F-BAR domain proteins that sculpture membranes: By evolution, or design?
https://reasonandscience.catsboard.com/t3038-f-bar-domain-proteins-that-sculpture-membranes-by-evolution-or-design
Remind some of the futuristic amazingly curved roof structures of buildings made by star architects like the Phaeno structure in Germany. The curves of the roof walls must be sustained by complex webs of scaffolds, which are designed to withstand the loads, forces, and different weather conditions to which the buildings are exposed. The dimensions and sizes and materials of the scaffolds and sustaining structures must be finely planned and calculations must be made in order for these structures to withstand the various environmental conditions. Nobody in its sane mind would imagine that such finely architectures structures could or would emerge by unplanned natural forces.
When you see the nice curvature of organelles in cells made by membranes, have you ever asked yourself how the different curved membrane shapes are accomplished ? It seems natural, and eventually we have never gave a second thought to that, but as it comes out, in the cell, nothing happens occasionally, or by chance, but everything must be finely orchestrated, and awe inspiring mechanisms need to be in place to accomplish every tiny bit of cell architecture, form and function.
Membrane Sculpting by F-BAR Domains Studied by Molecular Dynamics Simulations
January 31, 2013
Interplay between cellular membranes and their peripheral proteins drives many processes in eukaryotic cells. Proteins of the BAR domain family play a role in cellular morphogenesis, for example curving planar membranes into tubular membranes. F-BAR proteins form regular lattices on cylindrically deformed membrane surfaces. Such lattices, indeed, induce tubes of observed radii. F-BAR domain curves membranes via the so-called scaffolding mechanism. Plasticity of the F-BAR domain permits conformational change in response to membrane interaction, via partial unwinding of the domains 3-helix bundle structure.
To generate organelles, eukaryotic cells sculpt their membranes into compartments, often employing proteins as chaperones, for example, F-BAR domains. The latter induce formation of tubular and vesicular membranes.
F-BAR domains sculpt membranes through electrostatic interactions, driving the membrane to match the concave surface of the protein's banana-like shape. Lattice types are optimally attuned to producing high membrane curvature. The simulations reproduced also a process lasting 350 µs in which lattices of F-BAR domains form a complete tube out of an initially flat membrane.
Interplay between cellular membranes and their peripheral proteins drives many cellular processes, including cell division, growth, movement and cell-cell communication. During their lifetime and often with the help of membrane peripheral proteins, eukaryotic cells dynamically sculpt their various types of compartments.
BAR domain deficiency is related to a wide range of cancers and blood disorders. Resolved structures show that BAR domains form crescent-shaped homodimers, the monomers being composed of coiled-coil association of a 3-helix bundle structure. Three sub-families of BAR domains, namely N-BAR domains, FCH-BAR (F-BAR) domains and Inverse-BAR (I-BAR) domains, differ from each other in their structure and physiological function. In contrast to N-BAR domains that form a banana shaped dimer, F-BAR domains are elongated and only gently curved. A high density of positive charge is found on the part of the protein that is destined to interact with negatively-charged membranes. While N-BAR domains stabilize highly curved membrane structures, F-BAR domains stabilize membrane structures of small degree of curvature. N-BAR domains also have an N-terminal amphipathic helix, which aids membrane curvature stabilization by membrane insertion. Such helix is lacking in the case of F-BAR domains. Both N-BAR domains and F-BAR domains are found to induce formation of tubules in vitro.
According to the scaffolding mechanism, BAR domains bend membranes by attracting negatively-charged lipid headgroups to their positively-charged curved surface. During the scaffolding process, a BAR domain is considered to act as a rigid body, to which lipids are attracted via electrostatic interaction, transferring membrane binding energy into membrane bending energy. According to the membrane insertion mechanism, a BAR domain inserts its amphipathic groups like wedges into one leaflet of the membrane and, thereby, curves the membrane. N-BAR proteins use their N-helix as an amphipathic wedge, while for the F-BAR domain it is suspected that residue Phe117 inserts its bulky side chain into the membrane. Either mechanism involves strong membrane-protein interactions.
