HAROLD B. WHITE, III (1975): Coenzymes are complex organic molecules that are essential for many enzyme-catalyzed reactions. At least 52% of the nearly 1750 enzymes recently cataloged (IUPAC-IUB, 1972) require a coenzyme for activity. I propose that coenzymes are the surviving vestiges of nucleic acid enzymes which preceded the evolution of ribosomal protein synthesis. 34
If the RNA world were true, ribozymes would have had to catalyze a wide range of catalytic reactions which subsequently were substituted by proteins. About half would only obtain the necessary catalytic activity by recruiting and employing cofactors and coenzymes ( which, as we see, also depend on complex biosynthesis pathways, and alternative non-enzymatic emergence would be very unlikely). It is as if a Software engineer had to learn to become a mechanical engineer and assemble complex machines. There is no evidence that RNAs, made of just four different building blocks ( the four nucleobases), were ever able to catalyze the widely different enzymatic and metabolic reactions required for life to thrive. There is no evidence, that that somehow, prebiotic shuffling created a pool of millions of repetitive complex nucleotides, all with the repetitive configuration of purine and pyrimidine bases. There is no chemical logic that makes it appear plausible, that a gradual chemical evolutionary process would have promoted the emergence of an RNA world, followed by an RNA-peptide world. There is a wide unexplained gap between the RNA world, and the modern DNA - RNA - protein state of affairs in modern cells. Can this gap be bridged with new hypotheses and the advance in abiogenesis investigations? That prediction looks rather unlikely. Only time will tell.
Solving a chicken & egg problem?
Supposedly, the RNA world hypothesis solved a long-standing chicken & egg, or catch22 problem ( J.Wells [2006]: In Joseph Heller’s novel about World War II, Catch-22, an aviator could be excused from combat duty for being crazy. But a rule specified that he first had to request an excuse, and anyone who requested an excuse from combat duty was obviously not crazy, so such requests were invariably denied. The rule that made it impossible to be excused from combat duty was called “Catch-22.” ) In 1965, Sidney Fox wrote a scientific article, asking: How, when no life existed, did substances come into being which today are absolutely essential to living systems, yet which can only be formed by those systems? He was referring to a problem, outlined by Jordana Cepelewicz (2017): For scientists studying the origin of life, one of the greatest chicken-or-the-egg questions is: Which came first — proteins or nucleic acids like DNA and RNA? 33 This problem arises because DNA and RNA direct the synthesis of enzymes and proteins. But proteins synthesize the making of RNA and DNA.
Jessica C. Bowman (2015) claimed: The RNA World Hypothesis resolves the putative chicken and egg dilemma: which came first, polynucleotide or polypeptide? The simultaneous emergence from whole cloth of two functional biopolymers, one encoding the other, seems improbable. A single type of ancestral biopolymer (polynucleotide), performing multiple roles, appears to be characterized by high parsimony. A ‘‘Polymer Transition’’, a progression of biology from one polymer type (polynucleotide) to two polymer types (polynucleotide and polypeptide), is consistent with an expectation that ancient biology transitioned from simple to complex. 32
Under naturalism, the only possible explanation for the complexity seen in biochemistry is gradualism: From the simple to the complex. Gradually moving from chemistry to biology. A sudden appearance of all the complex interdependent intricacies observed in the living cannot be explained by a biochemical Big bang. It is untenable. Therefore, under the framework of philosophical naturalism, answers have to be found that confer with the gradualistic scenario. Only the model of intelligent design permits the hypothesis of instant creation by an intelligent agency. Giving up gradualism means giving up naturalism.
Eugene V Koonin (2007): The origin of the translation system is, arguably, the central and the hardest problem in the study of the origin of life, and one of the hardest in all evolutionary biology. The problem has a clear catch-22 aspect: high translation fidelity hardly can be achieved without a complex, highly evolved set of RNAs and proteins but an elaborate protein machinery could not evolve without an accurate translation system. 13
Paul C. W. Davies (2013): Because of the organizational structure of systems capable of processing algorithmic (instructional) information, it is not at all clear that a monomolecular system, where a single polymer plays the role of catalyst and informational carrier, is even logically consistent with the organization of information flow in living systems because there is no possibility of separating information storage from information processing (that being such a distinctive feature of modern life). As such, digital-first systems (as currently posed) represent a rather trivial form of information processing that fails to capture the logical structure of life as we know it. The real challenge of life's origin is thus to explain how instructional information control systems emerge naturally and spontaneously from mere molecular dynamics. 15
Self-replication in the RNA world
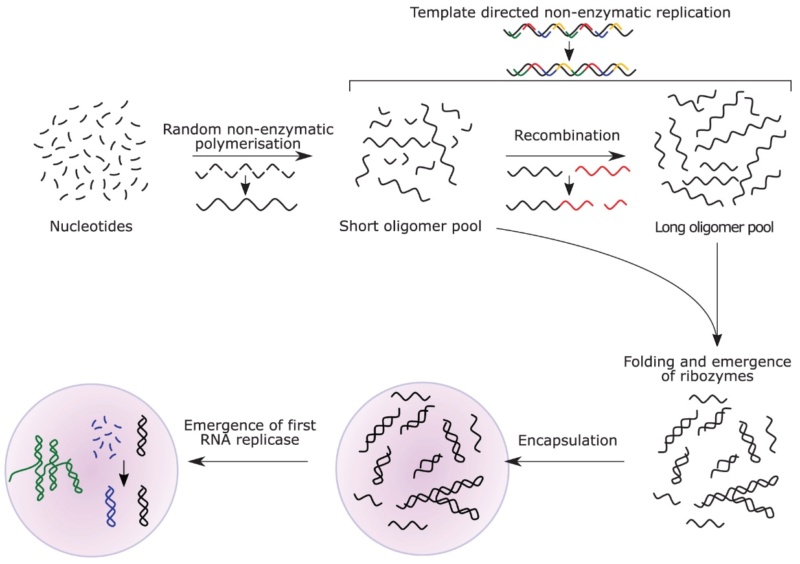
Despite the current celebrity status, there are several reasons that raise doubts that RNAs would self-assemble into ribozyme polymerases with function-bearing sequences, obtaining the right chemical structures with autocatalytic properties, and being apt to start self-replication.
