا معنی ارامی زبان میں "استاد" ہے)۔
This passage captures in my view the most emotional moment in the entire Bible. This scene begins in the early morning. Mary Magdalene's visit to the tomb, driven by her devotion and love for Jesus, sets the stage for this emotional encounter.
میرے خیال میں یہ آیت پوری بائبل کے سب سے زیادہ جذباتی لمحے کو بیان کرتی ہے۔ یہ منظر صبح سویرے شروع ہوتا ہے۔ مریم مگدلینی کا قبر پر جانا، جو کہ اس کی یسوع کے لیے محبت اور وفاداری سے محرک تھا، اس جذباتی ملاقات کے لیے منظر مقرر کرتا ہے۔
The emptiness of the tomb initially signifies loss and despair for Mary. In her grief, the empty tomb represents the finality of death and the apparent end of the journey she shared with Jesus.
قبر کی خالی ہونے کا ابتدائی معنی مریم کے لئے نقصان اور مایوسی ہے۔ اس کے غم میں، خالی قبر موت کی حتمیت اور اس سفر کے ظاہری اختتام کی نمائندگی کرتی ہے جو اس نے یسوع کے ساتھ بانٹی تھی۔
This moment of despair reflects a universal human experience of grief, where the absence of a loved one evokes sadness a feel of loss, and emptiness.
یہ مایوسی کا لمحہ انسانی غم کے ایک عالمی تجربے کی عکاسی کرتا ہے، جہاں کسی پیارے کی عدم موجودگی اداسی، نقصان کا احساس، اور خالی پن کو جنم دیتی ہے۔
However, the narrative quickly
shifts from despair to confusion and then to profound revelation. Mary's encounter with Jesus, whom she initially fails to recognize, speaks to the mysterious and transformative nature of the resurrection.
تاہم، کہانی جلد ہی مایوسی سے الجھن اور پھر گہرے انکشاف کی طرف منتقل ہوتی ہے۔ مریم کی یسوع سے ملاقات، جسے وہ شروع میں پہچان نہیں پاتی، قیامت کی پراسرار اور تبدیل کر دینے والی فطرت کو بیان کرتی ہے۔
It is only through the calling of her name that the familiar relationship is re-established, highlighting her recognition of the voice.
یہ صرف اس کے نام کے پکارے جانے کے ذریعے ہی ہے کہ واقفیت کا رشتہ دوبارہ قائم ہوتا ہے، جس سے اس کی آواز کی پہچان پر روشنی ڈالی جاتی ہے۔
When Mary recognizes Jesus and exclaims "Rabbouni," the emotional intensity of the scene reaches its apex. This exclamation is not merely a recognition but an expression of profound joy, relief, and reconnection.
جب مریم یسوع کو پہچانتی ہے اور "ربونی" کہتی ہے، تو منظر کی جذباتی شدت اپنے عروج پر پہنچ جاتی ہے۔ یہ اعلان صرف پہچان نہیں بلکہ گہری خوشی، راحت، اور دوبارہ جڑنے کا اظہار ہے۔
The use of "Rabbouni" underscores a personal and intimate relationship with Jesus, reflecting both respect and deep affection.
"ربونی" کے استعمال سے یسوع کے ساتھ ایک ذاتی اور قریبی تعلق کو زور دیا گیا ہے، جو کہ عزت اور گہری محبت دونوں کی عکاسی کرتا ہے۔
This encounter between Mary Magdalene and the resurrected Jesus symbolizes the passage from grief and loss to hope and renewal.
مریم مگدلینی اور قیامت پذیر یسوع کے درمیان یہ ملاقات غم اور نقصان سے امید اور تجدید کے سفر کی علامت ہے۔
Mary Magdalene's role in this narrative is significant. As a woman and a devoted follower, her witness to the resurrection and her commission to share the news with the disciples challenge contemporary cultural norms and elevate her role within the Christian tradition.
مریم مگدلینی کا کردار اس کہانی میں اہم ہے۔ ایک عورت اور ایک مخلص پیروکار کے طور پر، اس کی قیامت کی گواہی اور شاگردوں کے ساتھ خبریں بانٹنے کی ذمہ داری موجودہ ثقافتی اصولوں کو چیلنج کرتی ہے اور عیسائی روایت میں اس ک
ے کردار کو بلند کرتی ہے۔
This moment marks the beginning of her transformation from mourner to apostle to the apostles, underscoring the theme of empowerment and the spread of the new faith.