BAR domains are found to shape low-curvature liposomes into high-curvature tubules. Such extensive membrane remodeling requires collective action of multiple BAR domains. Striations observed on the surface of BAR domain-induced tubules suggest that the tubules are covered by an ordered arrangement of the proteins. Recent observations revealed that well-organized spirals of BAR domains form on the surface of membrane tubules. Differences in lattices formed by BAR domains may result in variations of membrane curvature and structure. However, it remains unclear how membrane curvature depends on the type of F-BAR domain lattice arrangement. Two further open questions are: How do individual F-BAR domains interact with a membrane to form local curvature? What dynamics is involved in membrane curvature formation by F-BAR domain lattices?
The F-BAR domain undergoes conformational change during membrane curvature generation
During the process of curvature generation, the F-BAR domain interacts with the membrane and undergoes a large conformational change involving its side helices
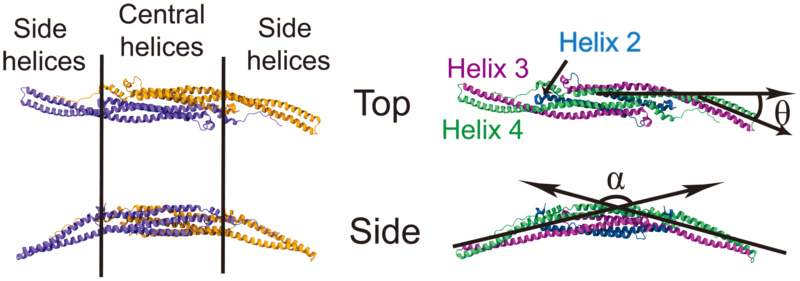
Conformation of F-BAR domain characterized through angles
(Left) The F-BAR domain is a dimer of two F-BAR proteins, the latter colored in purple and orange. (Right) Each F-BAR protein is composed of five helices forming a coiled-coil structure, with helix 2 to 4 colored blue, purple and green, respectively. is formed by the principal axes of the central part and the end part of helix 4 (green); is formed by the angle between the principal axes of the left and the right sides of helix 3 (purple).
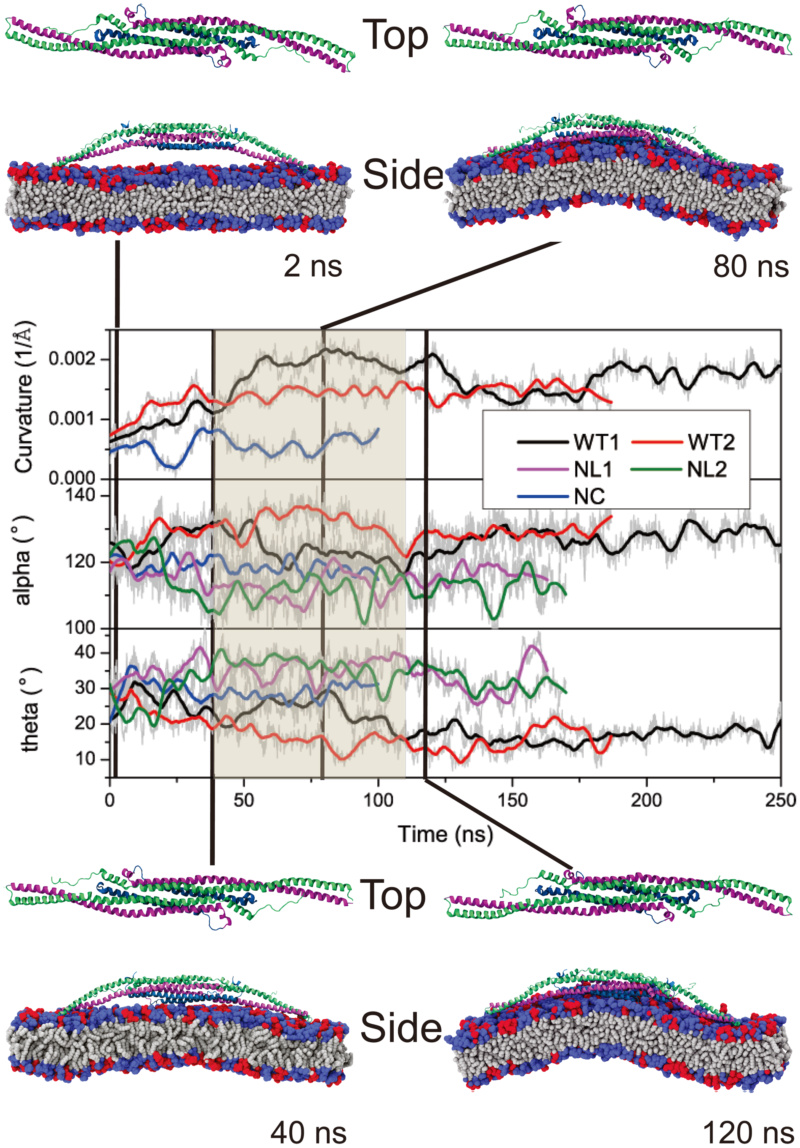
Change of membrane curvature and of angles .
.
As shown in Fig. above, both and
and  of WT1 change significantly upon interaction with the membrane;
of WT1 change significantly upon interaction with the membrane;  increases up to
increases up to  , then decreases to
, then decreases to  , fluctuating finally around
, fluctuating finally around  ;
;  decreases to
decreases to  , then increases back to
, then increases back to  , fluctuating finally around
, fluctuating finally around  . In control systems NL1, NL2 and NC,