Harold S Bernhardt (2012): The following objections have been raised to the RNA world hypothesis: (i) RNA is too complex a molecule to have arisen prebiotically; (ii) RNA is inherently unstable; (iii) catalysis is a relatively rare property of long RNA sequences only; and (iv) the catalytic repertoire of RNA is too limited. 12
Since the various problems of prebiotic RNA monomers have been outlined in the previous chapter, we will address now the issues with RNA self-replication. Steven Benner (2013): Catalysis and genetics place contradicting demands on any single molecular system asked to do both. For example, catalytic molecules should fold, to surround a transition state. Genetic molecules should not fold, to allow them to template the synthesis of their complements. Catalytic molecules should have many building blocks, to create versatile catalytic potential. Genetic molecules should have few building blocks, to ensure that they are copied with high fidelity 11
Jack W Szostak (2012): The first RNA World models were based on the concept of an RNA replicase - a ribozyme that was a good enough RNA polymerase that it could catalyze its own replication. Although several RNA polymerase ribozymes have been evolved in vitro, the creation of a true replicase remains a great experimental challenge. 6
Hannes Mutschler (2019): It has not yet been possible to demonstrate robust and continuous RNA self-replication from a realistic feedstock (i.e. activated mono- or short mixed-sequence oligonucleotides). In the case of ribozymes, only ‘simple’ ligation or recombination-based RNA replication from defined oligonucleotides has been demonstrated. Such systems have only a limited ability to transmit heritable information and so are not capable of open-ended evolution — the ability to indefinitely increase in complexity like living systems. Open-ended evolution requires that a replicase must at least be able to efficiently copy generic sequences longer than that required to encode its own function. RNA in isolation (including ribozymes) is simply not sufficient to catalyze its own replication, and substantial help from either other molecules or the environment is essential. 7
The annealing problem
Jordana Cepelewicz (2019): As a first step toward making a copy of itself, a single strand of RNA can take up complementary nucleotide building blocks from its surroundings and stitch them together. But the paired RNA strands then tend to bind to each other so tightly that they don’t unwind without help, which prevents them from acting as either catalysts or templates for further RNA strands. “It’s a real challenge,” Sutherland said. “It’s held the field back for a long time.” 8
Gerald F. Joyce (2018): Because the product of template copying is a double-stranded RNA, there must be some means of either strand separation or strand displacement synthesis. Transient temperature fluctuations could lead to thermal strand separation, but long RNA duplexes (≥30 base pairs) are difficult to denature thermally. 10
In modern cells, ribonuclease H enzymes destroy annealed RNAs but evidently, they were not around on prebiotic earth.
The Eigen paradox
Jaroslaw Synak (2022): Another challenge is the maintenance of genetic information in RNA sequences over many rounds of imperfect replication. In order to survive, RNA polymerase must be copied faster than it is hydrolyzed and accurately enough to preserve its function. In the early stages of molecular evolution, due to the lack of reliable replication mechanisms, the mutation rate was likely very high and the critical amount of information could not have been stored in long RNA sequences; on the other hand, the short ones could not be efficient enzymes. Maynard Smith estimated that the maximum length of the RNA replicase is approximately 100 nucleotides, assuming nonenzymatic replication with a copying fidelity of one base of up to 0.99. In order to further increase this length, the copying fidelity would have to be increased, which requires the presence of specific enzymes. This is known as Eigen’s paradox and is often equivalently formulated as: no enzymes without a large genome and no large 5
Indeed. But the problem is not only to maintain genetic information but for the first replicator to obtain it in the first place!
Natalia Szostak (2017): Researchers have performed many attempts to create RNA polymerase ribozyme, recently resulting in a cross-chiral RNA polymerase ribozyme and a system of cooperative RNA replicators, as well as RNA polymerase ribozyme that is able to synthesize structured functional RNAs, including aptamers and ribozymes. However, these molecules are too large to be maintained in a quasispecies population, as they exceed the 100 nucleotide error threshold, which is the maximum length polynucleotide molecule that can be accurately replicated without high fidelity polymerases. Eigen suggested hypercycles as a solution to the error threshold problem mentioned above. However, even if the traditional hypercycle model formulation based on ordinary differential equations is ecologically stable, it is proved to be evolutionarily unstable. To evolve life as we know it, separation of the roles performed by replicases, information storage, and replication of the information, into two molecules appears to be one of the crucial events that had to occur early in the stages leading to life. 4
A remarkable admission !!
