Light-independent reactions 1
https://reasonandscience.catsboard.com/t2164-the-calvin-benson-cycle
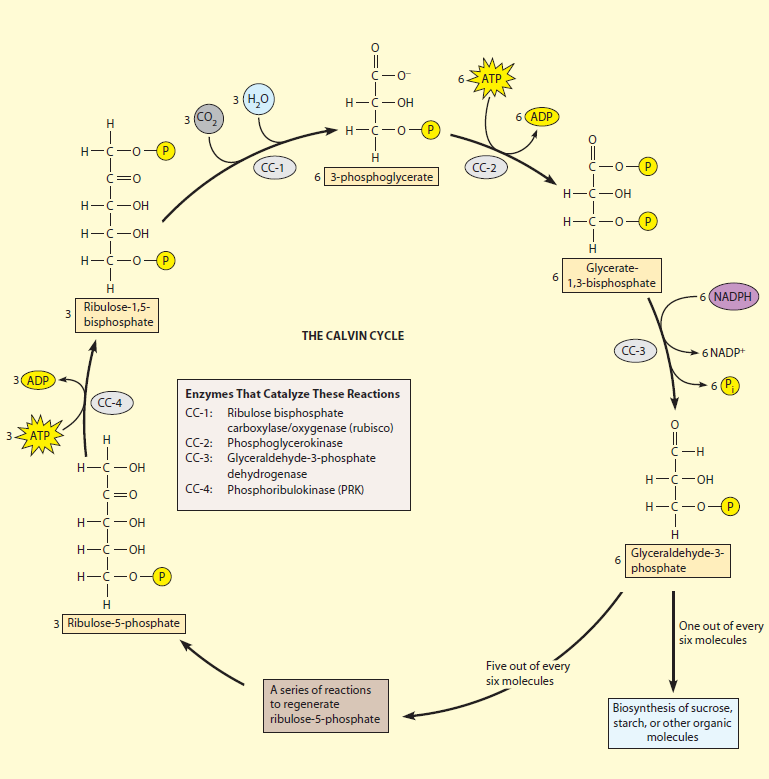
The light-independent reactions of photosynthesis are chemical reactions that convert carbon dioxide and other compounds into glucose. These reactions occur in the stroma, the fluid-filled area of a chloroplast outside of the thylakoid membranes. These reactions take the products (ATP and NADPH) of light-dependent reactions and perform further chemical processes on them. There are three phases to the light-independent reactions, collectively called the Calvin cycle: carbon fixation, reduction reactions, and ribulose 1,5-bisphosphate (RuBP) regeneration. Despite its name, this process occurs only when light is available. Plants do not carry out the Calvin cycle by night. They, instead, release sucrose into the phloem from their starch reserves. This process happens when light is available independent of the kind of photosynthesis (C3 carbon fixation, C4 carbon fixation, and Crassulacean Acid Metabolism); CAM plants store malic acid in their vacuoles every night and release it by day in order to make this process work.
The Calvin–Benson cycle is the primary photosynthetic carbon fixation pathway

Light-induced electron transport in oxygenic photosynthetic organisms generates ATP and NADPH, which are high-energy compounds of intermediate stability. They are stable against the sorts of rapid lossprocesses. However, they are not suitable for long-term storage of energy, such as building plant biomass, or for storage in seeds, tubers, or fruits. For these, it is necessary to convert the energy into a more stable and compact form. Most plants produce sugars or more complex carbohydrates such as starch for longterm
energy storage, although some plants produce large quantities of proteins or oils. Some of these conversions are carried out within the chloroplast, while in other cases they take place in the cell cytoplasm from building blocks that are exported from the chloroplast.
The following is a brief summary of each enzyme and its role in the regeneration of ribulose 1,5-bisphosphate in the order it appears in this specific phase.