. In control systems NL1, NL2 and NC,  and
and  do not show such changes and fluctuate around different average angles.
do not show such changes and fluctuate around different average angles.
Conformational change of F-BAR domain during interaction with the membrane.
When the F-BAR domain assumes a concave shape, the attached membrane undergoes induced-fit bending. Unlike N-BAR domains, which act like rigid bodies attracted to a membrane, the F-BAR domain and the membrane influence each others shape. Based on the conformation of the F-BAR domain and membrane curvature, the curvature generation process by the F-BAR domain can be separated into three phases. The curvature generation, in fact, is an induced-fit process, during which membrane binding energy is transfered into membrane bending energy through protein conformational change.
In phase 1, lasting from 0 to 40 ns, the F-BAR domain binds to the membrane and membrane curvature increases slowly, while increases and
increases and  decreases. During this phase, the side helices of the F-BAR domain straighten up and the domain adopts a shallow inner surface, to allow all positively charged residues along the concave surface to contact the negatively charged membrane (Fig. 1C); water molecules between the F-BAR domain and membrane are squeezed out; potential energy is stored in the newly formed F-BAR domain conformation.
decreases. During this phase, the side helices of the F-BAR domain straighten up and the domain adopts a shallow inner surface, to allow all positively charged residues along the concave surface to contact the negatively charged membrane (Fig. 1C); water molecules between the F-BAR domain and membrane are squeezed out; potential energy is stored in the newly formed F-BAR domain conformation.
In phase 2, lasting from 40 to 120 ns, membrane curvature is generated. During this phase, and
and  adjust and domain curvature increases. Potential energy stored in the F-BAR domain conformation is released into energy associated with membrane curvature.
adjust and domain curvature increases. Potential energy stored in the F-BAR domain conformation is released into energy associated with membrane curvature.
