Crosstalk between proteins, organelles and cells: By evolution, or design ?
Cells typically communicate using chemical signals. These chemical signals, which are proteins or other molecules produced by a sending cell, are often secreted from the cell and released into the extracellular space. There, they can float – like messages in a bottle – over to neighboring cells. 2
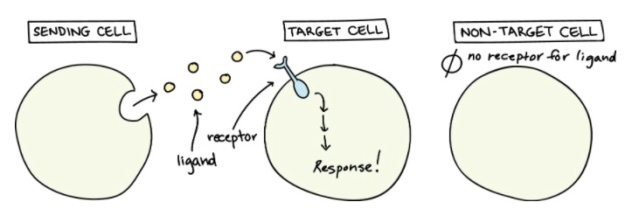
Not all cells can “hear” a particular chemical message. In order to detect a signal (that is, to be a target cell), a neighbor cell must have the right receptor for that signal. When a signaling molecule binds to its receptor, it alters the shape or activity of the receptor, triggering a change inside of the cell. Signaling molecules are often called ligands, a general term for molecules that bind specifically to other molecules (such as receptors).
The message carried by a ligand is often relayed through a chain of chemical messengers inside the cell. Ultimately, it leads to a change in the cell, such as alteration in the activity of a gene or even the induction of a whole process, such as cell division. Thus, the original intercellular (between-cells) signal is converted into an intracellular (within-cell) signal that triggers a response.
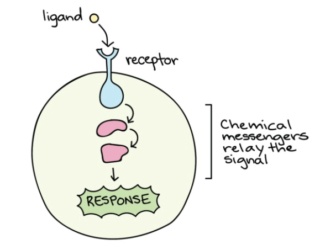
Inside a living cell, many important messages are communicated via interactions between proteins. For these signals to be accurately relayed, each protein must interact only with its specific partner, avoiding unwanted crosstalk with any similar proteins.
Crosstalk and the evolvability of intracellular communication 10 July 2017
https://www.nature.com/articles/ncomms16009
Metazoan signalling networks are complex, with extensive crosstalk between pathways. It is unclear what pressures drove the evolution of this architecture.
Comment: It comes to no surprise that the authors have no explanation of how evolution could account for communication in biology. Communication always requires a sender, a message, and a recipient. That is always an interdependent system, where all players have to be set up beforehand. If one of the participants is missing, communication cannot be established.
Werner R. Loewenstein: The Touchstone of Life Molecular Information, Cell Communication, and the Foundations of Life, page 176
To start, we will try to define the basic elements in this intercellular network in terms of information theory. That theory, specifically its branch, communication theory, specifies five stages for any communication system, regardless of its form: an information source, an encoder, a communication channel, a decoder (or receiver), and a user. The information source generates the information to be transmitted; the encoder transforms the information into a suitable message form for transmission over the communication channel; and the decoder performs the inverse operation of the encoder, or approximately so, for the user at the other end of the channel.
Signalling networks allow cells to process information from their environment and respond in appropriate ways to input signals. These networks are generally constructed from a set of interacting proteins; changes in the activity of these proteins across the network transmits the signal from the cell membrane to downstream elements, ultimately resulting (on average) in a particular phenotypic response (for example, proliferation, apoptosis or differentiation). Traditionally, these networks have been organized into ‘canonical pathways’ corresponding to sets of proteins that are involved in the transmission of a specific signal. The human signalling network includes pathways that are activated by insulin-like growth factor-I (IGF-I), Wnt or apoptotic signals. Although they are often studied separately, these pathways can demonstrate a high degree of ‘crosstalk,’ where proteins that are shared between two pathways cause one pathway’s activity to be modulated by the activity of another
Metazoan networks display incredible levels of crosstalk.
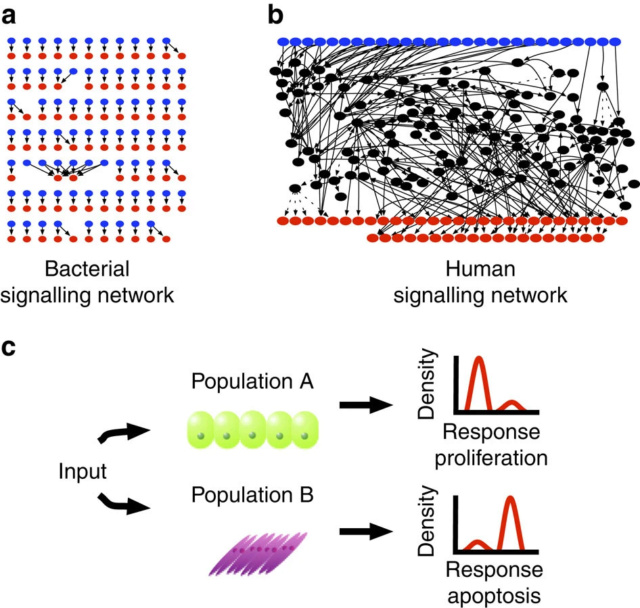
(a) Diagram of the bacterial TCS network. Blue nodes represent the inputs, sensor HKs, and red nodes represent the outputs, Response Regulators (RRs). Note the highly isolated nature of the TCS network: the majority of inputs only point to one, or at most two RR’s.
(b) Diagram of the human signalling network. This representation draws from six of the 29 canonical pathways from the KEGG pathways database3. Blue nodes are inputs, which are identified by searching for the keyword ‘Receptor’ in the UniProt entries of the genes associated with each node27. The red nodes are outputs, which are similarly identified by searching for the keyword ‘Transcription regulation’. Note that even with six canonical pathways the network is very interconnected and is difficult to globally comprehend.
(c) Two human cell populations from different cell lines are exposed to the same input. However, while the majority of Population A proliferates in response to the input, the majority of Population B undergoes apoptosis.
Molecular underpinnings of cytoskeletal cross-talk
Long considered independent, the microtubule and actin network systems are now known to engage in functional cross-talk to drive essential cellular processes such as whole-cell migration, mitotic spindle positioning, or cell-wide organelle transport. 3
It is becoming increasingly clear that proteins long-considered highly specific for one or the other cytoskeletal system do, in fact, make use of both filament types. How this functional duality of “shared proteins” has evolved and how their coadaptation enables cross-talk at the molecular level remain largely unknown.
1. https://news.mit.edu/2019/biologists-build-proteins-avoid-crosstalk-existing-molecules-1023
2. https://www.khanacademy.org/science/ap-biology/cell-communication-and-cell-cycle/cell-communication/a/introduction-to-cell-signaling#:~:text=Cells%20typically%20communicate%20using%20chemical,bottle%20–%20over%20to%20neighboring%20cells.
3. https://www.pnas.org/content/117/8/3944
Cells typically communicate using chemical signals. These chemical signals, which are proteins or other molecules produced by a sending cell, are often secreted from the cell and released into the extracellular space. There, they can float – like messages in a bottle – over to neighboring cells. 2

