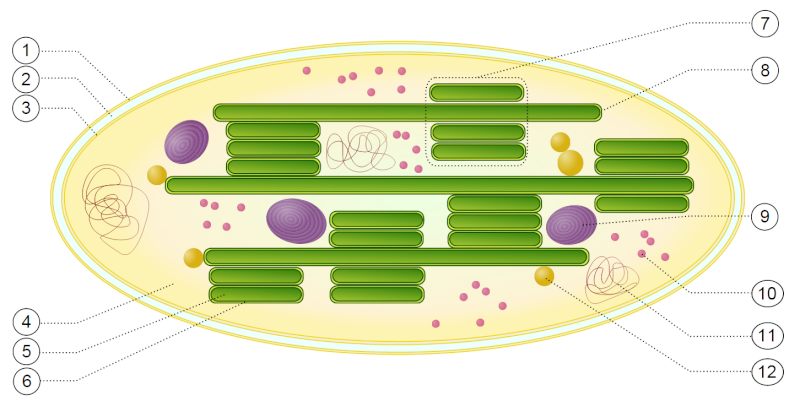
1 outer membrane
2 intermembrane space
3 inner membrane (1+2+3: envelope)
4 stroma (aqueous fluid)
5 thylakoid lumen (inside of thylakoid)
6 thylakoid membrane
7 granum (stack of thylakoids)
8 thylakoid (lamella)
9 starch
10 ribosome
11 plastidial DNA
12 plastoglobule (drop of lipids)
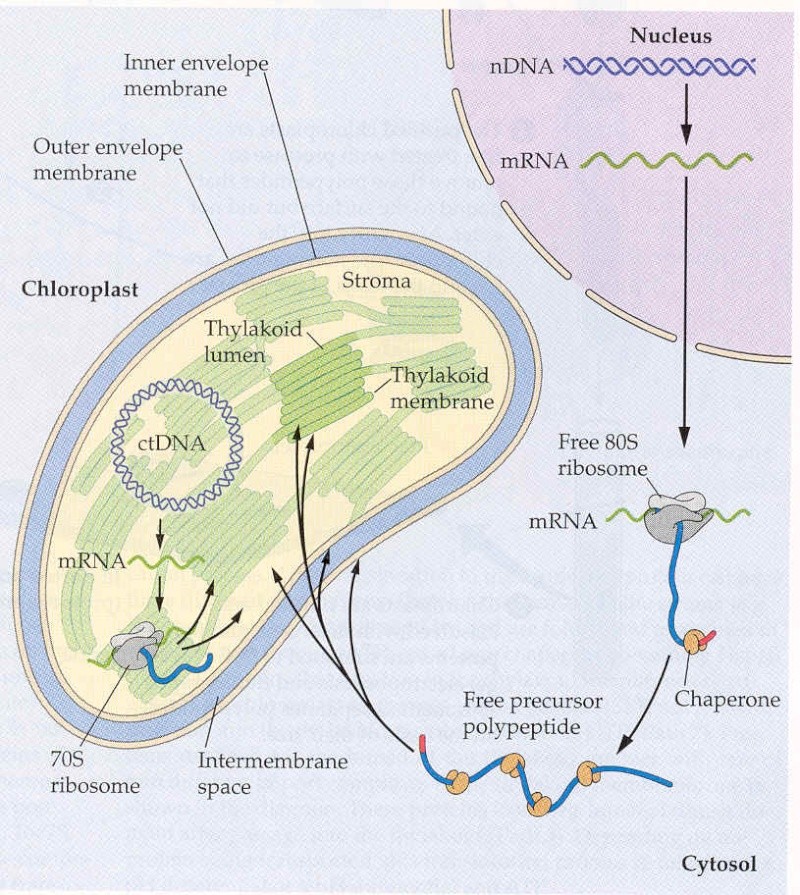
The most important consequence of the split genome in photosynthetic eukaryotes is the requirement that the vast majority of the proteins forming the chloroplast be coded for in the nucleus, synthesized on cytosolic ribosomes, and then imported into the chloroplast. Once inside the chloroplast, they must be targeted to the proper place and then, in many cases, assembled into large oligomeric complexes. There is a remarkably complex orchestration of the assembly and regulation of the chloroplast and the interaction between the chloroplast and the nucleus.