یہ لمحہ اس کے تبدیلی کی شروعات کو نشان زد کرتا ہے، غمزدہ سے رسول تک، اور رسولوں کے رسول تک، اختیار کی تھیم اور نئے عقیدے کے پھیلاؤ کو اجاگر کرتا ہے۔
In this narrative, the personal encounter with the divine, the transformation of understanding, and the commissioning to share the good news encapsulates the essence of the Christian message.
اس کہانی میں، الہی کے ساتھ ذاتی ملاقات، سمجھ میں تبدیلی، اور اچھی خبریں بانٹنے کی ذمہ داری عیسائی پیغام کے جوہر کو سموتی ہے۔
The emotional depth of this passage lies not only in the joy of the resurrection but in the profound changes it heralds for Mary Magdalene, the disciples, and ultimately, for us all.
اس آیت کی جذباتی گہرائی صرف قیامت کی خوشی میں نہیں ہے بلکہ مریم مگدلینی، شاگردوں، اور آخر کار، ہم سب کے لئے اس کے اعلان کردہ گہرے تبدیلیوں میں ہے۔
Mary Magdalene was dramatically healed by Jesus and followed him closely thereafter, making her recognition of him deeply personal and emotionally charged.
مریم مگدلینی کو یسوع نے نمایاں طور پر شفا دی اور اس کے بعد وہ اس کے قریب سے پیروی کرتی رہی، جس سے اس کی اس کی پہچان گہری ذاتی اور جذباتی طور پر چارج ہو گئی۔
The resurrection was a pivotal event. For Mary and other followers of Jesus, the crucifixion was a moment of despair and perceived defeat. Seeing Jesus alive again turned that despair into hope and joy, validating his teachings and promises.
قیامت ایک اہم واقعہ تھا۔ مریم اور یسوع کے دیگر پیروکاروں کے لئے، صلیب کی سزا مایوسی اور شکست کا لمحہ تھا۔ یسوع کو دوبارہ زندہ دیکھ کر وہ مایوسی امید اور خوشی میں بدل گئی، اس کی تعلیمات اور وعدوں کی تصدیق کرتے ہوئے۔
Mary Magdalene's prominent role in this event emphasizes the significant place of women in the early Christian community. Despite societal norms of the time, this narrative places a woman at the center of one of Christianity's most pivotal moments, illustrating the inclusive nature of Jesus' ministry.
مریم مگدلینی کا اس واقعہ میں نمایاں کردار عیسائی برادری میں عورتوں کے اہم مقام کو زور
دیتا ہے۔ اس وقت کے معاشرتی اصولوں کے باوجود، یہ کہانی عیسائیت کے سب سے اہم لمحوں میں سے ایک میں ایک عورت کو مرکزی کردار میں رکھتی ہے، جس سے یسوع کی وزارت کی شاملیت کی نوعیت کو واضح کیا گیا ہے۔
In a period where women were often regarded as second-class citizens and their testimonies were given little credence, the inclusion and elevation of women in key biblical narratives is significant.
ایک ایسے دور میں جب عورتوں کو اکثر دوسرے درجے کے شہریوں کے طور پر دیکھا جاتا تھا اور ان کی گواہی کو زیادہ اہمیت نہیں دی جاتی تھی، بائبل کی اہم کہانیوں میں عورتوں کی شمولیت اور ان کی بلندی اہم ہے۔
Throughout the Gospels, Jesus consistently breaks societal norms by valuing and respecting women. He interacts with women publicly, teaches them alongside men, and includes them in his ministry.
انجیلوں کے دوران، یسوع مستقل طور پر عورتوں کو قدر دے کر اور ان کا احترام کر کے معاشرتی اصولوں کو توڑتا ہے۔ وہ عورتوں سے عوامی طور پر بات چیت کرتا ہے، انہیں مردوں کے ساتھ ساتھ تعلیم دیتا ہے، اور انہیں اپنی وزارت میں شامل کرتا ہے۔
This was revolutionary in a context where women were often relegated to the background and excluded from religious and scholarly discussions.
یہ ایک ایسے تناظر میں انقلابی تھا جہاں عورتوں کو اکثر پس منظر میں رکھا جاتا تھا اور انہیں مذہبی اور علمی بحثوں سے خارج کر دیا جاتا تھا۔
In several pivotal biblical events, women are central figures. The resurrection narrative, where women (specifically Mary Magdalene) are the first to witness the empty tomb and the risen Christ, is a prime example.
