https://reasonandscience.catsboard.com/t3090-pelagibacter-ubique-is-the-best-candidate-to-investigate-origin-of-life-scenarios-here-is-why
The smallest free-living cell that is capable of synthesizing all amino acids on its own is the bacterium called Pelagibacter ubique, also known as SAR11. It is a marine bacterium that has a diameter of only about 0.1 to 0.2 micrometers, making it one of the smallest known organisms. Pelagibacter ubique is able to produce all 20 standard amino acids on its own, which is a unique feature among free-living bacteria. This is in contrast to many other bacterial species, which rely on external sources of amino acids for growth and survival. The ability of Pelagibacter ubique to synthesize all its own amino acids is thought to be a key adaptation for its survival in nutrient-poor environments, such as the open ocean. Pelagibacter ubique, like all living cells, has a gene regulatory network that controls the expression of its genes. The gene regulatory network of Pelagibacter ubique is relatively simple compared to that of other bacterial species, as it has one of the smallest known genomes of any free-living organism. However, even with its small genome size, Pelagibacter ubique has been shown to have a complex gene regulatory network that allows it to respond to changes in its environment, such as nutrient availability and temperature. The bacterium has been found to have a number of regulatory genes, including transcription factors and signal transduction proteins, which are involved in controlling the expression of its genes. These regulatory genes allow Pelagibacter ubique to adjust its metabolism and other cellular processes in response to changing environmental conditions. A gene regulatory network is essential for the function and survival of all living cells. The gene regulatory network controls the expression of genes, which in turn determines the specific proteins and other molecules that a cell produces, as well as when and in what amounts they are produced.
This regulation is critical for the proper functioning of cellular processes, such as metabolism, growth, and response to environmental cues. Without a gene regulatory network, a cell would not be able to properly respond to changing conditions, adapt to its environment, or carry out the functions necessary for its survival. The complexity of the gene regulatory network can vary greatly between different types of cells and organisms, but all living cells must have some level of gene regulation to maintain proper cellular function and survive. Yes, that's correct. A gene regulatory network and genes are interdependent, and neither has any function without the other. Genes are the basic units of heredity that contain the instructions for making the proteins and other molecules that a cell needs to function. However, the expression of genes must be properly regulated to ensure that the right molecules are produced at the right time and in the right amounts. This is where the gene regulatory network comes in - it is responsible for controlling the expression of genes and ensuring that the correct proteins and molecules are produced in response to the cell's needs and its environment. In other words, genes provide the raw material for the cell, while the gene regulatory network acts as the control system that coordinates the use of this raw material to ensure proper cellular function. Both are necessary for the cell to function properly and carry out the processes that are essential for life.
Yes, interdependence is a fundamental concept in biology. Organisms and their components, such as genes and regulatory networks, are highly interdependent and cannot function in isolation. Living organisms are made up of complex, interconnected systems of molecules, cells, tissues, and organs, and each part of the system relies on the others to function properly. For example, in order for a cell to carry out its functions, it requires a complex network of interacting molecules, including proteins, nucleic acids, lipids, and carbohydrates. Similarly, in order for an organism to survive and reproduce, it must be able to carry out a wide range of physiological and behavioral processes that are interdependent and require coordination. The study of biology seeks to understand these complex systems and the interdependencies that exist between their components, as well as the interactions between living organisms and their environment. By understanding these interdependencies, we can gain insights into how living systems work, and how they respond to changing conditions, which has important implications for fields such as medicine, agriculture, and ecology.
Life presumably started simple. This is plausible, but is an assumption. The smallest known bacteria are still rather large. One of the smallest known metabolically autonomous bacterial species is Pelagibacter ubique with about 1,400 genes (Giovannoni et al., 2005). Genome reduction of Mycoplasma mycoides by systematic deletion of individual genes resulted in a synthetic minimal genome of 473 genes (Hutchison et al., 2016). Can one consider simpler living entities?
https://www.frontiersin.org/articles/10.3389/fmicb.2019.00523/full
The first life form, the supposedly Last Universal Common Ancestor (LUCA), had to be already amazingly complex. One of the smallest free living creatures we know of, Pelagibacter Ubique, for example, which is also the most abundant organism on earth, a tiny bacterium, requires 1,3 million nucleotides, the building blocks of DNA. If we suppose that an unguided event ordered the right sequence, there would have to be 10^722000 attempts to get the right sequence. There are 10^80 atoms in the entire known universe.