There are at least four distinct targeting pathways of proteins to chloroplasts. Here we focus on the Toc–Tic pathway, which accounts for import of ∼90% of proteins targeted to the chloroplast . Toc and Tic are large protein complexes that are given the abbreviations Toc (translocon at the outer membrane of chloroplasts) or Tic (translocon at the inner membrane of chloroplasts). Almost all nuclear-encoded proteins that are destined for the chloroplast have an N-terminal extension to the translation product, compared with the mature protein or with an analogous protein found in cyanobacteria. This N-terminal extension, or transit sequence, serves to target the protein to the chloroplast and is then cleaved off by a processing protease. Proteins that are targeted to the lumen of the chloroplast have to cross three membranes (in many types of algae they have to cross as many as five!) to reach their site of function.
These proteins have a bipartite targeting domain, consisting of the normal chloroplast-targeting sequence plus an additional lumenal targeting domain. In these cases, the processing takes place in two steps,with the first step occurring on entry into the chloroplast, and the second step taking place upon transport into the thylakoid lumen. Considerable evidence supports the view that the chloroplast-targeting sequence is both necessary and sufficient to direct a particular protein to the chloroplast. If this domain is removed, the resultant protein is not targeted to the chloroplast, and if the targeting domain is transferred to a protein not normally targeted to the chloroplast, the presence of the domain is sufficient to direct that protein to the chloroplast. Remarkably, there does not seem to be a consensus chloroplast-targeting sequence, but in most cases these sequences are rich in the hydroxyl-containing amino acids serine and threonine and contain almost no acidic amino acids. The lumenal targeting domains are typical of signal sequences found in secreted proteins. In a few cases, proteins that are targeted to the outer chloroplast envelope membrane do not have a cleavable targeting targeting
sequence and are directly incorporated into the membrane.
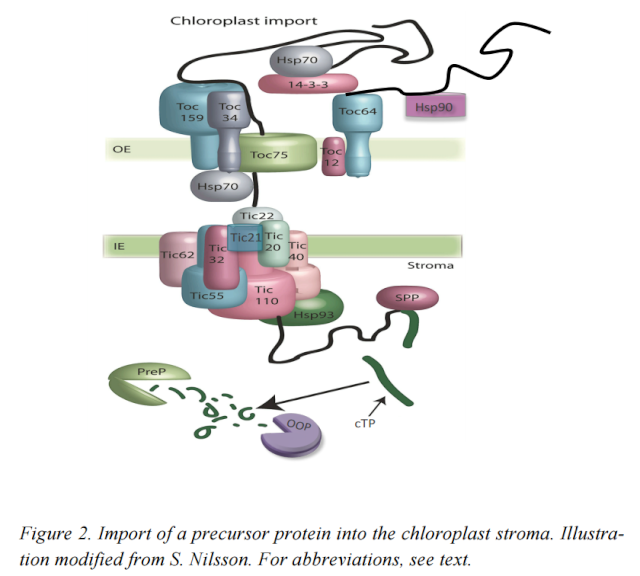
Figure below illustrates the pathway and components involved in the chloroplast Toc–Tic import machinery

Most nucleus-encoded plastid proteins are imported by TOC (translocon at the outer envelope membrane of chloroplasts) and TIC (translocon at the inner envelope membrane of chloroplasts) complexes that are located in the outer envelope membrane (OEM) and inner envelope membrane (IEM), respectively. 2 The core TOC machinery comprises two transit peptide receptors (TOC34 and TOC159) and channel-forming TOC75. The receptors are GTPases, accounting for the GTP requirement during the early stages of import. TOC159 additionally possesses an amino-terminal acidic (A) domain that may participate in transit peptide recognition. In Arabidopsis thaliana (and other plants), the receptors exist in multiple isoforms (TOC159: atTOC159, atTOC132 and atTOC120; TOC34: atTOC33 and atTOC34) with non-identical recognition specificities, which enables different, client-specific import pathways. SP1 is a RING finger (RNF) E3 ligase in the OEM that targets TOC proteins for ubiquitylation and turnover by the 26S proteasome (26SP), which results in the reorganization of the TOC machinery and balancing of client-specific pathways. A general TIC translocon of 1MDa, incorporating plastome-encoded TIC214 (also known as YCF1) and four nucleus-encoded components, may act upstream of a putative motor complex comprising TIC40 and TIC110. The latter complex recruits and regulates various stromal chaperones, which drive protein import and/or assist in protein folding at the expense of ATP hydrolysis. Additional IEM components may regulate import in response to internal redox signals (indicated by the question mark). HSP, heat shock protein. IMS, intermembrane space; SPP, stromal processing peptidase.