کئی اہم بائبل کے واقعات میں، عورتیں مرکزی کردار ہیں۔ قیامت کی کہانی، جہاں عورتیں (خاص طور پر مریم مگدلینی) خالی قبر اور قیامت پذیر مسیح کی پہلی گواہ ہیں، ایک بہترین مثال ہے۔
This is a powerful statement, especially considering that women's testimonies were often disregarded in legal and societal contexts at the time.
یہ ایک طاقتور بیان ہے، خاص طور پر اس وقت کے قانونی اور معاشرتی
تناظر میں عورتوں کی گواہی کو اکثر نظر انداز کیا جاتا تھا۔
Jesus' interactions with women often served to empower them. Examples include his conversation with the Samaritan woman at the well (John 4:1-26)
یسوع کی عورتوں کے ساتھ بات چیت اکثر انہیں بااختیار بنانے کے لئے ہوتی تھی۔ مثالوں میں اس کی سامری عورت کے ساتھ کنویں پر گفتگو شامل ہے (یوحنا ۴:۱-۲۶)
On that occasion, Jesus breaks cultural taboos to engage with her and reveal his identity as the Messiah, and his defense of Mary of Bethany's choice to sit and listen to his teachings (Luke 10:39-42).
اس موقع پر، یسوع ثقافتی ممنوعات کو توڑتا ہے تاکہ اس سے بات چیت کرے اور اپنی شناخت مسیح کے طور پر ظاہر کرے، اور بیتھانی کی مریم کے اس فیصلے کا دفاع کرے کہ وہ بیٹھ کر اس کی تعلیمات سنے (لوقا ۱۰:۳۹-۴۲)۔
The resurrection is central to Christian theology, symbolizing not just the defeat of death but also the potential for spiritual transformation. This event offers hope and a promise of new life beyond physical death, which is a cornerstone of our belief.
قیامت عیسائیت کے عقیدے کے لئے مرکزی ہے، جو صرف موت کی شکست کی علامت نہیں بلکہ روحانی تبدیلی کی صلاحیت کی بھی علامت ہے۔ یہ واقعہ امید اور جسمانی موت سے آگے نئی زندگی کا وعدہ پیش کرتا ہے، جو ہمارے عقیدے کا ایک بنیادی پتھر ہے۔
Jesus addressing Mary by name and her subsequent recognition of him underscores the personal nature of faith and the individual relationship that we believers can have with the Lord.
یسوع کا مریم کو نام سے مخاطب کرنا اور اس کے بعد اس کی ان کی پہچان عقیدے کی ذاتی نوعیت اور انفرادی تعلق کو اجاگر کرتا ہے جو ہم عقیدت مند خداوند کے ساتھ رکھ سکتے ہیں۔
Our faith in the Lord is deeply personal. Following her encounter, Mary Magdalene goes to the disciples to announce that she has seen the Lord. This act highlights the importance of proclamation in Christianity - the responsibility of believers to share their experiences and the message of Jesus Christ.
ہمارا عقیدہ خداوند میں گہرائی سے ذاتی ہے۔ اس ملاقات کے بعد، مریم مگدلینی شاگردوں کے پاس جاتی ہے تاکہ اعلان کرے کہ اس نے
خداوند کو دیکھا ہے۔ یہ عمل عیسائیت میں اعلان کی اہمیت کو اجاگر کرتا ہے - عقیدت مندوں کی ذمہ داری اپنے تجربات اور یسوع مسیح کے پیغام کو بانٹنے کی۔
The resurrection is God's seal of approval on the work of Christ. If Christ had remained dead, it would imply that His sacrifice was insufficient. However, the resurrection confirms that not only was Jesus' sacrifice sufficient, but it also demonstrates His power over death, affirming His divinity and the truth of His teachings.
قیامت مسیح کے کام پر خدا کی منظوری کی مہر ہے۔ اگر مسیح مردہ رہتے، تو اس کا مطلب ہوتا کہ ان کی قربانی ناکافی تھی۔ تاہم، قیامت اس بات کی تصدیق کرتی ہے کہ نہ صرف یسوع کی قربانی کافی تھی، بلکہ یہ ان کی موت پر قابو پانے کی طاقت کو بھی ظاہر کرتی ہے، ان کی الوہیت اور ان کی تعلیمات کی سچائی کی تصدیق کرتی ہے۔
For us believers, the resurrection is a source of hope and assurance. It promises us new life and a future resurrection, similar to Christ's. The same power that raised Jesus from the dead is at work in us who believe, ensuring our ultimate victory over sin and death.