The genome of P. ubique strain HTCC1062 was completely sequenced in 2005 showing that P. ubique has the smallest genome (1,308,759 bp) of any free living organism encoding only 1,354 open reading frames (1,389 genes total). The only species with smaller genomes are intracellular symbionts and parasites, such as Mycoplasma genitalium or Nanoarchaeum equitans It has the smallest number of open reading frames of any free living organism, and the shortest intergenic spacers, but it still has metabolic pathways for all 20 amino acids and most co-factors. 6
SAR11 Alphaproteobacteria (order Pelagibacterales) are the most abundant chemo-organotrophic bacteria in the oceans 1
“Sometimes we should try to ask the simplest questions. One of the simplest questions that my lab has concerned itself with over the last couple of decades is ‘how does a cell that is so simple—1.3 million base pairs, a little over 1000 genes—how does a cell that’s so simple succeed in capturing such a large part of all the organic matter entering the oceans?” 2
SAR11 is the most abundant organism on the planet! “If you go the ocean, and you go for a swim, and you accidentally swallow a mouthful of seawater, you’ve probably swallowed a million SAR11 cells.” 3
P. ubique is one of the smallest free-living cells. Tiny as it is, you may not be surprised to learn that P. ubique also has a small genome (1,308,759 bp), currently the smallest genome known for a free-living organism. The genome encodes 1,354 predicted proteins, 1 rRNA operon , and 32 tRNAs.
Genome data indicate that this bacterium possesses the complete pathways required to generate energy by respiration and also to harvest energy from the sun using proteorhodopsin, a light-dependent, retinylidine proton pump. P. ubique is also equipped with numerous high affinity nutrient transporters, consistent with a competitive strategy for nutrient scavenging. Most are ABC transporters, a large and ancient protein family noted for providing high substrate affinities at the cost of copious ATP. Clearly, the cells invest in transport to outcompete other microbes in nutrient acquisition and, based on their abundance and efficient assimilation of organic matter, they have succeeded.4.
Pelagibacter ubique, strain HTCC1062, is significantly known to be one of smallest and simplest, self-replicating, and free living cell. It is part of the SAR11 clade, which are small, heterotrophic alphaproteobacteria, equaling to ~25% of all microbial plankton cells. 5
Streamlining and Core Genome Conservation among Highly Divergent Members of the SAR11 Clade 7
SAR11 has evolved into perhaps a dozen or more specialized ecotypes that span evolutionary distances equivalent to a bacterial order. We found small genomes throughout the clade and a very high proportion of core genome genes (48 to 56%), indicating that small genome size is probably an ancestral characteristic. In their level of core genome conservation, the members of SAR11 are outliers, the most conserved free-living bacteria known. Variation among the genomes included genes for phosphorus metabolism, glycolysis, and C1 metabolism, suggesting that adaptive specialization in nutrient resource utilization is important to niche partitioning and ecotype divergence within the clade. The SAR11 clade is the most abundant group of marine microorganisms worldwide, making them key players in the global carbon cycle. Study of isolates in culture revealed atypical organic nutrient requirements that can be attributed to genome reduction, such as conditional auxotrophy for glycine and its precursors.