Sami EL Khatib (2021): Countless challenges are faced by an RNA self-replicating cycle; for it to be a fully chemically and enzymatically free reaction, the cycle loses rate and fidelity, so much that it does not even reach the critical threshold for the sustenance of life, meaning the RNA nucleotides break apart faster than the incorporation of nucleotides takes place, thus is the case when experimenting with modern substrates, that do not leak out of cells and are very polar with the regularly known triphosphate ester, this is advantageous to the modern cell where it uses enzymes to catalyze the release of di-phosphate, but not for the primitive cell as the substrates are found in the environment and require continuous dynamic exchange. The RNA world hypothesis has been criticized mostly because of the belief that long RNA sequences are needed for the catalytic function of RNA. These long sequences are enormous and are needed to isolate the catalytic and biding functions of the overall ribozyme. For example the best ribozyme replicase created so far, which is able to replicate an impressive 95-nucleotide stretch of RNA, is ~190 nucleotides in length, which is by far too large a number to have risen in any random assembly, thus in vitro selection experiments had to be designed where 10,000,000,000,000 – 1,000,000,000,000,000 of randomized RNA molecules are required as the starting point for the isolation of ribozymic and/or binding activity. This experiment clearly contradicts the probable prebiotic situation. 14
Eugene Koonin (2012): The primary incentive behind the theory of self-replicating systems that Manfred Eigen outlined was to develop a simple model explaining the origin of biological information and, hence, of life itself. Eigen’s theory revealed the existence of the fundamental limit on the fidelity of replication (the Eigen threshold): If the product of the error (mutation) rate and the information capacity (genome size) is below the Eigen threshold, there will be stable inheritance and hence evolution; however, if it is above the threshold, the mutational meltdown and extinction become inevitable (Eigen, 1971). The Eigen threshold lies somewhere between 1 and 10 mutations per round of replication; regardless of the exact value, staying above the threshold fidelity is required for sustainable replication and so is a prerequisite for the start of biological evolution. Indeed, the very origin of the first organisms presents at least an appearance of a paradox because a certain minimum level of complexity is required to make self-replication possible at all; high-fidelity replication requires additional functionalities that need even more information to be encoded. However, the replication fidelity at a given point in time limits the amount of information that can be encoded in the genome. What turns this seemingly vicious circle into the (seemingly) unending spiral of increasing complexity—the Darwin-Eigen cycle. The crucial question in the study of the origin of life is how the Darwin-Eigen cycle started—how was the minimum complexity that is required to achieve the minimally acceptable replication fidelity attained? In even the simplest modern systems, such as RNA viruses with the replication fidelity of only about 10^3 and viroids that replicate with the lowest fidelity among the known replicons (about 10^2), replication is catalyzed by complex protein polymerases. The replicase itself is produced by translation of the respective mRNA(s), which is mediated by the immensely complex ribosomal apparatus. Hence, the dramatic paradox of the origin of life is that, to attain the minimum complexity required for a biological system to start on the Darwin-Eigen spiral, a system of a far greater complexity appears to be required. How such a system could evolve is a puzzle that defeats conventional evolutionary thinking, all of which is about biological systems moving along the spiral; the solution is bound to be unusual.
When considering the origin of the first life forms, one faces the proverbial chicken-and-egg problem: What came first, DNA or protein, the gene or the product? In that form, the problem might be outright unsolvable due to the Darwin-Eigen paradox: To replicate and transcribe DNA, functionally active proteins are required, but production of these proteins requires accurate replication, transcription, and translation of nucleic acids. If one sticks to the triad of the Central Dogma, it is impossible to envisage what could be the starting material for the Darwin-Eigen cycle. Even removing DNA from the triad and postulating that the original genetic material consisted of RNA (thus reducing the triad to a dyad), although an important idea, does not help much because the paradox remains. For the evolution toward greater complexity to take off, the system needs to somehow get started on the Darwin-Eigen cycle before establishing the feedback between the (RNA) templates (the information component of the replicator system) and proteins (the executive component). The brilliantly ingenious and perhaps only possible solution was independently proposed by Carl Woese, Francis Crick, and Leslie Orgel in 1967–68: neither the chicken nor the egg, but what is in the middle—RNA alone. The unique property of RNA that makes it a credible—indeed, apparently, the best—candidate for the central role in the primordial replicating system is its ability to combine informational and catalytic functions. Thus, it was extremely tempting to propose that the first replicator systems—the first life forms—consisted solely of RNA molecules that functioned both as information carriers (genomes and genes) and as catalysts of diverse reactions, including, in particular, their own replication and precursor synthesis. This bold speculation has been spectacularly boosted by the discovery and subsequent study of ribozymes (RNA enzymes), which was pioneered by the discovery by Thomas Cech and colleagues in 1982 of the autocatalytic cleavage of the Tetrahymena rRNA intron, and by the demonstration in 1983 by Sydney Altman and colleagues that RNAse P is a ribozyme. Following these seminal discoveries, the study of ribozymes has evolved into a vast, expanding research area.
