https://reasonandscience.catsboard.com/t1437-chaperones
Molecular Chaperones Help Guide the Folding of Most Proteins
https://reasonandscience.catsboard.com/t1437-chaperones
Imagine machines, that fix machines, which help to fix other machines. All fully set up to operate in an independent, fully automated, robot-like manner. Evidently, such a process/system requires the highest sort of engineering artistry and foreknowledge of what could be wrong and how to fix the errors. That is what we see in life.
https://www.youtube.com/watch?v=--NcNeLc1mo
Proteins,marvelous pieces of chemical nanoengineering, in order to become functional, must fold from linear, to specific 3-D forms. Properly folded proteins are essential for life because they conduct most of the necessary functions in a cell. If it folds into the wrong shape, a protein is useless. If the first proteins fell into these death valleys, life on Earth would never have appeared. Spontaneous folding is quite rapid (milliseconds to seconds) for many proteins, but many large, critical proteins fail to find by themselves the right shape and, without help, would become only so much molecular waste.So when a protein misfolds, other proteins, named chaperonins, help proteins fold into the right shape. They have been shown to interact with up to 30% of the cell’s proteins, so their importance is real. Now, amazingly, even these very own proteins, named GroEL, which help misfolded proteins to fold properly, can also misfold. And it has been shown that GroES, a co-chaperone assists in folding GroEL. Amazing. Machines, that help other machines to assemble properly, are by themselves subject to errors, and life has inbuilt mechanisms to fix as well these machines that help fixing other machines !!
Most proteins probably do not fold correctly during their synthesis and require a special class of proteins called molecular chaperones to do so. Molecular chaperones are useful for cells because there are many different folding paths available to an unfolded or partially folded protein. Without chaperones, some of these pathways would not lead to the correctly folded (and most stable) form because the protein would become “kinetically trapped” in structures that are off-pathway. Some of these off-pathway configurations would aggregate and be left as irreversible dead ends of nonfunctional (and potentially dangerous) structures.
Molecular chaperones specifically recognize incorrect, off-pathway configurations by their exposure of hydrophobic surfaces, which in correctly folded proteins are typically buried in the interior. The binding of these exposed hydrophobic surfaces to each other is what causes off-pathway conformations to irreversibly aggregate. In some cases of inherited human diseases, aggregates do form and can cause severe symptoms and even death. Chaperones prevent this from happening in normal proteins by binding to the exposed hydrophobic surfaces using hydrophobic surfaces of their own. There are several types of chaperones; once bound to an incorrectly folded protein, they ultimately release it in a way that gives the protein another chance to fold correctly.