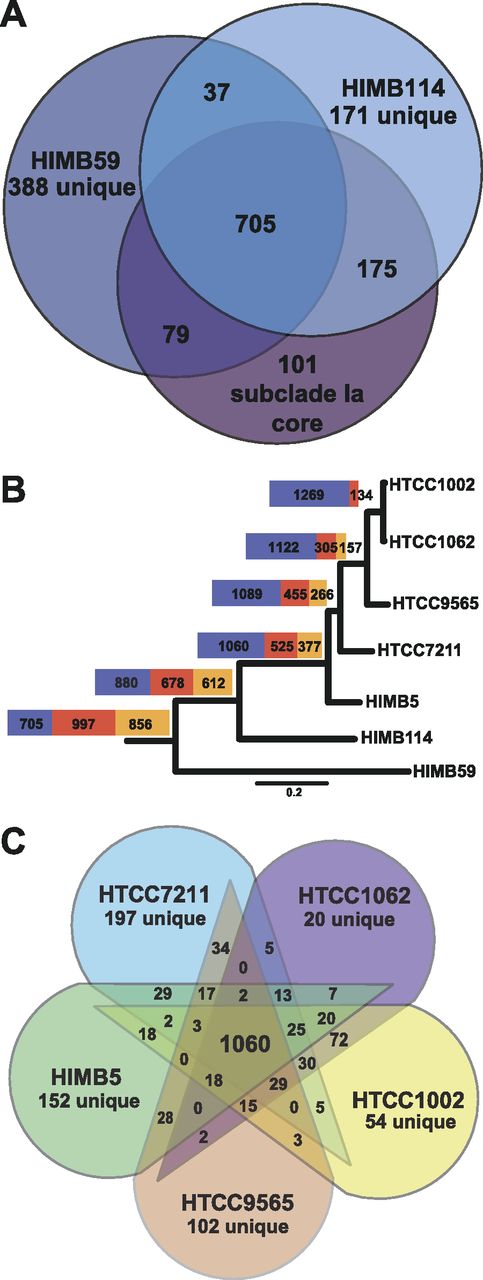
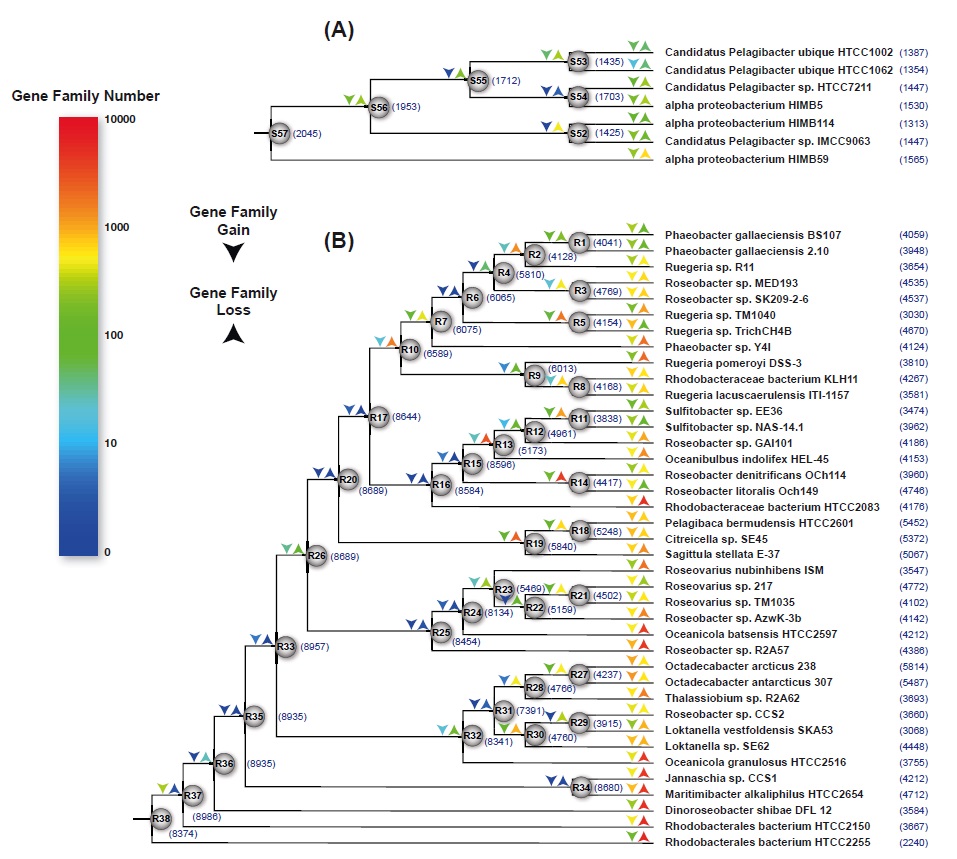
(A) Venn diagram showing the number of OCs shared between the SAR11 subclade Ia core genome, HIMB114, and HIMB59.