Import takes place in several stages, which can be manipulated experimentally to reveal the mechanisms involved. Some of these steps require energy in the form of either ATP or GTP, while others are energy-independent. The first step is the association of the protein that is to be transported with the outer chloroplast membrane. This is thought initially to involve energy-independent protein–lipid interactions of the transit peptide of the protein that is to be transportedwith themonogalactosyldiglyceride and digalactosyldiglyceride lipids that are the major lipid components of the chloroplast envelope. The next stage involves a series of energy-independent and energy-dependent protein–protein interactions.
Large multiprotein complexes are present in both the outer and the inner chloroplast membranes. The various proteins that are involved in the Toc and Tic complexes are identified by their molecular masses in kDa. The Toc complex consists of three membrane proteins:
Toc159
Toc75
Toc34
The Toc75 protein forms a narrow proteinconducting channel of 8–9 ˚A diameter, through which the unfolded protein is threaded through the outer membrane. The early stages of protein association and import through the Toc75 channel require GTP, while the later stages require ATP. Both the Toc159 and Toc34 proteins bind GTP. The GTP requirement may be primarily a gating function, to ensure that only the correct proteins enter the pathway. The functions of the Toc159 protein were earlier attributed to a Toc89 protein, which is a proteolytic fragment of the Toc159 protein. The ATP is probably utilized by molecular chaperones, Com70 and Hsp70 (heat shock protein of ). The Com70 protein is present on the cytoplasmic side of the chloroplast membrane and may help to stabilize the unfolded protein prior to translocation. In addition, Hsp70 is found in the intermembrane space between the inner and outer chloroplastmembranes, where it may help draw the proteins across the outer membrane. The Tic complex has a more transitory nature than does the stably assembled Toc complex and is assembled only when it associates with the Toc complex to form a supercomplex. The Tic/Toc supercomplex forms sites of adhesion between the
outer and inner chloroplast membranes, so that the imported proteins can be directly translocated across bothmembranes. The Tic complex consists of threemain proteins:
Tic22,
Tic110,
Tic20.
Tic22 is localized at the outer side of the inner membrane, and is the first component which the translocating protein associateswith after crossing the outer membrane. Tic20 is an integral membrane protein that is thought to be intimately involved in the translocation event itself. Tic110 is a transmembrane protein that has most of its mass on the stromal side of the innermembrane. It associates with another molecular chaperone, ClpC, which may help to pull the protein through the membrane in an ATP-dependent manner.
The next stop is the GroEL/GroES molecular chaperone, Cpn60, which helps the protein fold to its final conformation if it is a stromal protein. The stromal processing metalloprotease cleaves off the transit sequence. The protein can now have a number of possible fates. It can remain a stromal protein, be further translocated to the thylakoid lumen, or be assembled into a thylakoid membrane complex. Lumenal and thylakoid membrane targeting is surprisingly complex, with a minimum of four distinct pathways identified, each of which works with certain components but not with others
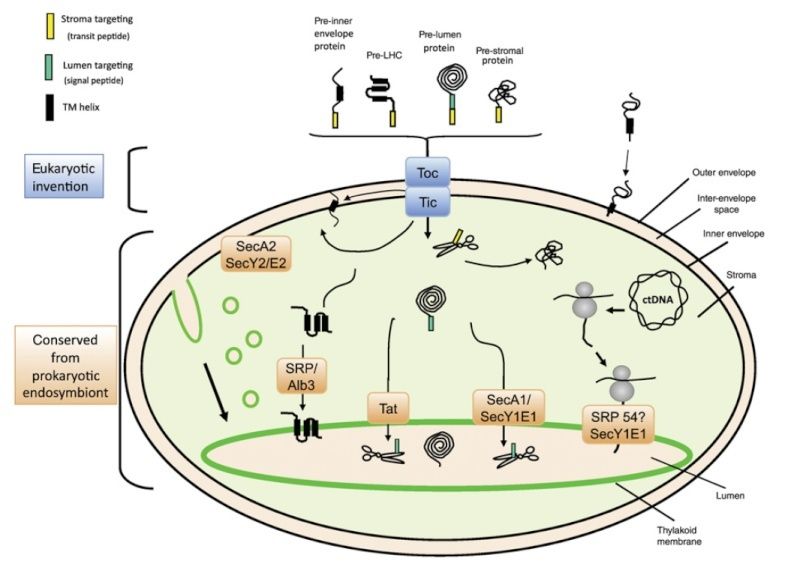
A model for the routing of lumenal targeted proteins and thylakoid integral membrane proteins. The four targeting pathways are shown, from left to right in the figure: the Sec pathway, which is ATP-dependent and ΔpH-stimulated; the signal recognition particle (SRP) pathway, which is GTP-dependent and ΔpH-stimulated; the ΔpH-dependent pathway; and the spontaneous pathway