Cells Utilize Several Types of Chaperones
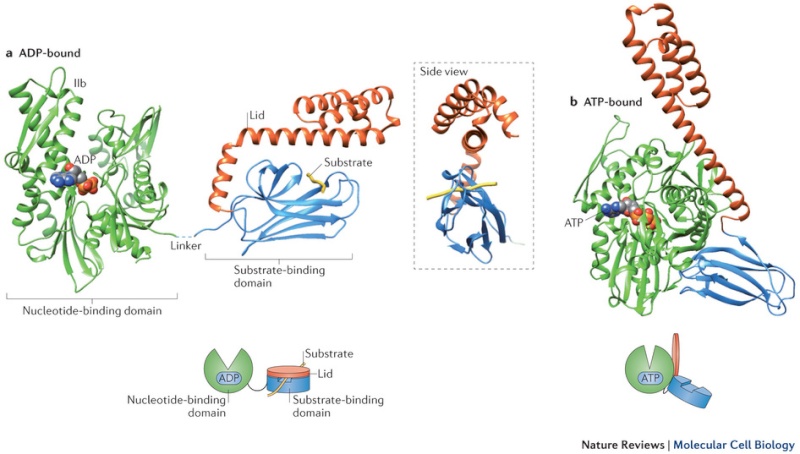
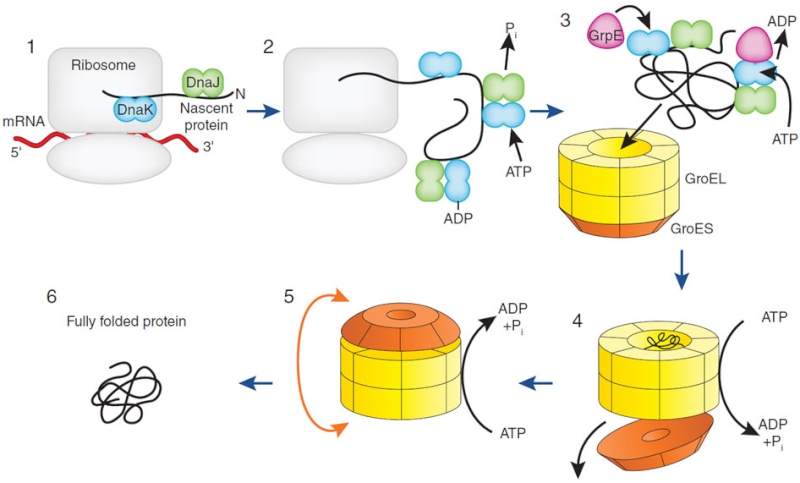
Many molecular chaperones are called heat-shock proteins (designated hsp), because they are synthesized in dramatically increased amounts after a brief exposure of cells to an elevated temperature (for example, 42°C for cells that normally live at 37°C). This reflects the operation of a feedback system that responds to an increase in misfolded proteins (such as those produced by elevated temperatures) by boosting the synthesis of the chaperones that help these proteins refold. There are several major families of molecular chaperones, including the hsp60 and hsp70 proteins. Different members of these families function in different organelles. Thus, mitochondria contain their own hsp60 and hsp70 molecules that are distinct from those that function in the cytosol; and a special hsp70 (called BIP) helps to fold proteins in the endoplasmic reticulum. The hsp60 and hsp70 proteins each work with their own small set of associated proteins when they help other proteins to fold. These hsps share an affinity for the exposed hydrophobic patches on incompletely folded proteins, and they hydrolyze ATP, often binding and releasing their protein substrate with each cycle of ATP hydrolysis. In other respects, the two types of hsp proteins function differently. The hsp70 machinery acts early in the life of many proteins (often before the protein leaves the ribosome), with each monomer of hsp70 binding to a string of about four or five hydrophobic amino acids ( see figure below )
The hsp70 family of molecular chaperones. These proteins act early, recognizing a small stretch of hydrophobic amino acids on a protein’s surface. Aided by a set of smaller hsp40 proteins (not shown), ATP-bound hsp70 molecules grasp their target protein and then hydrolyze ATP to ADP, undergoing conformational changes that cause the hsp70 molecules to associate even more tightly with the target. After the hsp40 dissociates, the rapid rebinding of ATP induces the dissociation of the hsp70 protein after ADP release. Repeated cycles of hsp binding and release help the target protein to refold.
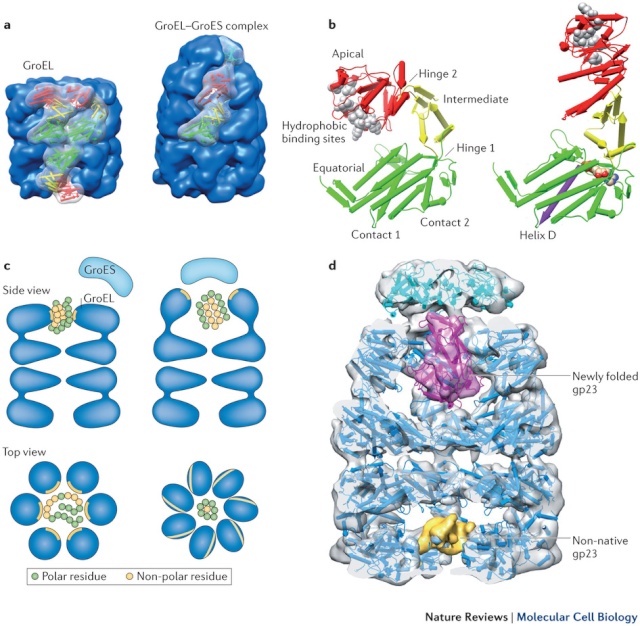
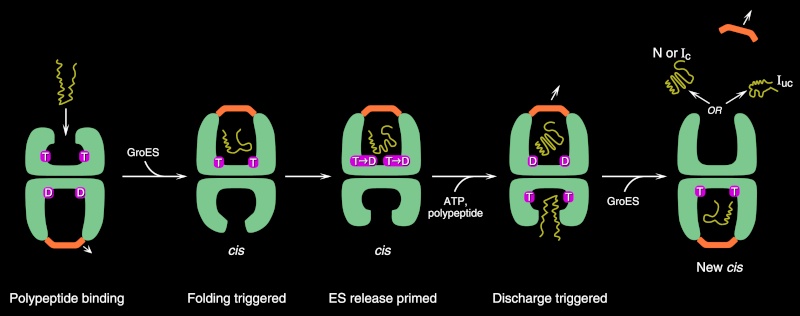
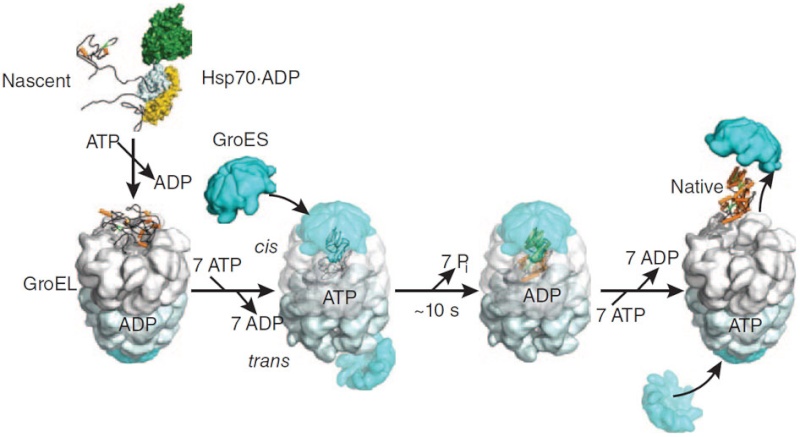
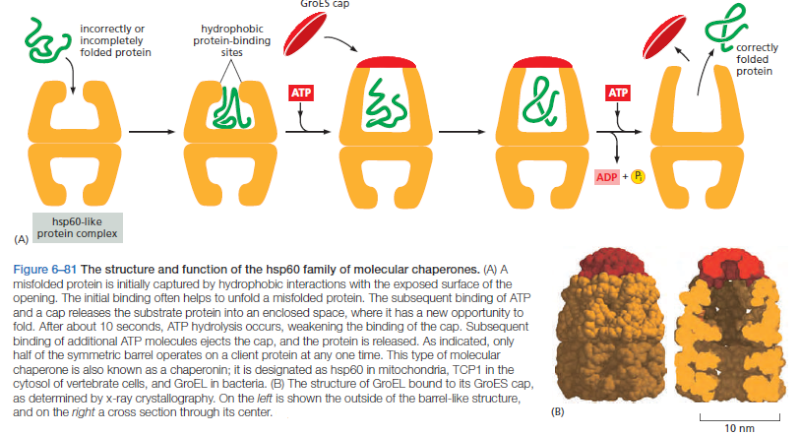
On binding ATP, hsp70 releases the protein into solution allowing it a chance to re-fold. In contrast, hsp60-like proteins form a large barrel-shaped structure that acts after a protein has been fully synthesized. This type of chaperone, sometimes called a chaperonin, forms an “isolation chamber” for the folding process (Figure below).