Not all cells can “hear” a particular chemical message. In order to detect a signal (that is, to be a target cell), a neighbor cell must have the right receptor for that signal. When a signaling molecule binds to its receptor, it alters the shape or activity of the receptor, triggering a change inside of the cell. Signaling molecules are often called ligands, a general term for molecules that bind specifically to other molecules (such as receptors).
The message carried by a ligand is often relayed through a chain of chemical messengers inside the cell. Ultimately, it leads to a change in the cell, such as alteration in the activity of a gene or even the induction of a whole process, such as cell division. Thus, the original intercellular (between-cells) signal is converted into an intracellular (within-cell) signal that triggers a response.

Inside a living cell, many important messages are communicated via interactions between proteins. For these signals to be accurately relayed, each protein must interact only with its specific partner, avoiding unwanted crosstalk with any similar proteins.
Crosstalk and the evolvability of intracellular communication 10 July 2017
https://www.nature.com/articles/ncomms16009
Metazoan signalling networks are complex, with extensive crosstalk between pathways. It is unclear what pressures drove the evolution of this architecture.
Comment: It comes to no surprise that the authors have no explanation of how evolution could account for communication in biology. Communication always requires a sender, a message, and a recipient. That is always an interdependent system, where all players have to be set up beforehand. If one of the participants is missing, communication cannot be established.
Werner R. Loewenstein: The Touchstone of Life Molecular Information, Cell Communication, and the Foundations of Life, page 176
To start, we will try to define the basic elements in this intercellular network in terms of information theory. That theory, specifically its branch, communication theory, specifies five stages for any communication system, regardless of its form: an information source, an encoder, a communication channel, a decoder (or receiver), and a user. The information source generates the information to be transmitted; the encoder transforms the information into a suitable message form for transmission over the communication channel; and the decoder performs the inverse operation of the encoder, or approximately so, for the user at the other end of the channel.
Signalling networks allow cells to process information from their environment and respond in appropriate ways to input signals. These networks are generally constructed from a set of interacting proteins; changes in the activity of these proteins across the network transmits the signal from the cell membrane to downstream elements, ultimately resulting (on average) in a particular phenotypic response (for example, proliferation, apoptosis or differentiation). Traditionally, these networks have been organized into ‘canonical pathways’ corresponding to sets of proteins that are involved in the transmission of a specific signal. The human signalling network includes pathways that are activated by insulin-like growth factor-I (IGF-I), Wnt or apoptotic signals. Although they are often studied separately, these pathways can demonstrate a high degree of ‘crosstalk,’ where proteins that are shared between two pathways cause one pathway’s activity to be modulated by the activity of another
Metazoan networks display incredible levels of crosstalk.

(a) Diagram of the bacterial TCS network. Blue nodes represent the inputs, sensor HKs, and red nodes represent the outputs, Response Regulators (RRs). Note the highly isolated nature of the TCS network: the majority of inputs only point to one, or at most two RR’s.
(b) Diagram of the human signalling network. This representation draws from six of the 29 canonical pathways from the KEGG pathways database3. Blue nodes are inputs, which are identified by searching for the keyword ‘Receptor’ in the UniProt entries of the genes associated with each node27. The red nodes are outputs, which are similarly identified by searching for the keyword ‘Transcription regulation’. Note that even with six canonical pathways the network is very interconnected and is difficult to globally comprehend.
(c) Two human cell populations from different cell lines are exposed to the same input. However, while the majority of Population A proliferates in response to the input, the majority of Population B undergoes apoptosis.
Molecular underpinnings of cytoskeletal cross-talk
Long considered independent, the microtubule and actin network systems are now known to engage in functional cross-talk to drive essential cellular processes such as whole-cell migration, mitotic spindle positioning, or cell-wide organelle transport. 3
It is becoming increasingly clear that proteins long-considered highly specific for one or the other cytoskeletal system do, in fact, make use of both filament types. How this functional duality of “shared proteins” has evolved and how their coadaptation enables cross-talk at the molecular level remain largely unknown.
1. https://news.mit.edu/2019/biologists-build-proteins-avoid-crosstalk-existing-molecules-1023
2. https://www.khanacademy.org/science/ap-biology/cell-communication-and-cell-cycle/cell-communication/a/introduction-to-cell-signaling#:~:text=Cells%20typically%20communicate%20using%20chemical,bottle%20–%20over%20to%20neighboring%20cells.
3. https://www.pnas.org/content/117/8/3944