One pathway is similar to the secretory (Sec) system found in many bacteria. It requires ATP hydrolysis and is responsible for translocating plastocyanin, the 33kDa protein of the PhotosystemII oxygen-evolving complex and the Photosystem I subunit that binds plastocyanin, the PsaF protein. The second pathway is similar to the signal recognition particles (SRP) that are involved in secretion by the endoplasmic reticulum in eukaryotes and many bacteria. It is a GTP dependent pathway and serves to insert the LHC antenna complexes into the thylakoid membrane. Both the Sec and the SRP pathways are stimulated by a pH difference across the thylakoid membrane, but the nucleotide triphosphates (NTP) are absolutely required. The third pathway operates only on ΔpH and has no NTP requirement. It is related to a bacterial pathway that has recently been discovered. An unusual aspect of the ΔpH pathway is that it can apparently translocate folded proteins without unfolding them. The 17 and 23 kDa oxygenevolving proteins utilize this pathway. The last pathwayfor protein import into thylakoids has no known energy or ΔpH requirement and appears to consist of spontaneous integration into the thylakoid membrane. For lumenal proteins with a second targeting sequence, it is cleaved off by a thylakoid protease in the lumen.
It is not clearwhy there is only a single dominant import pathway into the chloroplast – the Toc/Tic system – yet so many distinct protein import pathways into thylakoids. Some of the reasons for this pattern may lie in the specific aspects of particular proteins that utilize each pathway. For example, an integral membrane protein needs to avoid aggregation. This is accomplished by the SRP pathway. Similarly, the need to transport an already folded protein is compatible only with the ΔpH pathway .
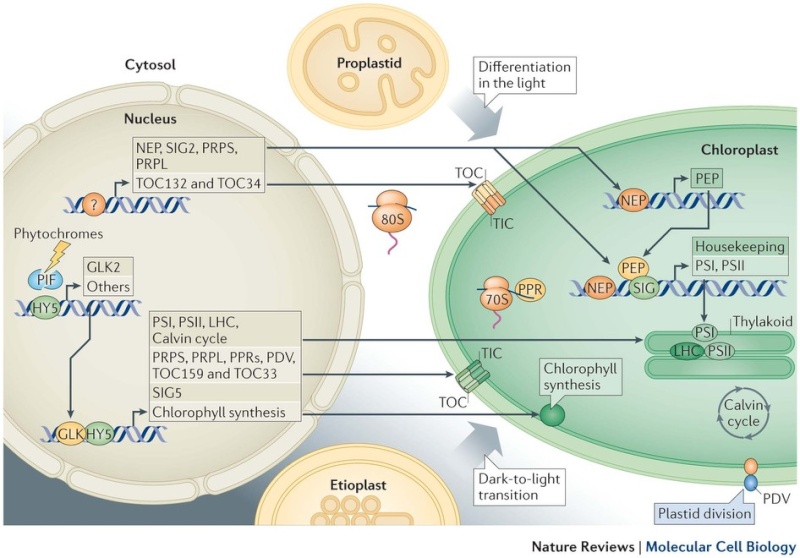
Developmentally, the bulk of information flows from the nucleus to plastids. Non-photosynthetic plant cells require basal operation of plastid housekeeping proteins that have been synthesized in the cytosol (by 80S ribosomes) and that have been imported through a housekeeping translocator (TOC132–TOC34, orange). Basal production of plastid ribosomal proteins (PRPS and PRPL, which are constituents of the 70S ribosome) also takes place. Factors driving and determining the extent of this initial plastid biogenesis are unknown (as indicated by the question mark). Plastid DNA expression is initiated by nucleus-encoded RNA polymerase (NEP); subsequently, plastid-encoded polymerase (PEP) becomes active. Proplastid-to-chloroplast differentiation during leaf development involves the expression of plastid genes that encode core components of the photosystems PSI and PSII, which is driven by PEP and involves regulators such as pentatricopeptide repeat proteins (PPRs) and abundant translational activity. Nucleus-encoded subunits, including those of the light-harvesting antenna complexes (LHC), assemble around these core components. Their import, in large quantities, uses a photosynthetic translocator (TOC159–TOC33; green), and is accompanied by extensive chloroplast growth and division (represented by the plastid division component PDV). Similar phenomena occur when chloroplasts develop from etioplasts following prolonged growth in darkness. Photosynthesis-related nuclear genes often possess G-box promoter elements. In the dark, the basic Leu zipper (bZIP) transcription factor HY5 is ubiquitylated and degraded, whereas basic helix–loop–helix (bHLH) PHYTOCHROME-INTERACTING FACTOR (PIF) regulators bind G-box elements and repress transcription. Photoreceptor activation causes PIF turnover and represses HY5 degradation; this enables its G-box binding and transcriptional activation. GLKs, which are Golden 2-like MYB transcription factors, are also major drivers of photosynthetic genes, with one (GLK2) being light-induced. Light- or circadian-mediated nuclear control of plastid DNA expression is exerted through σ-factors (SIG), which control PEP specificity (for example, SIG5).