ہم عقیدت مندوں کے لئے، قیامت امید اور یقین کا ذریعہ ہے۔ یہ ہمیں نئی زندگی اور مستقبل میں قیامت کا وعدہ دیتا ہے، جیسا کہ مسیح کا تھا۔ وہی طاقت جس نے یسوع کو مردوں میں سے اٹھایا تھا ہم میں کام کر رہی ہے جو عقیدہ رکھتے ہیں، ہماری گناہ اور موت پر آخری فتح کو یقینی بناتے ہوئے۔
How wonderful will our encounter be, when we see the first time the Lord, in which we put our hope for our entire life? Hallelujah.
ہماری ملاقات کتنی شاندار ہوگی، جب ہم پہلی بار خداوند کو دیکھیں گے، جس پر ہم نے اپنی پوری زندگی کی امید رکھی ہوتی ہے؟ ہلیلویاہ۔

The passage John 20:1-18 describes a deeply moving and significant moment in the Christian narrative. Mary Magdalene, one of Jesus' most devoted followers, visits Jesus' tomb early in the morning and finds it empty.
Distraught, she encounters Jesus but does not recognize him at first. When she does, she exclaims "Rabbouni!" (which means "Teacher" in Aramaic).
This passage captures in my view the most emotional moment in the entire Bible. This scene begins in the early morning. Mary Magdalene's visit to the tomb, driven by her devotion and love for Jesus, sets the stage for this emotional encounter.
The emptiness of the tomb initially signifies loss and despair for Mary. In her grief, the empty tomb represents the finality of death and the apparent end of the journey she shared with Jesus.
This moment of despair reflects a universal human experience of grief, where the absence of a loved one evokes sadness a feel of loss, and emptiness.
However, the narrative quickly shifts from despair to confusion and then to profound revelation. Mary's encounter with Jesus, whom she initially fails to recognize, speaks to the mysterious and transformative nature of the resurrection.
It is only through the calling of her name that the familiar relationship is re-established, highlighting her recognition of the voice.
When Mary recognizes Jesus and exclaims "Rabbouni," the emotional intensity of the scene reaches its apex. This exclamation is not merely a recognition but an expression of profound joy, relief, and reconnection.
The use of "Rabbouni" underscores a personal and intimate relationship with Jesus, reflecting both respect and deep affection.
This encounter between Mary Magdalene and the resurrected Jesus symbolizes the passage from grief and loss to hope and renewal.
Mary Magdalene's role in this narrative is significant. As a woman and a devoted follower, her witness to the resurrection and her commission to share the news with the disciples challenge contemporary cultural norms and elevate her role within the Christian tradition.
This moment marks the beginning of her transformation from mourner to apostle to the apostles, underscoring the theme of empowerment and the spread of the new faith.
In this narrative, the personal encounter with the divine, the transformation of understanding, and the commissioning to share the good news encapsulates the essence of the Christian message.
The emotional depth of this passage lies not only in the joy of the resurrection but in the profound changes it heralds for Mary Magdalene, the disciples, and ultimately, for us all.
Mary Magdalene was dramatically healed by Jesus and followed him closely thereafter, making her recognition of him deeply personal and emotionally charged. The resurrection was a pivotal event.
For Mary and other followers of Jesus, the crucifixion was a moment of despair and perceived defeat. Seeing Jesus alive again turned that despair into hope and joy, validating his teachings and promises.
Mary Magdalene's prominent role in this event emphasizes the significant place of women in the early Christian community. Despite societal norms of the time, this narrative places a woman at the center of one of Christianity's most pivotal moments, illustrating the inclusive nature of Jesus' ministry.
In a period where women were often regarded as second-class citizens and their testimonies were given little credence, the inclusion and elevation of women in key biblical narratives is significant.
Throughout the Gospels, Jesus consistently breaks societal norms by valuing and respecting women. He interacts with women publicly, teaches them alongside men, and includes them in his ministry.