1. RuBisCO catalyzes the carboxylation of ribulose-1,5-bisphosphate
2. phosphoglycerate kinase catalyzes the phosphorylation of 3-PGA by ATP
3. glyceraldehyde 3-phosphate dehydrogenase catalyzes the reduction of 1,3BPGA by NADPH
4. Triose phosphate isomerase: converts all G3P molecules into DHAP
5. Aldolase and fructose-1,6-bisphosphatase: converts G3P and DHAP into fructose 6-phosphate
6. Transketolase: removes two carbon molecules in fructose 6-phosphate to produce erythrose 4-phosphate (E4P); the two removed carbons are added to G3P to produce xylulose-5-phosphate (Xu5P)
7. Aldolase: converts E4P and a DHAP to sedoheptulose-1,7-bisphosphate
8. Sedoheptulase-1,7-bisphosphatase: cleaves the sedohetpulose-1,7-bisphosphate into sedoheptulase-7-phosphate (S7P)
9. Transketolase: removes two carbons from S7P and two carbons are transferred to one of the G3P molecules producing ribose-5-phosphate (R5P)and another Xu5P
10. Phosphopentose isomerase: converts the R5P into ribulose-5-phosphate (Ru5P)
11. Phosphopentose epimerase: converts the Xu5P into Ru5P
12. Phosphoribulokinase: phosphorylates Ru5P into ribulose-1,5-bisphosphate
https://bio.libretexts.org/Bookshelves/Microbiology/Book%3A_Microbiology_(Boundless)/5%3A_Microbial_Metabolism/5.12%3A_Biosynthesis/5.12E%3A_Regulation_of_the_Calvin_Cycle
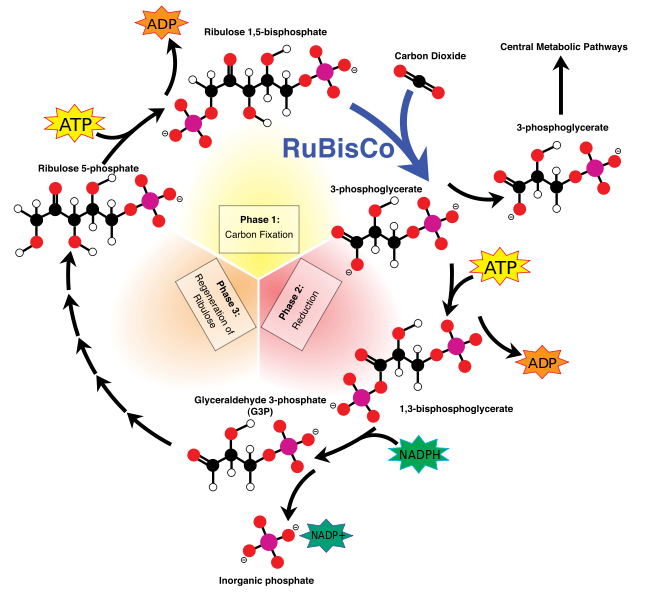
The metabolic pathway that incorporates carbon into plants by reduction of CO2 to sugars is known as the Calvin–Benson cycle, after the American chemists Melvin Calvin and Andrew Benson who, along with their associates, worked out the pathway in a brilliant series of experiments in the late 1940s and 1950s (Calvin, 1989, 1992; Benson, 2002). It is also sometimes called the reductive pentose phosphate, or RPP, cycle. The Calvin–Benson cycle is a complex series of chemical reactions that can seem intimidating at first. However, the overall cycle can be broken down into three phases, which are more easily understood . The three phases of carboxylation, reduction, and regeneration are schematically illustrated below.
and the reactions are summarized below:
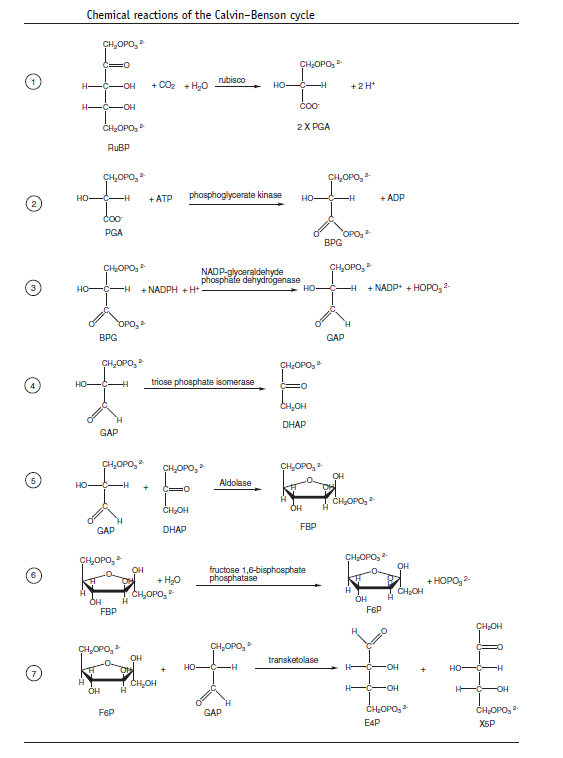
The carboxylation step generates PGA. The reduction phase uses the NADPH and some of the ATP produced by electron transport to reduce the PGA to triose phosphate. Most of the triose phosphate is used to regenerate RuBP in a complex set of reactions, while some is drawn off for starch synthesis in the chloroplast and sucrose synthesis in the cytoplasm.Most of the reactions that make up the Calvin–Benson cycle are identical to metabolic reactions that are well known in nonphotosynthetic tissues, including gluconeogenesis and the oxidative pentose phosphate cycle (Nelson and Cox, 2013). The enzymes that catalyze these steps are closely related to the ones that act in the nonphotosynthetic reactions, but are specialized for their role in the photosynthetic process, often in their method of regulation. The two unique reactions in the Calvin–Benson cycle that are not found in metabolic processes in most cells are the carboxylation reaction and the final step in the regeneration of RuBP.