Despite all invested effort, the in vitro evolved ribozymes remain (relatively) poor catalysts for most reactions; the lack of efficient, processive ribozyme polymerases seems particularly troubling. An estimate based on the functional tolerance of well-characterized ribozymes to mutations suggest that, at fidelity of 10^3 errors per nucleotide per replicase cycle (roughly, the fidelity of the RNA-dependent RNA polymerases of modern viruses), an RNA “organism” with about 100 “genes” the size of a tRNA (80 nucleotides) would be sustainable. Such a level of fidelity would require only an order of magnitude improvement over the most accurate ribozyme polymerases obtained by in vitro selection 3
Lack of prebiotic RNA repair mechanisms
Harris Bernstein (2020): Persistence and replication of even the simplest forms of RNA life must have depended on preserving the information content of the RNA genome from damage (a form of informational noise). Damage to the RNA genome likely occurred in a variety of ways including spontaneous hydrolysis, exposure to UV light and exposure to reactive chemicals. 9
Where did the energy come from?
Jack Szostak, interviewed by Suzan Mazur (2014): The problem is RNA falls apart. The activated nucleotides we use to do the non-enzymatic replication -- they react with water, so they fall apart. There needs to be a way to bring energy back into the system to essentially keep the battery charged. To keep all the nucleotides activated and to keep things running. 1
PHILIP BALL (2020): It’s an alluring picture – catalytic RNAs appear by chance on the early Earth as molecular replicators that gradually evolve into complex molecules capable of encoding proteins, metabolic systems and ultimately DNA. But it’s almost certainly wrong. For even an RNA-based replication process needs energy: it can’t shelve metabolism until later. And although relatively simple self-copying ribozymes have been made, they typically work only if provided with just the right oligonucleotide components to work on. What’s more, sustained cycles of replication and proliferation require special conditions to ensure that RNA templates can be separated from copies made on them. Perhaps the biggest problem is that self-replicating ribozymes are highly complex molecules that seem very unlikely to have randomly polymerized in a prebiotic soup. And the argument that they might have been delivered by molecular evolution merely puts the cart before the horse. The problem is all the harder once you acknowledge what a complex mess of chemicals any plausible prebiotic soup would have been. It’s nigh impossible to see how anything lifelike could come from it without mechanisms for both concentrating and segregating prebiotic molecules – to give RNA-making ribozymes any hope of copying themselves rather than just churning out junk, for example. In short, once you look at it closely, the RNA world raises as many questions as it answers. The best RNA polymerase the researchers obtained this way had a roughly 8% chance of inserting any nucleotide wrongly, and any such error increased the chance that the full chain encoded by the molecule would not be replicated. What’s more, making the original class I ligase was even more error-prone and inefficient – there was a 17% chance of an error on each nucleotide addition, plus a small chance of a spurious extra nucleotide being added at each position. These errors would be critical to the prospects of molecular evolution since there is a threshold error rate above which a replicating molecule loses any Darwinian advantage over the rest of the population – in other words, evolution depends on good enough replication. Fidelity of copying could thus be a problem, hitherto insufficiently recognized, for the appearance of a self-sustaining, evolving RNA-based system: that is, for an RNA world. Maybe this obstacle could have been overcome in time. But my hunch is that any prebiotic molecule will have been too inefficient, inaccurate, dilute and noise-ridden to have cleared the hurdle. 2
An RNA world could not explain the origin of the genetic code
Susan Lindquist (2010): An RNA-only world could not explain the emergence of the genetic code, which nearly all living organisms today use to translate genetic information into proteins. The code takes each of the 64 possible three-nucleotide RNA sequences and maps them to one of the 20 amino acids used to build proteins. 29
Without amino acids, there could not be an assignment. 64 trinucleotide codons are assigned to 20 amino acids. Both had to be present, in order for this assignment to occur. But having both would not be enough either. In reality, the entire system had to be created from the get-go, fully developed, and operational from the beginning. That led Fujio Egami (1981) to present a " working hypothesis on the interdependent genesis of nucleotide bases, protein amino acids, and the primitive genetic code: the primitive genetic code was dependent upon the concentration of different nucleotide bases and amino acids coexisting in the primeval environments and upon the selective affinity between bases and amino acids." 30
Charles W Carter, Jr (2017): Computational and structural modeling argue that some mutual, interdependent processes embedded information into proteins and nucleic acids.
While talking about evolutionary modification, Egami was probably not aware that an interdependent system cannot be the product of evolutionary change. An interdependent system must be right all at once, right from the start. Stepwise, gradual evolution of the system is not possible, since intermediate stages would confer no advantage, nor function. Mapping nucleotides to amino acids is only possible if all players are there. 31
The RNA-peptide world
The RNA-peptide world tries to build a bridge between the replication first, and metabolism first scenarios, advancing the RNA world and combining it with catalytic peptides and primitive metabolism.
Stephen D. Fried (2022): Diverse lines of research in molecular biology, bioinformatics, geochemistry, biophysics, and astrobiology provide clues about the progression and early evolution of proteins, and lend credence to the idea that early peptides served many central prebiotic roles before they were encodable by a polynucleotide template, in a putative ‘peptide-polynucleotide stage’. 23
The presupposition is that a result of chemical prebiotic conditions permitted the emergence of activated ribonucleotides and amino acids. The proposal hypothesizes that RNAs started to interact and get into a relationship with small peptides ( small amino acid strands) right from the beginning, rather than everything starting exclusively with RNAs, that later would transition to mutually beneficial interaction with amino acids. In modern cells, DNA that stores the genetic data using the genetic code is transcribed into messenger RNA (mRNA), subsequently translated in the ribosome apparatus into functional amino acid sequences, which form polypeptides, and in the end, proteins. The core problem is the origin of the codon-amino acid assignment through the genetic code. The RNA-peptide world attempts to address this current state of affairs, starting with an RNA-peptide world, which constitutes the first step to arrive at the end of the current solution, where the sophisticated translation is performed through the ribosome.