This was revolutionary in a context where women were often relegated to the background and excluded from religious and scholarly discussions. In several pivotal biblical events, women are central figures.
The resurrection narrative, where women (specifically Mary Magdalene) are the first to witness the empty tomb and the risen Christ, is a prime example. This is a powerful statement, especially considering that women's testimonies were often disregarded in legal and societal contexts at the time.
Jesus' interactions with women often served to empower them. Examples include his conversation with the Samaritan woman at the well (John 4:1-26)
On that occasion, Jesus breaks cultural taboos to engage with her and reveal his identity as the Messiah, and his defense of Mary of Bethany's choice to sit and listen to his teachings (Luke 10:39-42).
The resurrection is central to Christian theology, symbolizing not just the defeat of death but also the potential for spiritual transformation. This event offers hope and a promise of new life beyond physical death, which is a cornerstone of our belief.
Jesus addressing Mary by name and her subsequent recognition of him underscores the personal nature of faith and the individual relationship that we believers can have with the Lord.
Our faith in the Lord is deeply personal. Following her encounter, Mary Magdalene goes to the disciples to announce that she has seen the Lord. This act highlights the importance of proclamation in Christianity - the responsibility of believers to share their experiences and the message of Jesus Christ.
The resurrection is God's seal of approval on the work of Christ. If Christ had remained dead, it would imply that His sacrifice was insufficient. However, the resurrection confirms that not only was Jesus' sacrifice sufficient, but it also demonstrates His power over death, affirming His divinity and the truth of His teachings.
For us believers, the resurrection is a source of hope and assurance. It promises us new life and a future resurrection, similar to Christ's. The same power that raised Jesus from the dead is at work in us who believe, in ensuring our ultimate victory over sin and death.
How wonderful will our encounter be, when we see the first time the Lord, in which we put our hope for our entire life? Hallelujah.
==========================================================================================================================================
The passage John 20:1-18 describes a deeply moving and significant moment in the Christian narrative. Mary Magdalene, one of Jesus' most devoted followers, visits Jesus' tomb early in the morning and finds it empty.
یوحنا ۲۰:۱-۱۸ کی آیتیں ایک گہری اور اہم لمحے کو بیان کرتی ہیں جو عیسائیت کی کہانی میں بہت اہم ہے۔ مریم مگدلینی، جو یسوع کی سب سے مخلص پیروکاروں میں سے ایک تھی، صبح سویرے یسوع کی قبر پر جاتی ہے اور اسے خالی پاتی ہے۔
Distraught, she encounters Jesus but does not recognize him at first. When she does, she exclaims "Rabbouni!" (which means "Teacher" in Aramaic).
پریشان حال، وہ یسوع سے ملاقات کرتی ہے لیکن شروع میں اسے پہچان نہیں پاتی۔ جب وہ پہچانتی ہے، تو وہ "ربونی!" کہتی ہے (جس کا معنی ارامی زبان میں "استاد" ہے)۔
This passage captures in my view the most emotional moment in the entire Bible. This scene begins in the early morning. Mary Magdalene's visit to the tomb, driven by her devotion and love for Jesus, sets the stage for this emotional encounter.
میرے خیال میں یہ آیت پوری بائبل کے سب سے زیادہ جذباتی لمحے کو بیان کرتی ہے۔ یہ منظر صبح سویرے شروع ہوتا ہے۔ مریم مگدلینی کا قبر پر جانا، جو کہ اس کی یسوع کے لیے محبت اور وفاداری سے محرک تھا، اس جذباتی ملاقات کے لیے منظر مقرر کرتا ہے۔
The emptiness of the tomb initially signifies loss and despair for Mary. In her grief, the empty tomb represents the finality of death and the apparent end of the journey she shared with Jesus.
قبر کی خالی ہونے کا ابتدائی معنی مریم کے لئے نقصان اور مایوسی ہے۔ اس کے غم میں، خالی قبر موت کی حتمیت اور اس سفر کے ظاہری اختتام کی نمائندگی کرتی ہے جو اس نے یسوع کے ساتھ بانٹی تھی۔
This moment of despair reflects a universal human experience of grief, where the absence of a loved one evokes sadness a feel of loss, and emptiness.
