https://reasonandscience.catsboard.com/t2910-proteins-with-molybdenum-clusters-essential-for-life
Why, of all obscure elements known to man, would molybdenum be necessary for life? The biological versatility of Mo and W result not only from their redox-activity, ranging through oxidation states VI to IV, but because the
intermediate V valence state is also accessible, they can act as interfaces between one- and two-electron redox systems, which allows them to catalyze hydroxylation of carbon atoms using water as the ultimate source of oxygen.
Another factor which characterizes Mo (and W) enzymes is that, with the exception of bacterial nitrogenase, instead of having the metal itself directly coordinated to amino acid side chains of the protein, they contain a molybdenum pyranopterindithiolate cofactor (MoCo), which is the active component of their catalytic site. This cofactor coordinates the metal ion via a dithiolate side chain. The MoCo cofactor can exist in the fully oxidised (Mo(VI)) and fully reduced (Mo(IV)) forms, with some enzymes generating the (Mo(V)) form as a catalytic intermediate 17
The answer lies with the electrons that whirl around the molybdenum core. Like many other metals, molybdenum is eager to take an extra electron under its wings or give a spare one away. Life has eagerly exploited this ability to juggle electrons around. Cells not only incorporate molybdenum ions into their enzymes, but also zinc, copper, iron and nickel. Many of these metal containing proteins shuttle electrons between molecules, as if they are playing a massive game of hot potato, changing, breaking and building molecules along the way. Electrons truly are what makes life go round. Or, as the Hungarian Nobel prize winner Albert Szent-Györgyi put it: "Life is nothing but an electron looking for a place to rest". 15
Molybdenum is needed for at least three enzymes.
Sulfite oxidase catalyses the oxidation of sulfite to sulfate, necessary for metabolism of sulfur amino acids. 16
Xanthine oxidase catalyses oxidative hydroxylation of purines and pyridines including conversion of hypoxanthine to xanthine and xanthine to uric acid.
Aldehyde oxidase oxidises purines, pyrimidines, pteridines and is involved in nicotinic acid metabolism.
LUCA was supposedly an anaerobe, as long predicted by microbiologists. Its metabolism was replete with O2-sensitive enzymes. These include proteins rich in O2-sensitive iron-sulfur (FeS) clusters and enzymes that entail the generation of radicals (unpaired electrons) via S-adenosyl methionine (SAM) in their reaction mechanisms. That fits well with the 50-year-old but still modern view that FeS clusters represent very ancient cofactors in metabolism. 13
The Molybdo-enzyme superfamily as a whole, appear to have existed in LUCA. 14 A vast number of enzymes rely on metal cofactors for catalysis and/or redox conversions. CISM enzymes in LUCA likely performed energy conversion through the reduction of carbon dioxide, polysulfide or nitrate as well as from the oxidation of arsenite.
Numerous forms of life (including man) do not use proteins/enzymes with Tungsten metal atoms (W), and at least some species (Pyrococcales) do not use Molybdenum ( Mo) , but a cell that does not use either one is yet to be found. 9 Molybdenum (Mo) is a transition metal that plays an essential role in metabolism in the three domains of life. It is a trace element; living beings require it in small doses. The trace element molybdenum (Mo) is the catalytic component of important enzymes involved in global nitrogen, sulfur, and carbon metabolism in both prokaryotes and eukaryotes. 4 The trace element molybdenum (Mo) plays a critical role in several metabolic pathways and functions as a catalytic component of certain metalloenzymes that are essential for nearly all living organisms, including animals, plants, fungi and bacteria. In spite of that scarcity, molybdenum is essential to most organisms, from archaea and bacteria to higher plants and mammals, being found in the active site of enzymes that catalyze oxidation–reduction reactions involving carbon, nitrogen and sulfur atoms of key metabolites. Some of the molybdenum-dependent reactions constitute key steps in the global biogeochemical cycles of carbon, nitrogen, sulfur and oxygen, with particular emphasis on the atmospheric dinitrogen fixation (reduction) into organic ammonium (nitrogen cycle/nitrogenase enzyme).