In phase 3, lasting from 120 to 250 ns, the protein-membrane system relaxes. Membrane curvature decreases slightly and fluctuates around 0.0015A?−1; and
and  values are restored close to the native state values, indicating partial uncoiling of the coiled-coil structure (Fig. S6 in Text S1). However,
values are restored close to the native state values, indicating partial uncoiling of the coiled-coil structure (Fig. S6 in Text S1). However,  values in simulations NC, NL1 and NL2 are much lower than those in simulation WT1 and WT2, while
values in simulations NC, NL1 and NL2 are much lower than those in simulation WT1 and WT2, while  values show the reverse, indicating that the domains coiled-coil structure without interaction with the membrane becomes further coiled, which suggests that partial uncoiling of the domains coiled-coil structure provides the driving force for membrane curvature formation. Indeed, if one removes the membrane from the final conformation of simulation WT1, as is done in simulation WT1WAT, the conformation of the F-BAR domain is quickly restored to a near crystal conformation and
values show the reverse, indicating that the domains coiled-coil structure without interaction with the membrane becomes further coiled, which suggests that partial uncoiling of the domains coiled-coil structure provides the driving force for membrane curvature formation. Indeed, if one removes the membrane from the final conformation of simulation WT1, as is done in simulation WT1WAT, the conformation of the F-BAR domain is quickly restored to a near crystal conformation and  and
and  assume values similar to the ones they assume in simulation NL1 and NL2, indicating that the uncoiling is reversible (simulation WT1WAT, see Fig. S8 in Text S1).
assume values similar to the ones they assume in simulation NL1 and NL2, indicating that the uncoiling is reversible (simulation WT1WAT, see Fig. S8 in Text S1).
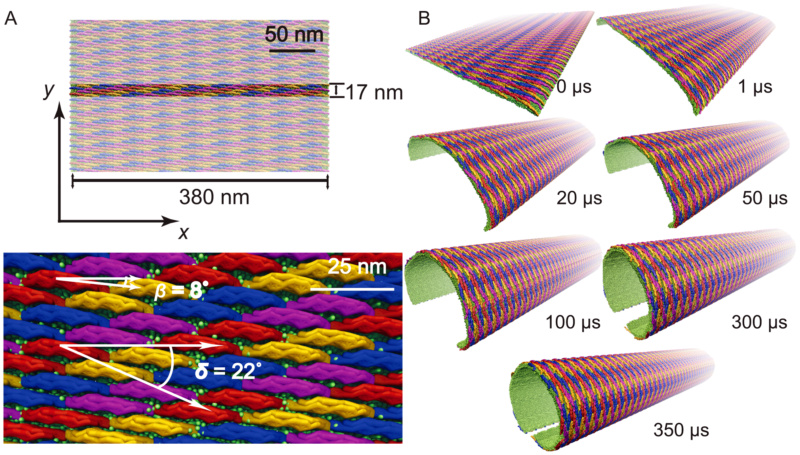
Membrane tubulation by lattices of F-BAR domains.
(A) Initial conformation of lattices of F-BAR domains on the membrane in a SBCG representation. A patch of membrane, 17 nm in width, is covered with 4 rows of F-BAR domains, each row containing 17 F-BAR domains. Each individual domain is tilted by with respect to the
with respect to the  -axis and each row of closest contact F-BAR domains is tilted by
-axis and each row of closest contact F-BAR domains is tilted by  with respect to the
with respect to the  -axis. Periodic boundary conditions are assumed in the
-axis. Periodic boundary conditions are assumed in the  -direction, so that the system can be regarded as a lipid patch of infinite length in this direction. (B) Membrane tube formation with lattices of F-BAR domains. Shown are snapshots of membrane structures during the
-direction, so that the system can be regarded as a lipid patch of infinite length in this direction. (B) Membrane tube formation with lattices of F-BAR domains. Shown are snapshots of membrane structures during the  simulation. Membrane lipids are shown in green; individual F-BAR domains are differentiated by color.
simulation. Membrane lipids are shown in green; individual F-BAR domains are differentiated by color.