یہ مایوسی کا لمحہ انسانی غم کے ایک عالمی تجربے کی عکاسی کرتا ہے، جہاں کسی پیارے کی عدم موجودگی اداسی، نقصان کا احساس، اور خالی پن کو جنم دیتی ہے۔
However, the narrative quickly
shifts from despair to confusion and then to profound revelation. Mary's encounter with Jesus, whom she initially fails to recognize, speaks to the mysterious and transformative nature of the resurrection.
تاہم، کہانی جلد ہی مایوسی سے الجھن اور پھر گہرے انکشاف کی طرف منتقل ہوتی ہے۔ مریم کی یسوع سے ملاقات، جسے وہ شروع میں پہچان نہیں پاتی، قیامت کی پراسرار اور تبدیل کر دینے والی فطرت کو بیان کرتی ہے۔
It is only through the calling of her name that the familiar relationship is re-established, highlighting her recognition of the voice.
یہ صرف اس کے نام کے پکارے جانے کے ذریعے ہی ہے کہ واقفیت کا رشتہ دوبارہ قائم ہوتا ہے، جس سے اس کی آواز کی پہچان پر روشنی ڈالی جاتی ہے۔
When Mary recognizes Jesus and exclaims "Rabbouni," the emotional intensity of the scene reaches its apex. This exclamation is not merely a recognition but an expression of profound joy, relief, and reconnection.
جب مریم یسوع کو پہچانتی ہے اور "ربونی" کہتی ہے، تو منظر کی جذباتی شدت اپنے عروج پر پہنچ جاتی ہے۔ یہ اعلان صرف پہچان نہیں بلکہ گہری خوشی، راحت، اور دوبارہ جڑنے کا اظہار ہے۔
The use of "Rabbouni" underscores a personal and intimate relationship with Jesus, reflecting both respect and deep affection.
"ربونی" کے استعمال سے یسوع کے ساتھ ایک ذاتی اور قریبی تعلق کو زور دیا گیا ہے، جو کہ عزت اور گہری محبت دونوں کی عکاسی کرتا ہے۔
This encounter between Mary Magdalene and the resurrected Jesus symbolizes the passage from grief and loss to hope and renewal.
مریم مگدلینی اور قیامت پذیر یسوع کے درمیان یہ ملاقات غم اور نقصان سے امید اور تجدید کے سفر کی علامت ہے۔
Mary Magdalene's role in this narrative is significant. As a woman and a devoted follower, her witness to the resurrection and her commission to share the news with the disciples challenge contemporary cultural norms and elevate her role within the Christian tradition.
مریم مگدلینی کا کردار اس کہانی میں اہم ہے۔ ایک عورت اور ایک مخلص پیروکار کے طور پر، اس کی قیامت کی گواہی اور شاگردوں کے ساتھ خبریں بانٹنے کی ذمہ داری موجودہ ثقافتی اصولوں کو چیلنج کرتی ہے اور عیسائی روایت میں اس ک
ے کردار کو بلند کرتی ہے۔
This moment marks the beginning of her transformation from mourner to apostle to the apostles, underscoring the theme of empowerment and the spread of the new faith.
یہ لمحہ اس کے تبدیلی کی شروعات کو نشان زد کرتا ہے، غمزدہ سے رسول تک، اور رسولوں کے رسول تک، اختیار کی تھیم اور نئے عقیدے کے پھیلاؤ کو اجاگر کرتا ہے۔
In this narrative, the personal encounter with the divine, the transformation of understanding, and the commissioning to share the good news encapsulates the essence of the Christian message.
اس کہانی میں، الہی کے ساتھ ذاتی ملاقات، سمجھ میں تبدیلی، اور اچھی خبریں بانٹنے کی ذمہ داری عیسائی پیغام کے جوہر کو سموتی ہے۔
The emotional depth of this passage lies not only in the joy of the resurrection but in the profound changes it heralds for Mary Magdalene, the disciples, and ultimately, for us all.
اس آیت کی جذباتی گہرائی صرف قیامت کی خوشی میں نہیں ہے بلکہ مریم مگدلینی، شاگردوں، اور آخر کار، ہم سب کے لئے اس کے اعلان کردہ گہرے تبدیلیوں میں ہے۔
Mary Magdalene was dramatically healed by Jesus and followed him closely thereafter, making her recognition of him deeply personal and emotionally charged.