While bacteria contain enzymes of all three enzyme families, enzymes of the DMSO reductase family are not present in eukaryotes. A significant number of bacteria and unicellular eukaryotes that do not require molybdenum, while all multi-cellular eukaryotes contain molybdoenzymes, which are essential for their viability.
Iron sulfur (Fe-S) clusters and the molybdenum cofactor (Moco) are present at enzyme sites, where the active metal facilitates electron transfer. They are required for respiration, DNA replication, transcription, translation, the biosynthesis of steroids, heme, catabolism of purines, hydroxylation of xenobiotics, and cellular sulfur metabolism. In the first known biochemical reactions on earth, molybdenum and iron-sulfur (Fe-S) clusters enabled electron transfers turning inorganic molecules into hydrogenated carbon molecules. 1 The majority of the Molybdo-enzyme superfamily as a whole, appear to have existed in LUCA. 2 Molybdenum (Mo) has during the last 2 decades been shown to constitute an essential cofactor in at least 3 distinct enzyme superfamilies, the most widespread of which is the so-called Complex Iron-Sulfur Molybdoenzyme (CISM) superfamily of molybdo-pterin containing enzymes.
The presence of the CISM superfamily in LUCA implies a vital role of its metal cofactors in early life. Mo's insolubility at neutral pH values, exacerbated by an anoxic atmosphere, suggested a low bioavailability of this element for early life. Certainly tungsten and most likely molybdenum ought to be added to the list of metals vital already to earliest life on Earth.
The only empirical way to deduce how life may have emerged is by taking the stance of assuming continuity of biology from its inception to the present day. 3
Involvement of molybdenum in purine metabolism is common to virtually all forms of life and only a small number of organisms use other mechanisms to oxidize xanthine (e.g. some yeasts), thus confirming the essential role of molybdenum for life on Earth. Molybdoenzymes are widespread in all domains of life and they catalyze key steps in carbon, sulfur and nitrogen metabolism.6 The physiological role of molybdoenzymes is fundamental since they are essential for most organisms and play a crucial role in fundamental biogeochemical cycles.
Mo-containing enzymes hold key positions both in the biogeochemical redox cycles of carbon, nitrogen and sulfur on Earth and in the metabolism of every the individual organism. 11
Molybdenum and Tungsten Enzymes Carola Schulzke, Martin L. Kirk, Russ Hille, page 6:
Both molybdo- and tungstoenzymes probably existed in the last universal common ancestor (LUCA).
There is a discrepancy of information, since in the coming text below, the authors claim that Molybden enzymes did substitute Tungsten enzymes at the great oxygenation event. What is it ?
The two cofactors that hold the metals in the enzymes active site would also have to have been present. This is particularly remarkable when we realize how elaborated the two cofactors are (particularly the nitrogenase one) and how “limited” their utilization compared to, for instance, porphyrin-related structures. Why do living organisms expend so much effort to use these metals in a (comparatively) small number of reactions? This effort (including synthesizing the protein machinery to scavenge the metals from the environment, producing and inserting the specialized cofactors and regulating the whole process) underscores how important both metals would have been, and still are to extant organisms, particularly in the case of molybdenum.
Another important aspect of molybdenum in biology can be seen in sulfite- oxidizing enzymes, which are used by almost all forms of life in the catabolism of sulfur-containing amino acids and other sulfur-containing compounds, oxidizing sulfite to sulfate.
More than 50 molybdenum-containing enzymes are known. The great majority are prokaryotic, with eukaryotes holding only restricted number of molybdoenzymes. When we think about the elements that are essential for life on Earth, we hardly ever consider molybdenum. The biological role of molybdenum can only be appreciated when put in perspective. Nitrogen is the fourth most abundant element in living organisms (only behind hydrogen, oxygen and carbon) and life on Earth depends on the nitrogen biogeochemical cycle to keep this element in forms that can be used by the organisms.Noteworthy, the “closing” of the nitrogen cycle, with the atmospheric dinitrogen fixation into ammonium depends virtually entirely on the trace element molybdenum : nitrogenase, a prokaryotic enzyme responsible for dinitrogen reduction to ammonium, requires one molybdenum atom in its active site.