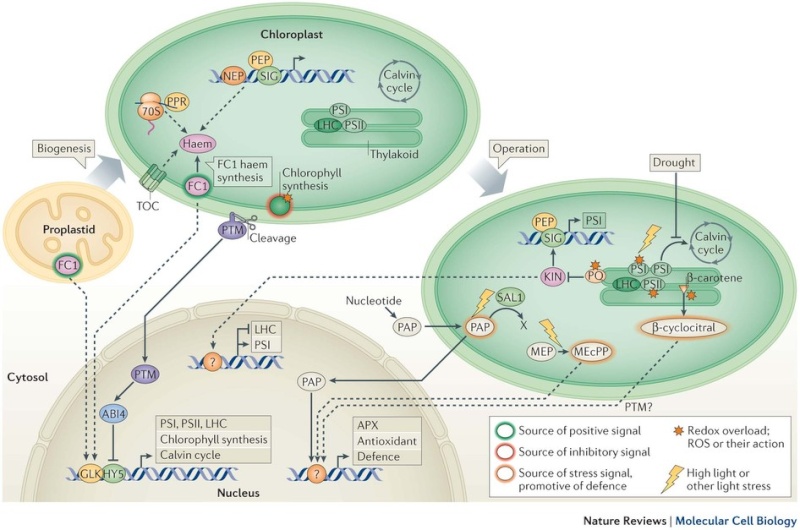
Signals that operate during chloroplast biogenesis report the readiness of the organelle to receive photosynthetic proteins. Haem synthesized by ferrochelatase 1 (FC1) is a positive signal that promotes the expression of nuclear genes encoding components of the photosynthetic machinery (probably indirectly; dashed arrow). Meanwhile, failure of some final steps of chlorophyll synthesis causes the repression of the same nuclear genes, possibly through a negative signal. Plastid photooxidation, and defective plastid transcription, protein synthesis or protein import may all have an impact on either or both of these positive (haem) and negative signals. Information from the negative, photooxidative signal, and from stresses which may impair the FC1-derived haem signal, is relayed to the nucleus (directly; solid arrow) via cleavage of the envelope-associated PTM (PHD WITH TRANSMEMBRANE DOMAINS) transcription factor. These phenomena affect nuclear gene expression, in part by reducing the activity of Golden 2-like MYB transcription factors (GLK) or by promoting the activity of ABI4 (ABSCISIC ACID INSENSITIVE 4; a competitor of G-box-binding factors such as HY5). During chloroplast photosynthetic operation, signals are triggered by environmental changes. Imbalances in the excitation of the photosystems PSI and PSII can cause an excessive reduction of plastoquinone (PQ); this signals plastid kinases (KIN) to orchestrate short-term responses, including modulating plastid DNA transcription (for example, KIN-dependent σ-factor 5 (SIG5) phosphorylation promotes PSI gene expression). Simultaneously, this regulates nuclear genes, which may include the genes that encode light-harvesting complex proteins (LHC). Nuclear genes that are coordinately repressed or activated by biogenic signals can be regulated in opposite directions by this mechanism. High light or loss of electron sinks in drought promote generation of reactive oxygen species (ROS), which leads to the production of β-cyclocitral, PAP (3′ phosphoadenosine- 5′ phosphate), MEcPP (methylerythritol cyclodiphosphate) and ABA (abscisic acid) (not shown). These factors activate antioxidant (including APX (ASCORBATE PEROXIDASE) and defence genes in the nucleus. Transcription factor targets of operational signals are largely unknown (represented by question marks). MEP, methyl-D-erythritol phosphate.
1) Molecular Mechanisms of Photosynthesis, Blankenship
Last edited by Admin on Wed Aug 19, 2015 9:30 am; edited 5 times in total