Charles W. Carter, Jr. (2015): In the RNA-world scenario, the necessary catalysts were initially entirely RNA-based and did not include genetically encoded proteins. IN the RNA-peptide world, the idea that coded peptides functioned catalytically in the early stages of the origin of life directly contradicts the second central tenet of the “RNA World” scenario. The important distinction between this scenario and the RNA World hypothesis is that the requisite specificity is low in the initial stages of the former but unacceptably high in the latter. Low specificity processes occur with greater frequency and hence are more likely to have occurred first. The unavailability of activated amino acids was the most critical barrier to the emergence of protein synthesis. 16
Dave Speijer (2015): Very small RNAs (versatile and stable due to base-pairing) and amino acids, as well as dipeptides, coevolved. The “RNA world” hypothesis is seen as one of the main contenders for a viable theory on the origin of life. Relatively small RNAs have catalytic power, RNA is everywhere in present-day life, the ribosome is seen as a ribozyme, and rRNA and tRNA are crucial for modern protein synthesis. However, this view is incomplete at best. The modern protein-RNA ribosome most probably is not a distorted form of a “pure RNA ribosome” evolution started out with. Though the oldest center of the ribosome seems “RNA only”, we cannot conclude from this that it ever functioned in an environment without amino acids and/or peptides. Very small RNAs (versatile and stable due to base-pairing) and amino acids, as well as dipeptides, coevolved. Remember, it is the amino group of aminoacylated tRNA that attacks peptidyl-tRNA, destroying the bond between peptide and tRNA. This activity of the amino acid part of aminoacyl-tRNA illustrates the centrality of amino acids in life. With the rise of the “RNA world” view of early life, the pendulum seems to have swung too much towards the ribozymatic part of early biochemistry. The necessary presence and activity of amino acids and peptides is in need of highlighting. We argue that an RNA world completely independent of amino acids never existed.
Indeed, I agree, that an RNA world never existed. But did an RNA-peptide world?
Speijer: The idea of an independent RNA world without oligopeptides or amino acids stabilizing structures and helping in catalysis does not seem a viable concept. On the other hand, the idea of catalytic protein existing without RNA storing the polypeptide sequences, which have catalytic activity, and organizing the production of these sequences, also does not seem a viable concept. Here we argue for a “coevolutionary” theory in which amino acids and (very small) peptides, as well as small RNAs, existed together and where their separate abilities not only reinforced each other’s survival but allowed life to more quickly climbing the ladder of complexity.
Every naturalistic approach works only from the simple to the complex in a slow, gradual manner. Even if not linear but with ups and downs, the outcome is always that there is more functional complexity at the end. That is as well Speijers proposal: "Starting with small molecules (easily) derived from prebiotic chemistry, we will try to reconstruct a possible history in which every stage of increased complexity arises from the previous more simple stage because specific nucleotide/amino acid (RNA/peptide) interactions allowed it do so." Observe how Speijer introduces teleonomy into the explanation when claiming: their separate abilities reinforced each other’s survival . As if RNAs and amino acids operated or behaved with the "aim" or purpose of arriving and keeping a state of affairs, that wasn't there yet. RNAs and amino acids on their own are not alive, and have no aim to enter the trajectory to "help" the arrival of the state of affairs of having something alive, part of the living. Molecules have no innate drive or "urge" to keep a specific state, to get out of thermodynamic equilibrium, that would favor a future outcome, the gradual complexification that would, in the end, result in the existence of self-replicating cells.
Speijer: We now come at a crucial and, we have to admit, somewhat theoretical juncture: coevolution is illustrated by the presumption that RNAs could not persist without peptide protection, that very short (very early) peptides were made more abundant by RNA-producing them, and that they co-evolve forming longer RNAs and peptides. This would constitute an RNA/peptide world of ribozymes and short oligopeptides. These oligopeptides had RNA protection functions (DADVDGD being the obvious ancestor sequence of the universal RNA polymerase active site sequence NADFDGD) This motif (Asn-Ala-Asp-Phe-Asp-Gly-Asp) is a specific stretch of amino acids that is central in all cellular life. RNA polymerases catalyze the transcription from DNA to mRNA. Dennis R. Salahub (2008): Most known RNA polymerases (RNAPs) share a universal heptapeptide, called the NADFDGD motif. The crystal structures of RNAPs indicate that in all cases this motif forms a loop with an embedded triad of aspartic acid residues. This conserved loop is the key part of the active site. 17
The odds to get this sequence randomly is one in 20^7 or one in 10^10, that is taking a pool of 20 selected amino acids used in life would have to be shuffled 10 billion times to get this specified functional sequence. Not forgetting, that it is incorporated in a much longer polymer sequence that also has to be functional, embedded, and working in a joint venture with other polymer subunit strands of RNA polymerase. A far fetch.
