What i most routinely see, is oponents of my posts arguing that intelligent design is a argument from ignorance. I disagree.
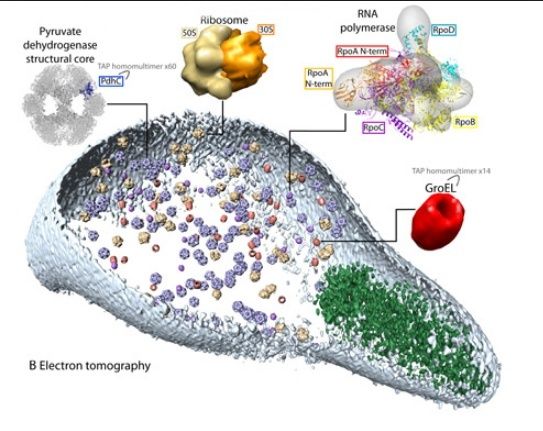
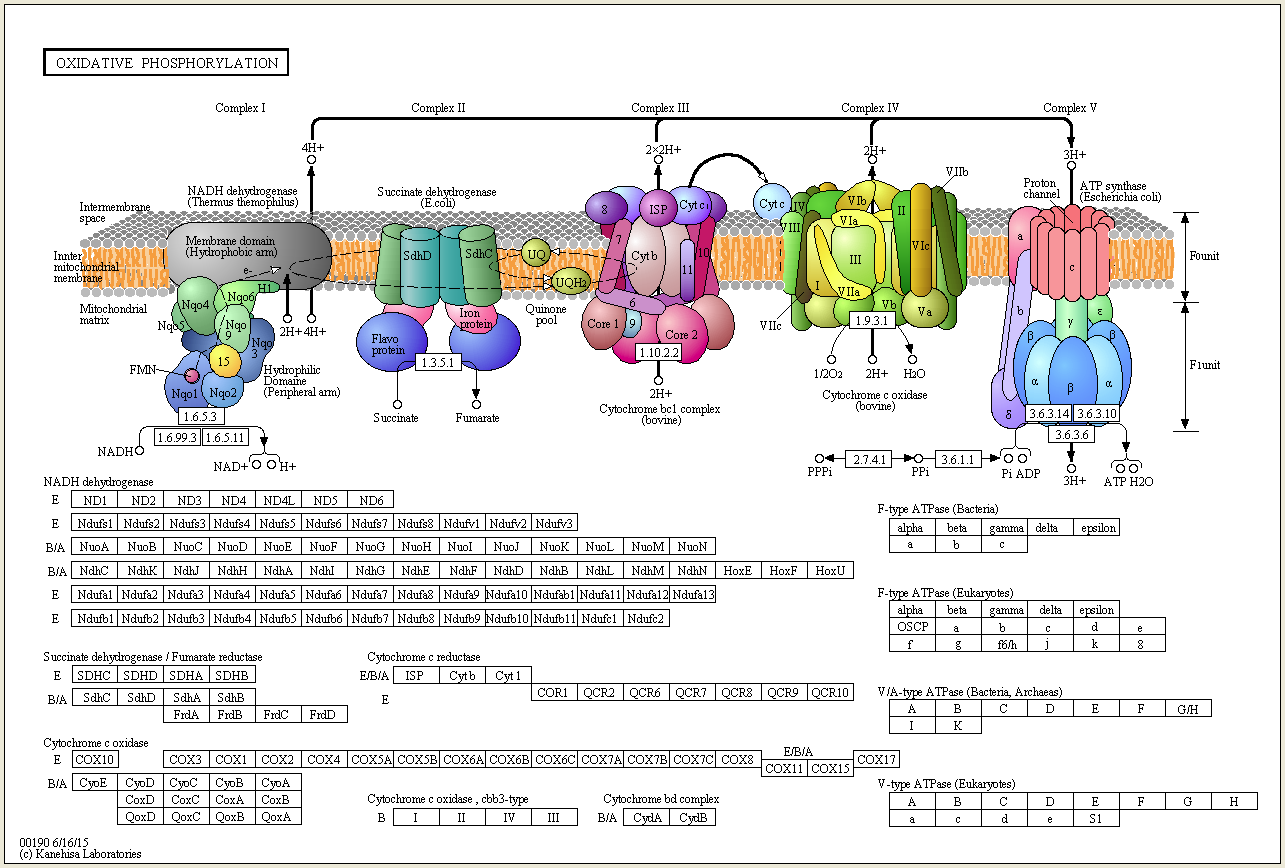
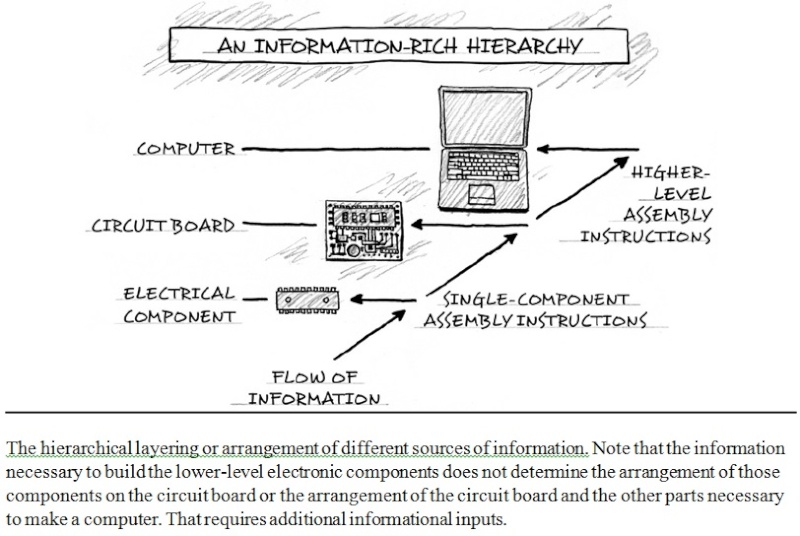
There are many parts, subunits of enzymes and proteins, co-factos etc. that have no apparent multiple functions and could not be co-opted or be available through horizontal gene transfer, or whatever. One example are the last 8 enzymes used in the biosynthesis pathway of chlorophyll http://reasonandscience.heavenforum.org/t1546-chlorophyll-biosynthesis-pathway for what reason would these enzymes emerge naturally , if by their own , and not duly embedded in the whole biosynthesis process, they have no use at all ? and even lets say chlorophyll : why would nature invent such extremely complex pathways to produce such a complex molecule, if, even if ready to go, it is not embedded in the whole process of photosynthesis ? and how could they be embedded in the system, if the light harvesting complex were not present ? please explain. Furthermore, i wonder how nature came up with the information to make the individual parts, the subunits, and providing the right assembly instructions for the individual parts, and the whole thing. As its known, body plans and 3d cell shape does not depend only on genetic information, but also epigenetic information and other unknown factors.
Darwins doubt, pg.268
What natural selection lacks, intelligent design—purposive, goal-directed selection—provides. Rational agents can arrange both matter and symbols with distant goals in mind. In using language, the human mind routinely "finds" or generates highly improbable linguistic sequences to convey an intended or preconceived idea. In the process of thought, functional objectives precede and constrain the selection of words, sounds, and symbols to generate functional (and meaningful) sequences from a vast ensemble of meaningless alternative possible combinations of sound or symbol. Similarly, the construction of complex technological objects and products, such as bridges, circuit boards, engines, and software, results from the application of goal-directed constraints. Indeed, in all functionally integrated complex systems where the cause is known by experience or observation, designing engineers or other intelligent agents applied constraints on the possible arrangements of matter to limit possibilities in order to produce improbable forms, sequences, or structures. Rational agents have repeatedly demonstrated the capacity to constrain possible outcomes to actualize improbable but initially unrealized future functions. Repeated experience affirms that intelligent agents (minds) uniquely possess such causal powers. Analysis of the problem of the origin of biological information, therefore, exposes a deficiency in the causal powers of natural selection and other undirected evolutionary mechanisms that corresponds precisely to powers