(B) The relative contribution of core (blue), shared non-core (orange), and unique (red) orthologs to the pan-genome at each level of divergence. The total size of each bar is proportional to the total number of orthologs in the pan-genome. The scale bar indicates 0.2 changes per position. The tree was redrawn based on the work of Thrash et al. (6). (C) Venn diagram showing the number of shared OCs among the five strains of SAR11 subclade Ia.
At ~600 genes, the core genome for SAR11 provides an estimate of the lower limit of genes essential for maintenance of the free-living state in marine environments.
Small genomes suggest specialization, but the members of SAR11, which have small genomes, cannot plausibly be characterized as specialists, being one of the most successful and widely distributed chemoheterotrophic groups in the ocean.
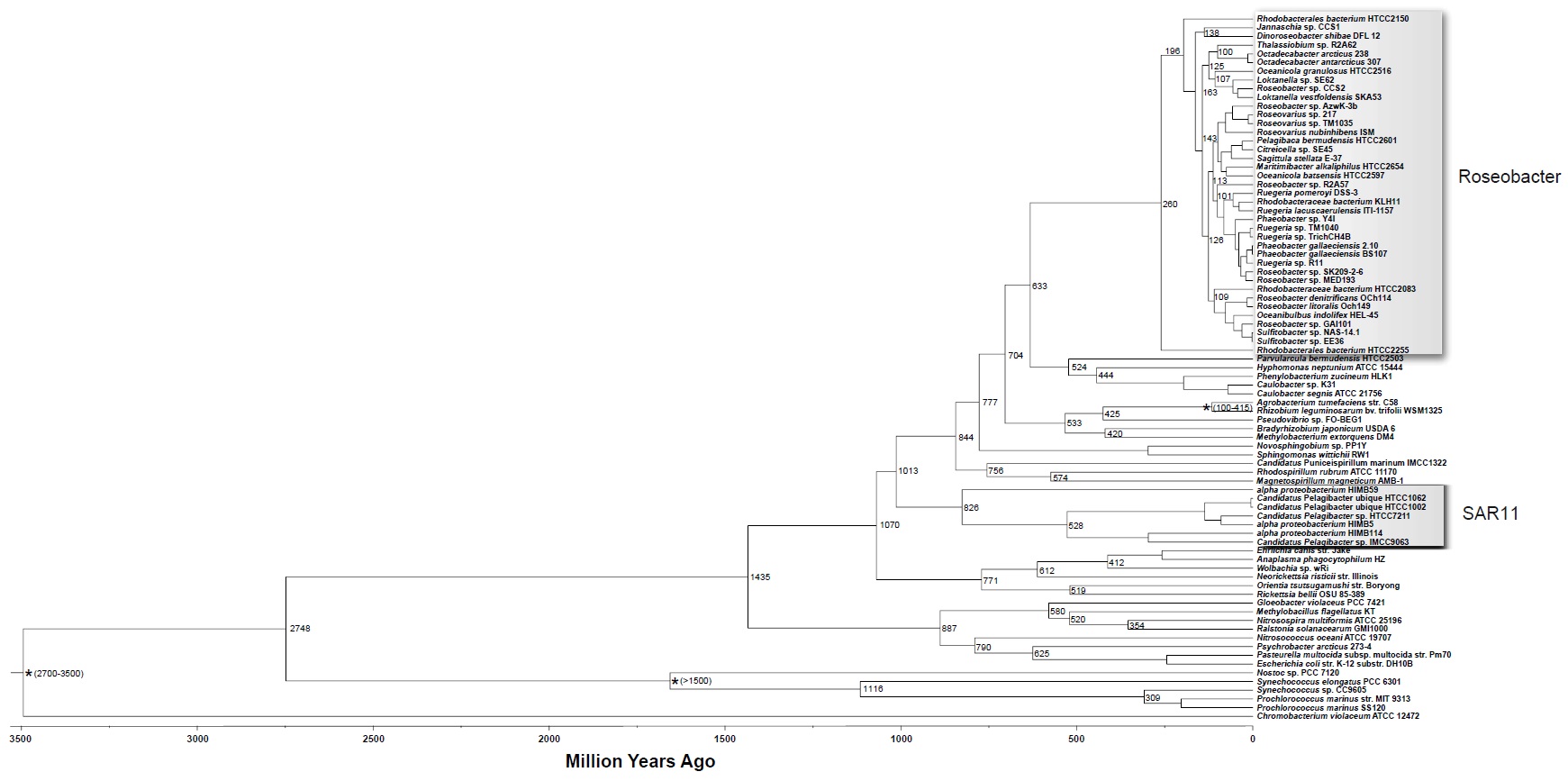
Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade14 June 2011 8
Mitochondria share a common ancestor with the Alphaproteobacteria, but determining their precise origins is challenging due to inherent difficulties in phylogenetically reconstructing ancient evolutionary events. The evidence supports a common origin of mitochondria and SAR11 as a sister group to the Rickettsiales. The simplest explanation of these data is that mitochondria evolved from a planktonic marine alphaproteobacterial lineage that participated in multiple inter-specific cell colonization events, in some cases yielding parasitic relationships, but in at least one case producing a symbiosis that characterizes modern eukaryotic life. Vital to the evolution of all known eukaryotic cells was the endosymbiotic event that resulted in the permanent acquisition of bacteria that through time were transformed into mitochondria. Even eukaryotes that were previously thought to lack mitochondria have now been shown to contain some remnants of that original endosymbiosis. current hypotheses, making use of mitochondrial genome and proteome data, agree that the mitochondrial ancestor was most closely related to Alphaproteobacteria. The sequencing of the first Rickettsia genome provided phylogenomic evidence for the previously established theory that the mitochondrial root was within the Rickettsiales.