Kunnev (2018): The hypothesis assumes that ribonucleotides would polymerize leading to very short RNAs from 2 to about 40 bases. The polymerization would incorporate random sequences and random 3D structures. The process would preserve mostly stable ones. Wet-Dry cycles could facilitate the process of RNA polymerization. Compartmentalization is another important factor since most of the described events are unlikely to occur in very low concentrations. Some level of environmental separation would be expected, for example, micro-chambers out of porous surface of rocks or lipid vesicles or both. Surface adsorption might have facilitated RNA-RNA interactions, RNA-lipids interactions and some beneficial chemical reactions. Thus, clay surfaces have been shown to promote encapsulation of RNA into vesicles and grow by incorporating fatty acid supplied as micelles and can divide without dilution of their contents. At temperatures between 1°C and to denaturation (about 55°C) temperature, short random RNA oligos would get stabilized via intra and intermolecular hybridization based on Watson-Crick base pairing, forming complexes of various 3D shape and size. Larger hybridized regions would confer greater stability and would be selected for. Highly self-complementary RNAs would be unlikely to exist, forcing intermolecular hybridization of short sequences and the emergence of complexes of several RNA oligos. The formation of RNA complexes also assumes a thermal cycle that would drive the process by sequential denaturation (~55–100°C) and re-annealing (<55°C) phases. Frequent repetition of the thermal cycle and stability selection would favor accumulation of complexes with higher degree of complementarity and higher GC content. Non-enzymatic aminoacylation between 2′ or 3′ positions of ribose and activated amino acids could occur. In addition, ribozymes capable of amino acid transfer from one RNA to another have been selected under laboratory conditions and similar molecules could have participated in aminoacylation of RNAs. Aminoacylated RNAs would be involved in complex formation, bringing some of the aminoacylated RNA 3′-ends in close proximity. This would promote peptide bond formation between two adjacent amino acids, most likely with the assistance of wet/dry natural cycles. All amino acids would have statistically equal probability to aminoacylate RNA. At that stage, any RNA molecule could be aminoacylated and could serve as a template. 18
That means, any available amino acid nearby could be involved in the reaction - inclusive amino acids not used in life, and they could be attached anywhere to the RNA molecule. There is also no restriction in regards of possible RNA configurations with any sort of nucleobases. There is no mechanism that would prevent other than the nucleobases used in life to be involved in the reactions. It would result simply in a disordered random accumulation of RNA-peptides.
Kunnev: We presume that following this initial stage all components of the translation system would co-evolve in a stepwise way. Specialization of ribosomal Large Subunit—LSU will start with evolution of peptidyl transferase center (PTC). The evolution of peptides to proteins would occur from small motif to domains and finally— folded proteins.
Felix Müller (2022): The ability to grow peptides on RNA with the help of non-canonical vestige nucleosides offers the possibility of an early co-evolution of covalently connected RNAs and peptides, which then could have dissociated at a higher level of sophistication to create the dualistic nucleic acid–protein world that is the hallmark of all life on Earth. It is difficult to imagine how an RNA world with complex RNA molecules could have emerged without the help of proteins and it is hard to envision how such an RNA world transitions into the modern dualistic RNA and protein world, in which RNA predominantly encodes information whereas proteins are the key catalysts of life.22
This story, when it comes to elucidating the trajectory from these small RNA-peptides, to fully developed proteins is very "sketchy" and superficial. This is a common modus operandi to uphold a story, that by looking closer, does not withstand scrutiny.
Charles Carter, structural biologist (2017): For life to take hold, the mystery polymer would have had to coordinate the rates of chemical reactions that could differ in speed by as much as 20 orders of magnitude. 24
Marcel Filoche (2019): Enzymes speed up biochemical reactions at the core of life by as much as 15 orders of magnitude. Yet, despite considerable advances, the fine dynamical determinants at the microscopic level of their catalytic proficiency are still elusive. Rate-promoting vibrations in the picosecond range, specifically encoded in the 3D protein structure, are localized vibrations optimally coupled to the chemical reaction coordinates at the active site. Remarkably, our theory also exposes a hitherto unknown deep connection between the unique localization fingerprint and a distinct partition of the 3D fold into independent, foldspanning subdomains that govern long-range communication. The universality of these features is demonstrated on a pool of more than 900 enzyme structures, comprising a total of more than 10,000 experimentally annotated catalytic sites. Our theory provides a unified microscopic rationale for the subtle structure-dynamics-function link in proteins. The intricate networks of metabolic cascades that power living organisms ultimately rest on the exquisite ability of enzymes to increase the rate of chemical reactions by many orders of magnitude. Although many molecular machines contain intrinsically disordered domains, the 3D fold is central to enzyme functioning. In particular, increasing evidence is accumulating in the literature in favor of the existence of specific fold-encoded motions believed to govern the relevant collective coordinate(s) that are coupled to the chemical transformation. These motions typically correspond to localized vibrations of the protein scaffold that contribute to the catalytic reaction, i.e., modes that, if impeded, would lead to a deterioration of the catalytic efficiency.