Starting at 41.13, this video showes nicely the amazing movement and functioning of this machine :
To enter a chamber, a substrate protein is first captured via the hydrophobic entrance to the chamber. The protein is then released into the interior of the chamber, which is lined with hydrophilic surfaces, and the chamber is sealed with a lid, a step requiring ATP. Here, the substrate is allowed to fold into its finalconformation in isolation, where there are no other proteins with which to aggregate. When ATP is hydrolyzed, the lid pops off, and the substrate protein, whether folded or not, is released from the chamber. Chaperones often need many cycles of ATP hydrolysis to fold a single polypeptide chain correctly. This energy is used to perform mechanical movements of the hsp60 and hsp70 “machines,” converting them from binding forms to releasing forms. Just as we saw for transcription, splicing, and translation, the expenditure of free energy can be used by cells to improve the accuracy of a biological process. In the case of protein folding, ATP hydrolysis allows chaperones to recognize a wide variety of misfolded structures, to halt any further misfolding, and to recommence the folding of a protein in an orderly way.
Question: Could this recognition be a mechanism that arose by natural means? If so, why would chance, or evolution or whatever natural mechanism be supposed, produce such a device, that performs in such an accurate way production control and repair and optimization? Is that not a goal-driven event, that has to be programmed and invented? Or does mindless matter have any goal-driven purposes? That's hard to fathom.
Although our discussion focuses on only two types of chaperones, the cell has a variety of others. The enormous diversity of proteins in cells presumably requires
a wide range of chaperones with versatile surveillance and correction capabilities
Exposed Hydrophobic Regions Provide Critical Signals for Protein Quality Control
If radioactive amino acids are added to cells for a brief period, the newly synthesized proteins can be followed as they mature into their final functional forms.
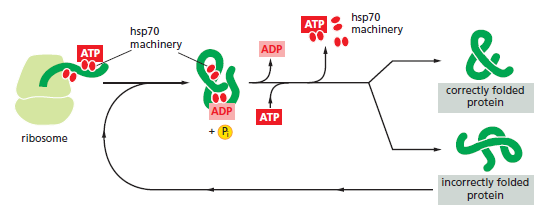
This type of experiment demonstrates that the hsp70 proteins act first, beginning when a protein is still being synthesized on a ribosome, and the hsp60-like proteins act only later to help fold completed proteins. We have seen that the cell distinguishes misfolded proteins, which require additional rounds of ATP-catalyzed refolding, from those with correct structures through the recognition of hydrophobic surfaces. Usually, if a protein has a sizable exposed patch of hydrophobic amino acids on its surface, it is abnormal: it has either failed to fold correctly after leaving the ribosome, suffered an accident that partly unfolded it at a later time, or failed to find its normal partner subunit in a larger protein complex. Such a protein is not merely useless to the cell, it can be dangerous.