مریم مگدلینی کو یسوع نے نمایاں طور پر شفا دی اور اس کے بعد وہ اس کے قریب سے پیروی کرتی رہی، جس سے اس کی اس کی پہچان گہری ذاتی اور جذباتی طور پر چارج ہو گئی۔
The resurrection was a pivotal event. For Mary and other followers of Jesus, the crucifixion was a moment of despair and perceived defeat. Seeing Jesus alive again turned that despair into hope and joy, validating his teachings and promises.
قیامت ایک اہم واقعہ تھا۔ مریم اور یسوع کے دیگر پیروکاروں کے لئے، صلیب کی سزا مایوسی اور شکست کا لمحہ تھا۔ یسوع کو دوبارہ زندہ دیکھ کر وہ مایوسی امید اور خوشی میں بدل گئی، اس کی تعلیمات اور وعدوں کی تصدیق کرتے ہوئے۔
Mary Magdalene's prominent role in this event emphasizes the significant place of women in the early Christian community. Despite societal norms of the time, this narrative places a woman at the center of one of Christianity's most pivotal moments, illustrating the inclusive nature of Jesus' ministry.
مریم مگدلینی کا اس واقعہ میں نمایاں کردار عیسائی برادری میں عورتوں کے اہم مقام کو زور
دیتا ہے۔ اس وقت کے معاشرتی اصولوں کے باوجود، یہ کہانی عیسائیت کے سب سے اہم لمحوں میں سے ایک میں ایک عورت کو مرکزی کردار میں رکھتی ہے، جس سے یسوع کی وزارت کی شاملیت کی نوعیت کو واضح کیا گیا ہے۔
In a period where women were often regarded as second-class citizens and their testimonies were given little credence, the inclusion and elevation of women in key biblical narratives is significant.
ایک ایسے دور میں جب عورتوں کو اکثر دوسرے درجے کے شہریوں کے طور پر دیکھا جاتا تھا اور ان کی گواہی کو زیادہ اہمیت نہیں دی جاتی تھی، بائبل کی اہم کہانیوں میں عورتوں کی شمولیت اور ان کی بلندی اہم ہے۔
Throughout the Gospels, Jesus consistently breaks societal norms by valuing and respecting women. He interacts with women publicly, teaches them alongside men, and includes them in his ministry.
انجیلوں کے دوران، یسوع مستقل طور پر عورتوں کو قدر دے کر اور ان کا احترام کر کے معاشرتی اصولوں کو توڑتا ہے۔ وہ عورتوں سے عوامی طور پر بات چیت کرتا ہے، انہیں مردوں کے ساتھ ساتھ تعلیم دیتا ہے، اور انہیں اپنی وزارت میں شامل کرتا ہے۔
This was revolutionary in a context where women were often relegated to the background and excluded from religious and scholarly discussions.
یہ ایک ایسے تناظر میں انقلابی تھا جہاں عورتوں کو اکثر پس منظر میں رکھا جاتا تھا اور انہیں مذہبی اور علمی بحثوں سے خارج کر دیا جاتا تھا۔
In several pivotal biblical events, women are central figures. The resurrection narrative, where women (specifically Mary Magdalene) are the first to witness the empty tomb and the risen Christ, is a prime example.
کئی اہم بائبل کے واقعات میں، عورتیں مرکزی کردار ہیں۔ قیامت کی کہانی، جہاں عورتیں (خاص طور پر مریم مگدلینی) خالی قبر اور قیامت پذیر مسیح کی پہلی گواہ ہیں، ایک بہترین مثال ہے۔
This is a powerful statement, especially considering that women's testimonies were often disregarded in legal and societal contexts at the time.
یہ ایک طاقتور بیان ہے، خاص طور پر اس وقت کے قانونی اور معاشرتی
تناظر میں عورتوں کی گواہی کو اکثر نظر انداز کیا جاتا تھا۔
Jesus' interactions with women often served to empower them. Examples include his conversation with the Samaritan woman at the well (John 4:1-26)
یسوع کی عورتوں کے ساتھ بات چیت اکثر انہیں بااختیار بنانے کے لئے ہوتی تھی۔ مثالوں میں اس کی سامری عورت کے ساتھ کنویں پر گفتگو شامل ہے (یوحنا ۴:۱-۲۶)
On that occasion, Jesus breaks cultural taboos to engage with her and reveal his identity as the Messiah, and his defense of Mary of Bethany's choice to sit and listen to his teachings (Luke 10:39-42).