RuBisCO , the carboxylation enzyme
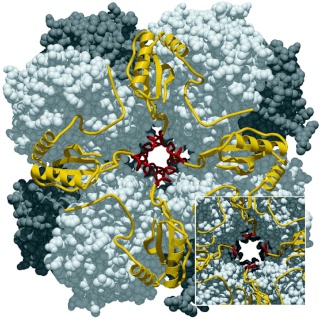
A remarkable enzyme called ribulose-bisphosphate carboxylase/oxygenase, or RuBisCO, carries out the carboxylation step in carbon fixation. RuBisCO is a complex enzyme that is highly regulated and exhibits two distinct activities: carboxylation and oxygenation. RuBisCO fromhigher plants andmost photosynthetic bacteria consists of eight copies each of large (L) subunits and small (S) subunits , giving an L8S8 quaternary structure The L subunits contain the catalytic site, which is shared between two L subunits, so the minimal active complex is an L2 dimer. Some purple photosynthetic bacteria contain two distinct types of RuBisCO complexes: both an L2 dimer and an L8S8 complex. The function of the S subunit is not well understood, but it may stabilize the larger L8S8 complex or in some way increase the catalytic efficiency. In the majority of eukaryotic photosynthetic organisms, the S subunits are nuclearencoded, whereas the L subunits are chloroplastencoded. Chaperonins, large protein-folding complexes, are essential for RuBisCO assembly. The energetics of carboxylation are very strongly exergonic, so carboxylation is essentially an irreversible reaction. The mechanism of the reaction is thought to involve spontaneous formation of an enediol form of RuBP. The enediol formation is followed by carboxylation,to give an unstable intermediate, 2-carboxy- 3-ketoarabinitol-1,5-bisphosphate, followed by its spontaneous breakdown into twomolecules of PGA, one of which contains the carbon atom that was derived from atmospheric CO2.
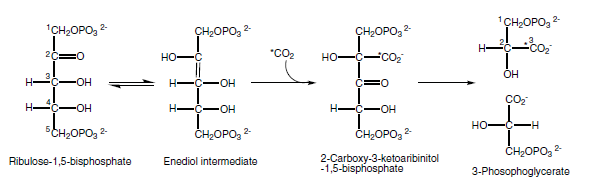
The chemical steps of carboxylation in RuBisCO. RuBP binds to the active site. Enediol formation takes place, promoted by the carbamate group on an active site lysine. CO2 is then added to form the 2-carboxy-3-ketoarabinitol-1,5-bisphosphate intermediate, which is then hydrolyzed to form two molecules of PGA. The newly added C atom is in the 3 position of one of the PGA molecules.
Before RuBisCO can become active catalytically, a lysine residue in the active site must be modified by carbamylation. In this process, a CO2 molecule (not one that will be fixed into photosynthate) reacts with the E-amino group of a specific lysine in the L subunit,
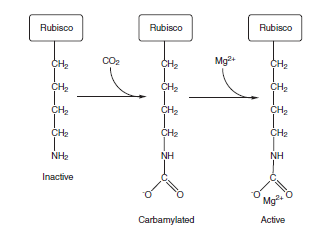
The carbamylation of lysine in RuBisCO. A CO2 molecule reacts with the E-amino group of a lysine residue in the active site in RuBisCO, which then binds Mg2+ to form the active enzyme. The negatively charged carbamylate group acts as a general base in the formation of the enediol.