The sequencing of the first Rickettsia genome provided phylogenomic evidence for the previously established theory that the mitochondrial root was within the Rickettsiales5,10,12,13 (and references therein). This theory was intellectually satisfying because all known Rickettsia have extremely reduced genomes and obligate intracellular lifestyles, although there was evidence to suggest that the genome reduction in mitochondria and Rickettsia probably happened independently.
SAR11 genomes bear distinctly different evolutionary signatures from other Rickettsiales genomes, indicating that selection for a streamlined genome, rather than genetic drift, has been the path of genome reduction in these organisms. For example, the sequence of Candidatus Pelagibacter ubique HTCC1062 has few recent gene duplications, phage, transposons, or pseudogenes, has extremely small intergenic regions and is optimized for a highly efficient lifestyle, whereas organisms like Rickettsia and those of the Anaplasmataceae contain large numbers of duplications, putatively inactive genes, larger intergenic regions, and/or mobile elements, indicative of the different selective pressures of obligate intracellular life.
Modern SAR11 cells have small genomes imposed by selection for efficient replication in nutrient limited systems17, which have evolved unusual nutritional requirements that would pre-adapt them to closer dependence on other organisms.
1. https://sci-hub.ren/10.1038/nmicrobiol.2016.65
2. https://asm.org/Podcasts/MTM/Episodes/SAR11-and-Other-Marine-Microbes-with-Steve-Giovann
3. https://twitter.com/asmicrobiology/status/1209160770136813568
4. https://schaechter.asmblog.org/schaechter/2015/02/the-most-abundant-small-things-considered.html
5. https://microbewiki.kenyon.edu/index.php/Pelagibacter_ubique
6. https://alchetron.com/Pelagibacter-ubique
7. https://mbio.asm.org/content/3/5/e00252-12
8. https://www.nature.com/articles/srep00013
Last edited by Otangelo on Fri Feb 17, 2023 12:35 pm; edited 10 times in total