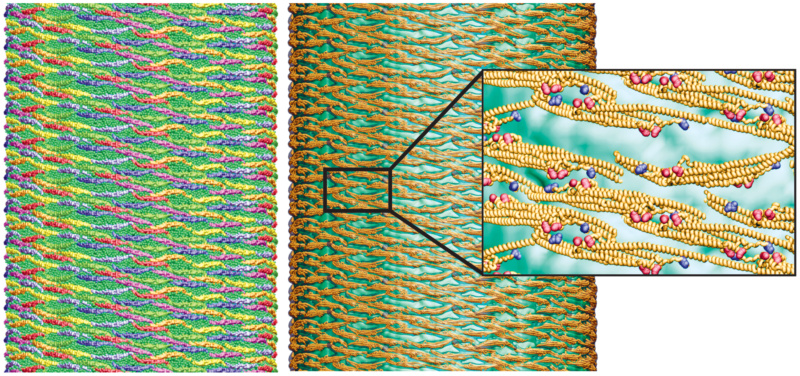
All-atom structure of F-BAR domain lattice on the formed membrane tube.
(Left) Coarse-grained tube structure from simulation TUBULATION, (Right) All-atom structure constructed from the SBCG structure. (Insert right) Close up view of all-atom structure, rendered in so-called cartoon representation. Residues Lys66, Arg47 are shown in van der Waals representation and colored in red; Glu285, Asp161 are represented in the same way, but colored in blue.

1. https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1002892
https://reasonandscience.catsboard.com/t3038-f-bar-domain-proteins-that-sculpture-membranes-by-evolution-or-design
Remind some of the futuristic amazingly curved roof structures of buildings made by star architects like the Phaeno structure in Germany. The curves of the roof walls must be sustained by complex webs of scaffolds, which are designed to withstand the loads, forces, and different weather conditions to which the buildings are exposed. The dimensions and sizes and materials of the scaffolds and sustaining structures must be finely planned and calculations must be made in order for these structures to withstand the various environmental conditions. Nobody in its sane mind would imagine that such finely architectures structures could or would emerge by unplanned natural forces.
When you see the nice curvature of organelles in cells made by membranes, have you ever asked yourself how the different curved membrane shapes are accomplished ? It seems natural, and eventually we have never gave a second thought to that, but as it comes out, in the cell, nothing happens occasionally, or by chance, but everything must be finely orchestrated, and awe inspiring mechanisms need to be in place to accomplish every tiny bit of cell architecture, form and function.
Membrane Sculpting by F-BAR Domains Studied by Molecular Dynamics Simulations
January 31, 2013
Interplay between cellular membranes and their peripheral proteins drives many processes in eukaryotic cells. Proteins of the BAR domain family play a role in cellular morphogenesis, for example curving planar membranes into tubular membranes. F-BAR proteins form regular lattices on cylindrically deformed membrane surfaces. Such lattices, indeed, induce tubes of observed radii. F-BAR domain curves membranes via the so-called scaffolding mechanism. Plasticity of the F-BAR domain permits conformational change in response to membrane interaction, via partial unwinding of the domains 3-helix bundle structure.
To generate organelles, eukaryotic cells sculpt their membranes into compartments, often employing proteins as chaperones, for example, F-BAR domains. The latter induce formation of tubular and vesicular membranes.
F-BAR domains sculpt membranes through electrostatic interactions, driving the membrane to match the concave surface of the protein's banana-like shape. Lattice types are optimally attuned to producing high membrane curvature. The simulations reproduced also a process lasting 350 µs in which lattices of F-BAR domains form a complete tube out of an initially flat membrane.
Interplay between cellular membranes and their peripheral proteins drives many cellular processes, including cell division, growth, movement and cell-cell communication. During their lifetime and often with the help of membrane peripheral proteins, eukaryotic cells dynamically sculpt their various types of compartments.
BAR domain deficiency is related to a wide range of cancers and blood disorders. Resolved structures show that BAR domains form crescent-shaped homodimers, the monomers being composed of coiled-coil association of a 3-helix bundle structure. Three sub-families of BAR domains, namely N-BAR domains, FCH-BAR (F-BAR) domains and Inverse-BAR (I-BAR) domains, differ from each other in their structure and physiological function. In contrast to N-BAR domains that form a banana shaped dimer, F-BAR domains are elongated and only gently curved. A high density of positive charge is found on the part of the protein that is destined to interact with negatively-charged membranes. While N-BAR domains stabilize highly curved membrane structures, F-BAR domains stabilize membrane structures of small degree of curvature. N-BAR domains also have an N-terminal amphipathic helix, which aids membrane curvature stabilization by membrane insertion. Such helix is lacking in the case of F-BAR domains. Both N-BAR domains and F-BAR domains are found to induce formation of tubules in vitro.