A Mg2+ion coordinates to the carbamylated residue to generate the active form of the enzyme. The carbamate is thought to act as a general base, facilitating the formation of the enediol intermediate. RuBP binds very tightly to RuBisCO, changing the conformation of RuBisCO in a way that effectively blocks access to the active site carbamylated group. If RuBP binds before carbamylation takes place, the enzyme becomes “stuck” in an inactive state. Another enzyme, RuBisCO activase, promotes carbamylation of RuBisCO, probably by altering the structure of RuBisCO in a way that weakens the binding of RuBP to the nonactivated enzyme. Because this process requires ATP, the activity of RuBisCO changes with light intensity, so the rate of carboxylation is coordinately regulated with electron transport activity. In addition to RuBP RuBisCO activase also facilitates removal of other compounds that bind to the active site of RuBisCO, including carboxyarabinitol-1-phosphate, a naturally occurring inhibitor, as well as xylulose 1,5-bisphosphate, an unreactive isomer of RuBP that can form on the enzyme and inhibit its activity. The most important alternate reaction that RuBisCO carries out involves the addition of O2 instead of CO2 and is therefore called oxygenation, which leads to photorespiration. Oxygenation is a significant drain on a plant’s resources, because inmost plants it takes place approximately a third of the time instead of carboxylation. This process is shown below:
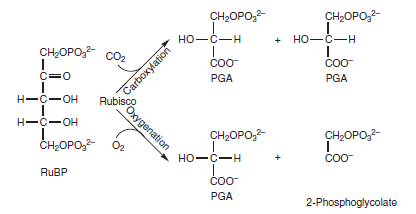
The carboxylation and oxygenation reactions of RuBisCO. RuBP reacts with CO2 to form two molecules of PGA. Alternatively, RuBP reacts with O2 to form one molecule of PGA and one molecule of 2-phosphoglycolate. The 2-phosphoglycolate is recovered by a complex series of reactions as described in Figure below:
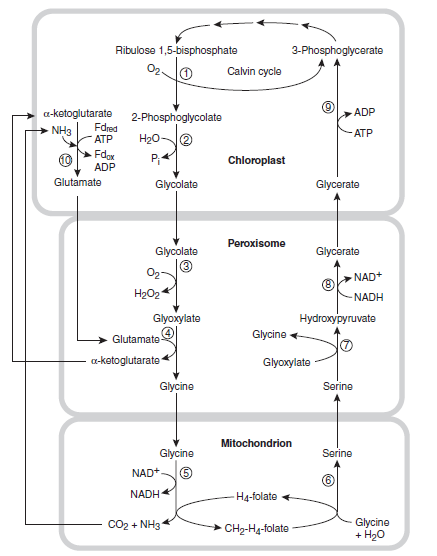
The photorespiratory cycle. The 2-phosphoglycolate formed in the oxygenation reaction is converted to glycolate, exported from the chloroplast, and is imported into the peroxisome, where it is metabolized into glycine. The glycine is exported from the peroxisome and taken up by the mitochondrion, where two molecules are combined and decarboxylated to form one molecule of serine. The serine is then transported to the peroxisome, where it is converted to glycerate and then reimported into the chloroplast and phosphorylated to form PGA.
Because CO2 and O2 are competitive substrates, the ratio of carboxylation to oxygenation reactions is determined by the catalytic rates and affinities of RuBisCO for the two substrates, weighted by the concentrations of O2 and CO2 in air. The enzyme turnover number kcat is the number of molecules of substrate that can be processed by the enzyme at saturating substrate concentration. The KM constant, or Michaelis constant, is the concentration of substrate at which the enzyme shows half the rate that it does at saturating substrate concentrations. The specificity factor is the kinetic preference of the enzyme for CO2 over O2. To obtain the specificity factor for a given substrate, it is first necessary to determine the ratio of kcat and KM for that substrate. The significance of kcat/KM in enzyme kinetics is that it is equal to the effective secondorder rate constant for the reaction of the enzyme with the substrate to form a product and is considered to be a measure of the catalytic efficiency of the enzyme . The specificity factor is then obtained by dividing the kcat/KM values for carboxylation by the corresponding values for oxygenation:
RuBisCO has a turnover number of only a fewCO2 molecules fixed per second for each enzyme molecule, even at saturating concentrations of CO2 . This means that in order to have a high rate of CO2 fixation, a high concentration of RuBisCO is needed. Remarkably, under typical conditions the concentration of RuBisCO in a chloroplast (∼4 mM) is asmuch as 1000 times higher than the concentration of one substrate, CO2, and is comparable with the concentration of the other substrate, RuBP. Consequently, as much as 50% of the soluble protein in a leaf is RuBisCO. It is very likely that RuBisCO is the most abundant protein on Earth.