that agents are uniquely known to possess. Intelligent agents have foresight. Such agents can determine or select functional goals before they are physically instantiated. They can devise or select material means to accomplish those ends from among an array of possibilities. They can then actualize those goals in accord with a preconceived design plan or set of functional requirements. Rational agents can constrain combinatorial space with distant information-rich outcomes in mind. The causal powers that natural selection lacks—by definition—are associated with the attributes of consciousness and rationality—with purposive intelligence. Thus, by invoking intelligent design to overcome a vast combinatorial search problem and to explain the origin of new specified information, contemporary advocates of intelligent design are not positing an arbitrary explanatory element unmotivated by a consideration of the evidence.
Irreducible complexity is not based on a negative, namely that there is no evidence for a naturalistic pathway. Rather than that, it makes a positive claim, which can be falsified, upon : (a) gene knockout, (b) reverse engineering, (c) examining homologous systems, and (d) sequencing the genome of the biochemical structure. ( Dennis Jones ) Gene knockout has been done several times, providing evidence that the organism was unable to replace given gene or protein by natural means. 1 The absence of evidence that evolution is not capable to replace given part is empirical evidence, that falsifies the claim of the ToE. Its therefore not justified to claim the inference is a argument of ignorance. Quit the contrary is the case. As for example, if i ask you : can you change a us$100 bill ? and you answer: sorry, i have no smaller bills. You open your wallet, and and its confirmed, no change in your wallet, then you have proven that you have indeed no smaller bills. You have proven a negative, which is not a argument of ignorance, since you checked and got a empirical proof.
If proponents of intelligent design were arguing in the preceding manner, they would be guilty of arguing from ignorance. But the argument takes the following form:
Premise One: Despite a thorough search, no material causes have been discovered that demonstrate the power to produce large amounts of specified information, irreducible and interdependent biological systems.
Premise Two: Intelligent causes have demonstrated the power to produce large amounts of specified information, irreducible and interdependent systems of all sorts.
Conclusion: Intelligent design constitutes the best, most causally adequate, explanation for the information and irreducible complexity in the cell, and interdependence of proteins, organelles, and bodyparts, and even of animals and plants, aka moths and flowers, for example.
http://reasonandscience.heavenforum.org/t2099-irreducible-complexity-is-not-a-argument-from-ignorance
Last edited by Admin on Wed Mar 08, 2017 2:01 am; edited 1 time in total