اس موقع پر، یسوع ثقافتی ممنوعات کو توڑتا ہے تاکہ اس سے بات چیت کرے اور اپنی شناخت مسیح کے طور پر ظاہر کرے، اور بیتھانی کی مریم کے اس فیصلے کا دفاع کرے کہ وہ بیٹھ کر اس کی تعلیمات سنے (لوقا ۱۰:۳۹-۴۲)۔
The resurrection is central to Christian theology, symbolizing not just the defeat of death but also the potential for spiritual transformation. This event offers hope and a promise of new life beyond physical death, which is a cornerstone of our belief.
قیامت عیسائیت کے عقیدے کے لئے مرکزی ہے، جو صرف موت کی شکست کی علامت نہیں بلکہ روحانی تبدیلی کی صلاحیت کی بھی علامت ہے۔ یہ واقعہ امید اور جسمانی موت سے آگے نئی زندگی کا وعدہ پیش کرتا ہے، جو ہمارے عقیدے کا ایک بنیادی پتھر ہے۔
Jesus addressing Mary by name and her subsequent recognition of him underscores the personal nature of faith and the individual relationship that we believers can have with the Lord.
یسوع کا مریم کو نام سے مخاطب کرنا اور اس کے بعد اس کی ان کی پہچان عقیدے کی ذاتی نوعیت اور انفرادی تعلق کو اجاگر کرتا ہے جو ہم عقیدت مند خداوند کے ساتھ رکھ سکتے ہیں۔
Our faith in the Lord is deeply personal. Following her encounter, Mary Magdalene goes to the disciples to announce that she has seen the Lord. This act highlights the importance of proclamation in Christianity - the responsibility of believers to share their experiences and the message of Jesus Christ.
ہمارا عقیدہ خداوند میں گہرائی سے ذاتی ہے۔ اس ملاقات کے بعد، مریم مگدلینی شاگردوں کے پاس جاتی ہے تاکہ اعلان کرے کہ اس نے
خداوند کو دیکھا ہے۔ یہ عمل عیسائیت میں اعلان کی اہمیت کو اجاگر کرتا ہے - عقیدت مندوں کی ذمہ داری اپنے تجربات اور یسوع مسیح کے پیغام کو بانٹنے کی۔
The resurrection is God's seal of approval on the work of Christ. If Christ had remained dead, it would imply that His sacrifice was insufficient. However, the resurrection confirms that not only was Jesus' sacrifice sufficient, but it also demonstrates His power over death, affirming His divinity and the truth of His teachings.
قیامت مسیح کے کام پر خدا کی منظوری کی مہر ہے۔ اگر مسیح مردہ رہتے، تو اس کا مطلب ہوتا کہ ان کی قربانی ناکافی تھی۔ تاہم، قیامت اس بات کی تصدیق کرتی ہے کہ نہ صرف یسوع کی قربانی کافی تھی، بلکہ یہ ان کی موت پر قابو پانے کی طاقت کو بھی ظاہر کرتی ہے، ان کی الوہیت اور ان کی تعلیمات کی سچائی کی تصدیق کرتی ہے۔
For us believers, the resurrection is a source of hope and assurance. It promises us new life and a future resurrection, similar to Christ's. The same power that raised Jesus from the dead is at work in us who believe, ensuring our ultimate victory over sin and death.
ہم عقیدت مندوں کے لئے، قیامت امید اور یقین کا ذریعہ ہے۔ یہ ہمیں نئی زندگی اور مستقبل میں قیامت کا وعدہ دیتا ہے، جیسا کہ مسیح کا تھا۔ وہی طاقت جس نے یسوع کو مردوں میں سے اٹھایا تھا ہم میں کام کر رہی ہے جو عقیدہ رکھتے ہیں، ہماری گناہ اور موت پر آخری فتح کو یقینی بناتے ہوئے۔
How wonderful will our encounter be, when we see the first time the Lord, in which we put our hope for our entire life? Hallelujah.
ہماری ملاقات کتنی شاندار ہوگی، جب ہم پہلی بار خداوند کو دیکھیں گے، جس پر ہم نے اپنی پوری زندگی کی امید رکھی ہوتی ہے؟ ہلیلویاہ۔

Last edited by Otangelo on Fri Feb 16, 2024 6:06 pm; edited 1 time in total
ElShamah - Reason & Science: Defending ID and the Christian Worldview » Various issues » My articles
Similar topics
Permissions in this forum:
You cannot reply to topics in this forum