The early Earth is thought to have been essentially anaerobic when RuBisCO was first selected by evolution as the carboxylation enzyme. As oxygen levels increased as a result of oxygenic photosynthesis, evolutionary selection pressure improved the performance of the enzyme.
Maybe the author will still elucidate us how exactly he thinks that could have happened.....
The selection of RuBisCO as the carboxylation enzyme for oxygenic photosynthesis is something of an enigma. Of all the known carboxylation enzymes (e.g., phosphoenolpyruvate carboxylase used in C4 and CAM), RuBisCO seems the least well adapted to cope with an oxygen-containing atmosphere
1) https://en.wikipedia.org/wiki/Light-independent_reactions
https://reasonandscience.catsboard.com/t2164-the-calvin-benson-cycle
The light-independent reactions of photosynthesis are chemical reactions that convert carbon dioxide and other compounds into glucose. These reactions occur in the stroma, the fluid-filled area of a chloroplast outside of the thylakoid membranes. These reactions take the products (ATP and NADPH) of light-dependent reactions and perform further chemical processes on them. There are three phases to the light-independent reactions, collectively called the Calvin cycle: carbon fixation, reduction reactions, and ribulose 1,5-bisphosphate (RuBP) regeneration. Despite its name, this process occurs only when light is available. Plants do not carry out the Calvin cycle by night. They, instead, release sucrose into the phloem from their starch reserves. This process happens when light is available independent of the kind of photosynthesis (C3 carbon fixation, C4 carbon fixation, and Crassulacean Acid Metabolism); CAM plants store malic acid in their vacuoles every night and release it by day in order to make this process work.
The Calvin–Benson cycle is the primary photosynthetic carbon fixation pathway

Light-induced electron transport in oxygenic photosynthetic organisms generates ATP and NADPH, which are high-energy compounds of intermediate stability. They are stable against the sorts of rapid lossprocesses. However, they are not suitable for long-term storage of energy, such as building plant biomass, or for storage in seeds, tubers, or fruits. For these, it is necessary to convert the energy into a more stable and compact form. Most plants produce sugars or more complex carbohydrates such as starch for longterm
energy storage, although some plants produce large quantities of proteins or oils. Some of these conversions are carried out within the chloroplast, while in other cases they take place in the cell cytoplasm from building blocks that are exported from the chloroplast.
The following is a brief summary of each enzyme and its role in the regeneration of ribulose 1,5-bisphosphate in the order it appears in this specific phase.
1. RuBisCO catalyzes the carboxylation of ribulose-1,5-bisphosphate
2. phosphoglycerate kinase catalyzes the phosphorylation of 3-PGA by ATP
3. glyceraldehyde 3-phosphate dehydrogenase catalyzes the reduction of 1,3BPGA by NADPH
4. Triose phosphate isomerase: converts all G3P molecules into DHAP
5. Aldolase and fructose-1,6-bisphosphatase: converts G3P and DHAP into fructose 6-phosphate
6. Transketolase: removes two carbon molecules in fructose 6-phosphate to produce erythrose 4-phosphate (E4P); the two removed carbons are added to G3P to produce xylulose-5-phosphate (Xu5P)
7. Aldolase: converts E4P and a DHAP to sedoheptulose-1,7-bisphosphate
8. Sedoheptulase-1,7-bisphosphatase: cleaves the sedohetpulose-1,7-bisphosphate into sedoheptulase-7-phosphate (S7P)
9. Transketolase: removes two carbons from S7P and two carbons are transferred to one of the G3P molecules producing ribose-5-phosphate (R5P)and another Xu5P
10. Phosphopentose isomerase: converts the R5P into ribulose-5-phosphate (Ru5P)
11. Phosphopentose epimerase: converts the Xu5P into Ru5P
12. Phosphoribulokinase: phosphorylates Ru5P into ribulose-1,5-bisphosphate
https://bio.libretexts.org/Bookshelves/Microbiology/Book%3A_Microbiology_(Boundless)/5%3A_Microbial_Metabolism/5.12%3A_Biosynthesis/5.12E%3A_Regulation_of_the_Calvin_Cycle
The metabolic pathway that incorporates carbon into plants by reduction of CO2 to sugars is known as the Calvin–Benson cycle, after the American chemists Melvin Calvin and Andrew Benson who, along with their associates, worked out the pathway in a brilliant series of experiments in the late 1940s and 1950s (Calvin, 1989, 1992; Benson, 2002). It is also sometimes called the reductive pentose phosphate, or RPP, cycle. The Calvin–Benson cycle is a complex series of chemical reactions that can seem intimidating at first. However, the overall cycle can be broken down into three phases, which are more easily understood . The three phases of carboxylation, reduction, and regeneration are schematically illustrated below.