The more the function of a machine depends on its precise setup and arrangement respecting very limited tolerances, the more efforts have to be undertaken to achieve the required precision, demanding engineering solutions where nothing can be left to chance. That is precisely the case with proteins. There is an extraordinarily limited tolerance upon which proteins have to be engineered and designed, a requirement to achieve the necessary catalytic functions. That sets the bar for the cause to instantiate this state of affairs very high, for which random events are entirely inadequate!! The situation becomes even worse when we consider what Mathieu E. Rebeaud described as (2021): the challenge of reaching and maintaining properly folded and functional proteomes. Most proteins must fold to their native structure in order to function, and their folding is largely imprinted in their primary amino acid sequence. However, many proteins, especially large multidomain polypeptides, or certain protein types such as all-beta or repeat proteins, tend to misfold and aggregate into inactive species that may also be toxic. Life met this challenge by evolving employing molecular chaperones that can minimize protein misfolding and aggregation, even under stressful out-of-equilibrium conditions favoring aggregation. 25
Hays S. Rye (2013): Protein folding is a spontaneous process that is essential for life, yet the concentrated and complex interior of a cell is an inherently hostile environment for the efficient folding of many proteins. Some proteins—constrained by sequence, topology, size, and function—simply cannot fold by themselves and are instead prone to misfolding and aggregation. This problem is so deeply entrenched that a specialized family of proteins, known as molecular chaperones assists in protein folding. The bacterial chaperonin GroEL, along with its co-chaperonin GroES, is probably the best-studied example of this family of protein-folding machine. 27
Chaperones do bear no function unless there are misfolded proteins, that need to be re-folded in order to function. But non-functional proteins accumulating in the cell would be toxic waste and eventually kill the cell. So this creates another chicken & egg problem. What came first: Protein synthesis, or chaperones helping proteins to fold correctly? Consider as well, that, as Jörg Martin puts it (2000): The intracellular assembly of GroEL-type chaperonins appears to be a chaperone-dependent process itself and requires functional preformed chaperonin complexes !! 26 There are machines in the cell, that help other machines to be folded correctly, and these machines are also employed to help other machines to fold in order to be able to operate properly! Amazing!
Thorsten Hugel (2020): In a living cell, protein function is regulated in several ways, including post-translational modifications (PTMs), protein-protein interaction, or by the global environment (e.g. crowding or phase separation). While site-specific PTMs act very locally on the protein, specific protein interactions typically affect larger (sub-)domains, and global changes affect the whole protein non-specifically. Herein, we directly observe protein regulation under three different degrees of localization, and present the effects on the Hsp90 chaperone system at the levels of conformational steady states, kinetics and protein function. Interestingly using single-molecule FRET, we find that similar functional and conformational steady states are caused by completely different underlying kinetics. We disentangle specific and non-specific effects that control Hsp90’s ATPase function, which has remained a puzzle up to now. Lastly, we introduce a new mechanistic concept: functional stimulation through conformational confinement. Our results demonstrate how cellular protein regulation works by fine-tuning the conformational state space of proteins. 28
Susan Lindquist (2010): Cells also require a ubiquitin-proteasome system, targeting terminally misfolded proteins for degradation, and with translocation machineries to get proteins to their proper locations. These protein folding agents constitute a large, diverse, and structurally unrelated group. Many are upregulated in response to heat and are therefore termed heat shock proteins (HSPs). HSP90 is one of the most conserved HSPs, present from bacteria to mammals, and is an essential component of the protective heat shock response. The role of HSP90, however, extends well beyond stress tolerance. Even in nonstressed cells, HSP90 is highly abundant and associates with a wide array of proteins (known as clients) that depend on its chaperoning function to acquire their active conformations. 20% of yeast proteins are influenced by Hsp90 function, making it the most highly connected protein in the yeast genome, and GroES mediates the folding of ~10% of proteins in E. coli.29
Short RNA-peptides, or peptides on their own, are not functional and are useless in a supposed "proto-cell" unless they have the right size and sequence, able to fold into the functional 3D conformation. In face of this evidence, supposing and theorizing intermediate states and transitions of growing size and complexity over long periods of time until a functional state of affairs is achieved, is untenable. It opposes the evidence just described. Sophisticated exquisite mechanisms have to be instantiated from the get-go, to guarantee the right setup and folding of proteins of the full length. Such a hypothesized transition is never to work and going to happen. These RNA-peptides would simply lay around, and then sooner or later disintegrate. These explanations not including an intelligent agent are entirely inadequate to account for the origin of this kind of these high-tech engineering marvel implementations on a molecular scale!
George Church, Professor of Genetics, described the ribosome as "the most complicated thing that is present in all organisms". The peptidyl transferase center (PTC) is the core of the ribosome, where peptide bond formation occurs, which is a central catalytic reaction in life, where proteins are synthesized, and is as such of particular importance. The process is so intriguingly complex, that a science paper in 2015 had to admit that: "The detailed mechanism of peptidyl transfer, as well as the atoms and functional groups involved in this process are still in limbo." 19 The PTC is a ribozyme, which means it is composed of ribosomal RNAs ( rRNAs). Francisco Prosdocimi (2020): The PTC region has been considered crucial in the understanding about the origins of life. It has been described as the most significant trigger that engendered a mutualistic behavior between nucleic acids and peptides, allowing the emergence of biological systems. The emergence of this proto-PTC is a prerequisite to couple a chemical symbiosis between RNAs and peptides. Of 1434 complete sequences of 23S ribosomal RNAs analyzed, it was demonstrated that site A2451 from the 23S rRNA, which is the catalytic site of the PTC, is essential for the peptide bond to occur, and is absolutely preserved in each and every analyzed sequence. The PTC is known to be a flexible and efficient catalyst as it is capable of recognizing different, specific substrates (20 different amino acids bind to aminoacyl-tRNAs) and polymerizing proteins at a similar rate. 20
Sávio T.Farias (2014): Studies reveal that the PTC has a symmetrical structure comprising approximately 180 nucleotides. Molecular structure models suggest that the catalytic portion of the 23S rRNA entities of the symmetrical region possesses the common stem-elbow-stem (SES) structural motif. 21
Let's suppose that this structure would have emerged in an RNA-peptide world. Let's also not consider, that finding a functional sequence of 180 RNAs would vastly exceed the resources in sequence space, exhausting the maximum number of possible events in a universe that is 18 Billion years old (10^16 seconds) where every atom (10^80) is changing its state at the maximum rate of 10^40 times per second is 10^139. If we had such a core PTC, it would have no function whatsoever, unless all other players would be in place to perform translation from RNA to amino acids, having as well the genetic code implemented, and the entire chain from DNA to mRNA, to then coming to the events in translation. All these proposals, the RNA world, and the RNA-peptide world are based on silly pipe dreams - that they call theories when they are not more than ideas, based on fertile minds, and not results based on scientific evidence, experimentation, and tests in the lab. These are just invented scenarios - out of the need to keep an explanatory framework based on philosophical naturalism to find answers that do not require invoking a supernatural entity. All these proposals have been shown to be inadequate and doomed to failure. Biological cells are too complicated, sophisticated, integrated, and functional in order to warrant the belief that they could have originated by unguided means - the ribosome is a prime example to conclude this.