According to the scaffolding mechanism, BAR domains bend membranes by attracting negatively-charged lipid headgroups to their positively-charged curved surface. During the scaffolding process, a BAR domain is considered to act as a rigid body, to which lipids are attracted via electrostatic interaction, transferring membrane binding energy into membrane bending energy. According to the membrane insertion mechanism, a BAR domain inserts its amphipathic groups like wedges into one leaflet of the membrane and, thereby, curves the membrane. N-BAR proteins use their N-helix as an amphipathic wedge, while for the F-BAR domain it is suspected that residue Phe117 inserts its bulky side chain into the membrane. Either mechanism involves strong membrane-protein interactions.
BAR domains are found to shape low-curvature liposomes into high-curvature tubules. Such extensive membrane remodeling requires collective action of multiple BAR domains. Striations observed on the surface of BAR domain-induced tubules suggest that the tubules are covered by an ordered arrangement of the proteins. Recent observations revealed that well-organized spirals of BAR domains form on the surface of membrane tubules. Differences in lattices formed by BAR domains may result in variations of membrane curvature and structure. However, it remains unclear how membrane curvature depends on the type of F-BAR domain lattice arrangement. Two further open questions are: How do individual F-BAR domains interact with a membrane to form local curvature? What dynamics is involved in membrane curvature formation by F-BAR domain lattices?
The F-BAR domain undergoes conformational change during membrane curvature generation
During the process of curvature generation, the F-BAR domain interacts with the membrane and undergoes a large conformational change involving its side helices

Conformation of F-BAR domain characterized through angles
(Left) The F-BAR domain is a dimer of two F-BAR proteins, the latter colored in purple and orange. (Right) Each F-BAR protein is composed of five helices forming a coiled-coil structure, with helix 2 to 4 colored blue, purple and green, respectively. is formed by the principal axes of the central part and the end part of helix 4 (green); is formed by the angle between the principal axes of the left and the right sides of helix 3 (purple).

Change of membrane curvature and of angles
As shown in Fig. above, both
Conformational change of F-BAR domain during interaction with the membrane.
When the F-BAR domain assumes a concave shape, the attached membrane undergoes induced-fit bending. Unlike N-BAR domains, which act like rigid bodies attracted to a membrane, the F-BAR domain and the membrane influence each others shape. Based on the conformation of the F-BAR domain and membrane curvature, the curvature generation process by the F-BAR domain can be separated into three phases. The curvature generation, in fact, is an induced-fit process, during which membrane binding energy is transfered into membrane bending energy through protein conformational change.
In phase 1, lasting from 0 to 40 ns, the F-BAR domain binds to the membrane and membrane curvature increases slowly, while
In phase 2, lasting from 40 to 120 ns, membrane curvature is generated. During this phase,
In phase 3, lasting from 120 to 250 ns, the protein-membrane system relaxes. Membrane curvature decreases slightly and fluctuates around 0.0015A?−1;

Membrane tubulation by lattices of F-BAR domains.
(A) Initial conformation of lattices of F-BAR domains on the membrane in a SBCG representation. A patch of membrane, 17 nm in width, is covered with 4 rows of F-BAR domains, each row containing 17 F-BAR domains. Each individual domain is tilted by

All-atom structure of F-BAR domain lattice on the formed membrane tube.
(Left) Coarse-grained tube structure from simulation TUBULATION, (Right) All-atom structure constructed from the SBCG structure. (Insert right) Close up view of all-atom structure, rendered in so-called cartoon representation. Residues Lys66, Arg47 are shown in van der Waals representation and colored in red; Glu285, Asp161 are represented in the same way, but colored in blue.

1. https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1002892