Proteins that rapidly fold correctly on their own do not display such patterns and generally bypass the chaperones. For the others, the chaperones can carry out
“protein repair” by giving them additional chances to fold while, at the same time, preventing their aggregation.
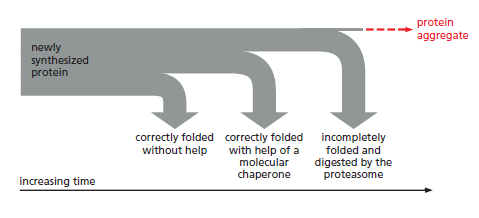
The processes that monitor protein quality following protein synthesis. A newly synthesized protein sometimes folds correctly and assembles on its own with its partner proteins, in which case the quality control mechanisms leave it alone. Incompletely folded proteins are helped to properly fold by molecular chaperones: first by a family of hsp70 proteins, and then, in some cases, by hsp60-like proteins. For both types of chaperones, the substrate proteins are recognized by an abnormally exposed patch of hydrophobic amino acids on their surface. These “protein-rescue” processes compete with another mechanism that, upon recognizing an abnormally exposed hydrophobic patch, marks the protein for destruction by the proteasome. The combined activity of all of these processes is needed to prevent massive protein aggregation in a cell, which can occur when many hydrophobic regions on proteins clump together nonspecifically.
The figure above outlines all of the quality-control choices that a cell makes for a difficult-to-fold, newly synthesized protein. As indicated, when attempts to refold
a protein fail, an additional mechanism is called into play that completely destroys the protein by proteolysis. This proteolytic pathway begins with the recognition of an abnormal hydrophobic patch on a protein’s surface, and it ends with the delivery of the entire protein to a protein-destruction machine, a complex protease
known as the proteasome. As described next, this process depends on an elaborate protein-marking system that also carries out other central functions in the cell by destroying selected normal proteins.
Chaperones are a marvel of molecular engineering:
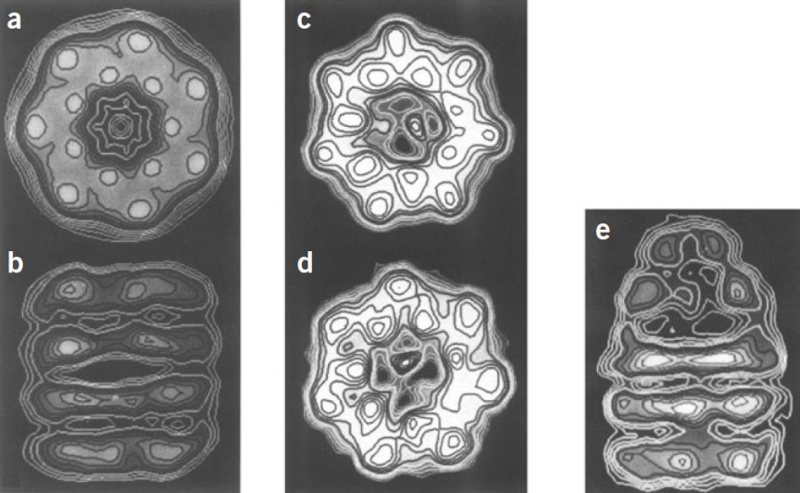
Molecular chaperones caught on film 1
'The movements we see in the videos suggest how the subunits can first bind the protein inside the cavity, potentially stretch it to undo incorrectly folded molecules, and then release it to allow it to find its own structure in the aqueous environment inside the cavity, while at the same time binding the GroES lid to prevent its premature escape,' Saibil explains. 'If the protein still displays hydrophobic patches after it is released from the chaperone, it will bind to a new GroEL ring and run through the cycle again. Once it is correctly folded it will be stable in the cell environment and will carry out its biological function.'
1) http://www.rsc.org/chemistryworld/News/2012/March/groel-groes-chaperone-caught-on-film.asp
more:
http://www.cryst.bbk.ac.uk/PPS2/course/section12/ellis.html
http://www.stanford.edu/group/frydman/web/
http://www4.utsouthwestern.edu/chuang_lab/project3.html
Last edited by Otangelo on Wed Sep 28, 2022 12:43 pm; edited 20 times in total