The biologically available form of Mo is the oxyanion molybdate (MoO42−) from which organisms take up Mo along their daily life. Although only a minor constituent of the Earth’s crust, Mo is readily available due to its presence as a trace element in aquatic environments (as molybdate, MoO24−). The metal itself is biologically inactive unless it is complexed by a special cofactor. Mo is bound to a pterin, thus forming the molybdenum cofactor (Moco) which is the active compound at the catalytic site of all other Mo-enzymes.
The choice of molybdenum versus tungsten
Mo's insolubility at neutral pH values, exacerbated by an anoxic atmosphere1, suggested a low bioavailability of this element for early life1,3. Mo-isotope analyses on samples from the Archaean era indeed show substantially lower levels than during Phanerozoic times 22
Two scenarios can reconcile the results of molecular phylogeny and paleogeochemistry.
1. The ancestral CISM enzyme exclusively used W which was later replaced by Mo.
2. CISM-catalyzed reactions in early life used Mo supplied by alkaline hydrothermal vents, proposed as cradles for life23. The exclusiveness for Mo of many CISM-members as well as findings that primary productivity involving Mo has been comparable to the present since the geological record began at 3.8 Ga 24 lead us to favor the second scenario.
In oxic aqueous systems, these metals exist in the form of their tetrahedral MoO42− or WO42− oxyanions, and are therefore easily mobilizable into enzyme systems with the important caveat that they must be distinguished during enzyme maturation to ensure proper redox function and therefore catalysis. It is claimed that prior to approximately 2.3 billion years ago the earth’s biosphere was essentially anoxic, high in sulfur, and highly reducing.Mo and W have broadly equivalent natural abundance in the earth’s crust, but sulfides of W are soluble in water whereas those of Mo are not.
Most microbes cannot distinguish tungstate from molybdate, and substitution of one for the other usually affects enzymic activity. Tungstate similarly inactivates many enzymes with molybdopterin cofactors. In a few cases, molybdenum replaces tungsten functionally and vice versa. For example, tungsten can replace molybdenum in the catalytic center of Rhodobacter capsulatus dimethylsufoxide reductase to give an active enzyme that accesses the same range of oxidation states as the molybdenum enzyme 10 Tungstoenzymes predominantly occur in thermophilic and hyperthermophilic organisms in the specialised niche of oceanic hydrothermal vents
Moco is very labile and sensitive to air oxidation.
As a result,W would have been bioavailable in the primordial earth’s biosphere whereas Mo would not. Following the emergence of biological water oxidation, photosynthetic organisms supposedly caused a dramatic increase in ambient redox potential that paralleled the increase in atmospheric oxygen, and this resulted in the appearance and bioavailability of Mo in the form of MoO42−. As a consequence, the bioavailability of the two elements were reversed, with Mo being present at a concentration in sea water at 100 times that of W. It is claimed that this idea is supported by the significant correlation between archaeal life and W-biochemistry. The name archaeon (previously: archaebacterium) is of course intended to transmit the notion that these forms of life are thought to be the most similar to ‘primitive’ life as it must have existed in early geological times not long after the appearance of the first living cell. Most archaea are anaerobes (or perhaps microaerophiles) and the link may simply be one of mutual exclusion of molecular oxygen and tungsten biochemistry.
Both elements are able to assemble into mononuclear molybdoenzymes in an essentially identical manner. This presents a critical biological problem, because redox reactions catalyzed by W typically occur at much lower potentials than those catalyzed by Mo, and the active sites of the enzymes have to modulate the redox properties of their cognate metal ion. Thus, assembly of W in place of Mo in a bona fide molybdoenzyme would elicit dramatic changes in catalytic efficacy.