1. Suzan Mazur: The Origin of Life Circus: A How To Make Life Extravaganza November 30, 2014
2. PHILIP BALL: Flaws in the RNA world 12 FEBRUARY 2020
3. Eugene Koonin: The Logic of Chance: The Nature and Origin of Biological Evolution 31 agosto 2011
4. Natalia Szostak: Simulating the origins of life: The dual role of RNA replicases as an obstacle to evolution July 10, 2017
5. Jaroslaw Synak: RNA World Modeling: A Comparison of Two Complementary Approaches 11 April 2022
6. Jack W Szostak: The eightfold path to non-enzymatic RNA replication 03 February 2012
7. Hannes Mutschler: The difficult case of an RNA-only origin of life AUGUST 28 2019
8. Jordana Cepelewicz: Origin-of-Life Study Points to Chemical Chimeras, Not RNA September 16, 2019
9. Harris Bernstein: Origin of DNA Repair in the RNA World October 12th, 2020
10. Gerald F. Joyce: Protocells and RNA Self-Replication 2018
11. Steven A. Benner: The ‘‘Strong’’ RNA World Hypothesis: Fifty Years Old 2013 Apr;13
12. Harold S Bernhardt:[url=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3495036/#:~:text=However%2C the following objections have,of RNA is too limited.] The RNA world hypothesis: the worst theory of the early evolution of life[/url] (except for all the others) 2012 Jul 13
13. Eugene V Koonin: On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization 2007 May 31
14. Sami EL Khatib: Assumption and Criticism on RNA World Hypothesis from Ribozymes to Functional Cells March 12, 2021
15. Paul C. W. Davies: The algorithmic origins of life 2013 Feb 6
16. Charles W. Carter, Jr. What RNA World? Why a Peptide/RNA Partnership Merits Renewed Experimental Attention 23 January 2015
17. Dennis R. Salahub: Characterization of the active site of yeast RNA polymerase II by DFT and ReaxFF calculations 08 April 2008
18. Dimiter Kunnev: Possible Emergence of Sequence Specific RNA Aminoacylation via Peptide Intermediary to Initiate Darwinian Evolution and Code Through Origin of Life 2018 Oct 2;8
19. Hadieh Monajemi: The P-site A76 2′-OH acts as a peptidyl shuttle in a stepwise peptidyl transfer mechanism 2015
20. Francisco Prosdocimi: The Ancient History of Peptidyl Transferase Center Formation as Told by Conservation and Information Analyses 2020 Aug 5
21. Sávio T.Farias: Origin and evolution of the Peptidyl Transferase Center from proto-tRNAs 2014
22. Felix Müller: A prebiotically plausible scenario of an RNA–peptide world 11 May 2022
23. Stephen D. Fried: Peptides before and during the nucleotide world: an origins story emphasizing cooperation between proteins and nucleic acids 09 February 2022
24. Jordana Cepelewicz: The End of the RNA World Is Near, Biochemists Argue December 19, 2017
25. Mathieu E. Rebeaud: On the evolution of chaperones and cochaperones and the expansion of proteomes across the Tree of Life May 17, 2021
26. Jörg Martin: Assembly and Disassembly of GroEL and GroES Complexes 2000
27. Hays S. Rye: GroEL-Mediated Protein Folding: Making the Impossible, Possible 2013 Sep 25
28. Thorsten Hugel: Controlling protein function by fine-tuning conformational flexibility 2020 Jul 22
29. Susan Lindquist: HSP90 at the hub of protein homeostasis: emerging mechanistic insights 2010 Jul;11
30. F Egami: A working hypothesis on the interdependent genesis of nucleotide bases, protein amino acids, and primitive genetic code 1981 Sep;11
31. Charles W Carter, Jr: Interdependence, Reflexivity, Fidelity, Impedance Matching, and the Evolution of Genetic Coding 24 October 2017
32. Jessica C. Bowman: The Ribosome Challenge to the RNA World 20 February 2015
33. Jordana Cepelewicz: Life’s First Molecule Was Protein, Not RNA, New Model Suggests November 2, 2017
34. Coenzymes & Cofactors