and the reactions are summarized below:

The carboxylation step generates PGA. The reduction phase uses the NADPH and some of the ATP produced by electron transport to reduce the PGA to triose phosphate. Most of the triose phosphate is used to regenerate RuBP in a complex set of reactions, while some is drawn off for starch synthesis in the chloroplast and sucrose synthesis in the cytoplasm.Most of the reactions that make up the Calvin–Benson cycle are identical to metabolic reactions that are well known in nonphotosynthetic tissues, including gluconeogenesis and the oxidative pentose phosphate cycle (Nelson and Cox, 2013). The enzymes that catalyze these steps are closely related to the ones that act in the nonphotosynthetic reactions, but are specialized for their role in the photosynthetic process, often in their method of regulation. The two unique reactions in the Calvin–Benson cycle that are not found in metabolic processes in most cells are the carboxylation reaction and the final step in the regeneration of RuBP.
RuBisCO , the carboxylation enzyme
A remarkable enzyme called ribulose-bisphosphate carboxylase/oxygenase, or RuBisCO, carries out the carboxylation step in carbon fixation. RuBisCO is a complex enzyme that is highly regulated and exhibits two distinct activities: carboxylation and oxygenation. RuBisCO fromhigher plants andmost photosynthetic bacteria consists of eight copies each of large (L) subunits and small (S) subunits , giving an L8S8 quaternary structure The L subunits contain the catalytic site, which is shared between two L subunits, so the minimal active complex is an L2 dimer. Some purple photosynthetic bacteria contain two distinct types of RuBisCO complexes: both an L2 dimer and an L8S8 complex. The function of the S subunit is not well understood, but it may stabilize the larger L8S8 complex or in some way increase the catalytic efficiency. In the majority of eukaryotic photosynthetic organisms, the S subunits are nuclearencoded, whereas the L subunits are chloroplastencoded. Chaperonins, large protein-folding complexes, are essential for RuBisCO assembly. The energetics of carboxylation are very strongly exergonic, so carboxylation is essentially an irreversible reaction. The mechanism of the reaction is thought to involve spontaneous formation of an enediol form of RuBP. The enediol formation is followed by carboxylation,to give an unstable intermediate, 2-carboxy- 3-ketoarabinitol-1,5-bisphosphate, followed by its spontaneous breakdown into twomolecules of PGA, one of which contains the carbon atom that was derived from atmospheric CO2.

The chemical steps of carboxylation in RuBisCO. RuBP binds to the active site. Enediol formation takes place, promoted by the carbamate group on an active site lysine. CO2 is then added to form the 2-carboxy-3-ketoarabinitol-1,5-bisphosphate intermediate, which is then hydrolyzed to form two molecules of PGA. The newly added C atom is in the 3 position of one of the PGA molecules.
Before RuBisCO can become active catalytically, a lysine residue in the active site must be modified by carbamylation. In this process, a CO2 molecule (not one that will be fixed into photosynthate) reacts with the E-amino group of a specific lysine in the L subunit,

The carbamylation of lysine in RuBisCO. A CO2 molecule reacts with the E-amino group of a lysine residue in the active site in RuBisCO, which then binds Mg2+ to form the active enzyme. The negatively charged carbamylate group acts as a general base in the formation of the enediol.