The toxicity of W towards molybdoenzymes and the inferred toxicity of Mo towards tungstoenzymes present cells with a serious problem: toxicity of the antagonist oxyanion via incorrect metal insertion. Mo and W have the same atomic radii (1.75 ˚A and 1.78A˚ , respectively), the same electronegativity, the same free energy of solvation (−226.8 kcal mol−1 and −230.1 kcal mol−1, respectively) and the same covalent solution radii (2.75 ˚A and 2.83A˚ , respectively). In order to distinguish them, exquisitely discriminating systems are necessary at the levels of metal uptake into bacterial cells.
Molybdo and tungsto-enzymes coexist in some bacteria, which must be able to discriminate between the two anions.
In some microorganisms (mostly thermophilic archaea), tungsten (W) is also coordinated by pyranopterin (Wco). W can still today be selectively transported into prokaryotic cells by certain transporters. Some organisms can distinguish between tungstate and molybdate. In the thermophilic methanogenic Archaea Methanobacterium wolfei and M. thermoautotrophicum, the enzyme formylmethanofuran dehydrogenase catalyzes the first step in methane formation from carbon dioxide. Both organisms synthesise distinct tungsten- and molybdenum-containing enzymes. Expression of the tungstoenzyme is constitutive. Molybdate induces expression of the molybdoenzyme, whereas tungstate does not. Some organisms have a high-affinity ABC transport system that specifically transports tungstate in the presence of molybdate. In the Gram-positive anaerobe Eubacterium acidaminophilum, which contains at least two tungstoenzymes, an equimolar amount of molybdate does not affect the transport of tungstate.
The basis for this extraordinary discrimination between tungstate and molybdate is not yet known, but it could depend on differences in pK between molybdate and tungstate, as the latter is more basic.
In order to understand, how the transition from Tungsten to Molybdenum could have ocurred, it has to be elucidated how the two metals are recognized and imported into the Cell.
The bioinorganic chemistry of tungsten 5
Tungsten is the bioelement with the highest atomic number,74. Tungsten is widely distributed in biology, however, it is not a universal bioelement. For some species tungsten is essential: their life depends on the presence of the element. Molybdenum is in many ways the twin element of tungsten. Also in biology the coordination chemistries of W and Mo are similar in structural and functional aspects. It has been suggested that in an evolutionary sense tungsten is an ‘old’ element on its way to be replaced by ‘modern’ molybdenum.
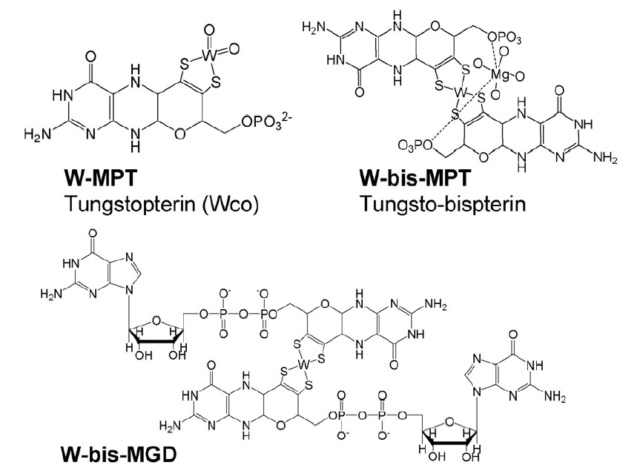
The structure of tungsten-containing pterin b cofactors and intermediates: tungsten-containing metal binding pterin (W-MPT) (I), tungsto-bispterin (W-bis- MPT) (II), tungsto-bispterin guanine dinucleotide (W-bis-MGD) (III)
In general, the biochemistry of a metal in a monocellular organism encompasses several processes: sequestering and transport over the cytoplasmic membrane, storage and release, metal-cofactor biosynthesis, metalloenzyme catalysis, and metal-controlled regulation of transcription and/or translation.