A Mg2+ion coordinates to the carbamylated residue to generate the active form of the enzyme. The carbamate is thought to act as a general base, facilitating the formation of the enediol intermediate. RuBP binds very tightly to RuBisCO, changing the conformation of RuBisCO in a way that effectively blocks access to the active site carbamylated group. If RuBP binds before carbamylation takes place, the enzyme becomes “stuck” in an inactive state. Another enzyme, RuBisCO activase, promotes carbamylation of RuBisCO, probably by altering the structure of RuBisCO in a way that weakens the binding of RuBP to the nonactivated enzyme. Because this process requires ATP, the activity of RuBisCO changes with light intensity, so the rate of carboxylation is coordinately regulated with electron transport activity. In addition to RuBP RuBisCO activase also facilitates removal of other compounds that bind to the active site of RuBisCO, including carboxyarabinitol-1-phosphate, a naturally occurring inhibitor, as well as xylulose 1,5-bisphosphate, an unreactive isomer of RuBP that can form on the enzyme and inhibit its activity. The most important alternate reaction that RuBisCO carries out involves the addition of O2 instead of CO2 and is therefore called oxygenation, which leads to photorespiration. Oxygenation is a significant drain on a plant’s resources, because inmost plants it takes place approximately a third of the time instead of carboxylation. This process is shown below:

The carboxylation and oxygenation reactions of RuBisCO. RuBP reacts with CO2 to form two molecules of PGA. Alternatively, RuBP reacts with O2 to form one molecule of PGA and one molecule of 2-phosphoglycolate. The 2-phosphoglycolate is recovered by a complex series of reactions as described in Figure below:

The photorespiratory cycle. The 2-phosphoglycolate formed in the oxygenation reaction is converted to glycolate, exported from the chloroplast, and is imported into the peroxisome, where it is metabolized into glycine. The glycine is exported from the peroxisome and taken up by the mitochondrion, where two molecules are combined and decarboxylated to form one molecule of serine. The serine is then transported to the peroxisome, where it is converted to glycerate and then reimported into the chloroplast and phosphorylated to form PGA.
Because CO2 and O2 are competitive substrates, the ratio of carboxylation to oxygenation reactions is determined by the catalytic rates and affinities of RuBisCO for the two substrates, weighted by the concentrations of O2 and CO2 in air. The enzyme turnover number kcat is the number of molecules of substrate that can be processed by the enzyme at saturating substrate concentration. The KM constant, or Michaelis constant, is the concentration of substrate at which the enzyme shows half the rate that it does at saturating substrate concentrations. The specificity factor is the kinetic preference of the enzyme for CO2 over O2. To obtain the specificity factor for a given substrate, it is first necessary to determine the ratio of kcat and KM for that substrate. The significance of kcat/KM in enzyme kinetics is that it is equal to the effective secondorder rate constant for the reaction of the enzyme with the substrate to form a product and is considered to be a measure of the catalytic efficiency of the enzyme . The specificity factor is then obtained by dividing the kcat/KM values for carboxylation by the corresponding values for oxygenation:
RuBisCO has a turnover number of only a fewCO2 molecules fixed per second for each enzyme molecule, even at saturating concentrations of CO2 . This means that in order to have a high rate of CO2 fixation, a high concentration of RuBisCO is needed. Remarkably, under typical conditions the concentration of RuBisCO in a chloroplast (∼4 mM) is asmuch as 1000 times higher than the concentration of one substrate, CO2, and is comparable with the concentration of the other substrate, RuBP. Consequently, as much as 50% of the soluble protein in a leaf is RuBisCO. It is very likely that RuBisCO is the most abundant protein on Earth.
The early Earth is thought to have been essentially anaerobic when RuBisCO was first selected by evolution as the carboxylation enzyme. As oxygen levels increased as a result of oxygenic photosynthesis, evolutionary selection pressure improved the performance of the enzyme.
Maybe the author will still elucidate us how exactly he thinks that could have happened.....
The selection of RuBisCO as the carboxylation enzyme for oxygenic photosynthesis is something of an enigma. Of all the known carboxylation enzymes (e.g., phosphoenolpyruvate carboxylase used in C4 and CAM), RuBisCO seems the least well adapted to cope with an oxygen-containing atmosphere
1) https://en.wikipedia.org/wiki/Light-independent_reactions
Last edited by Admin on Wed Jul 22, 2020 5:36 pm; edited 6 times in total