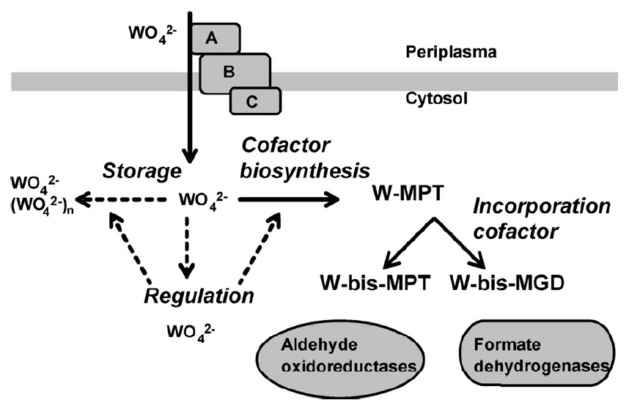
A schematic overview of the stages in cellular metabolism of tungsten (in various chemical forms): uptake, storage, regulation, cofactor biosynthesis, and incorporation in enzymes. Dashed arrows correspond to hypothetical processes based on cellular processes known for molybdate.
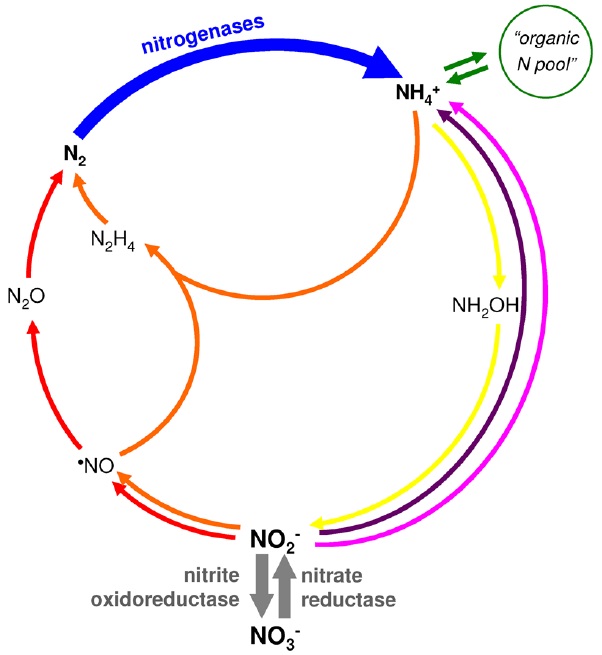
Biochemical cycle of nitrogen.
Dinitrogen fixation, blue arrow; “organic nitrogen pool”, green arrows; assimilatory ammonification, pink arrow; dissimilatory nitrate reduction to ammonium, violet arrow; nitrification, yellow arrows; denitrification, red arrows; anaerobic ammonium oxidation (AnAmmOx), orange arrows. The steps catalyzed by molybdenum-containing enzymes are highlighted with thick arrows, nitrogenase (blue), nitrate reductase and nitrite oxidoreductase (grey).
With this wide perspective in mind, the molybdenum biological role certainly assumes another dimension. In fact the lack of molybdenum, while hampering the existence of an efficient nitrogenase, would have been one of the limiting factors for life to emerge. The involvement of molybdenum in the nitrogen cycle is not restricted to the dinitrogen fixation, as the element is also essential for the reduction of nitrate to nitrite and for the oxidation of nitrite to nitrate, both processes being exclusively dependent (as far as is presently known) on the molybdenum- containing enzymes nitrate reductases (from both prokaryotic and eukaryotic sources) and nitrite oxidoreductases (from prokaryotes only).
Noteworthy, molybdenum is also essential for nitrite reduction to nitric oxide for biological signalling purposes. Nitric oxide is a signalling molecule involved in several physiological processes, in both prokaryotes and eukaryotes, and nitrite is presently recognized as a nitric oxide source particularly relevant to cell signalling and survival under challenging conditions.
The primitive carbon cycle would have also been dependent on molybdenum, as the metal (together with tungsten)would have been essential for the earliest, strictly anaerobic, organisms to handle aldehydes and carboxylic acids, catalyzing their interconversion.
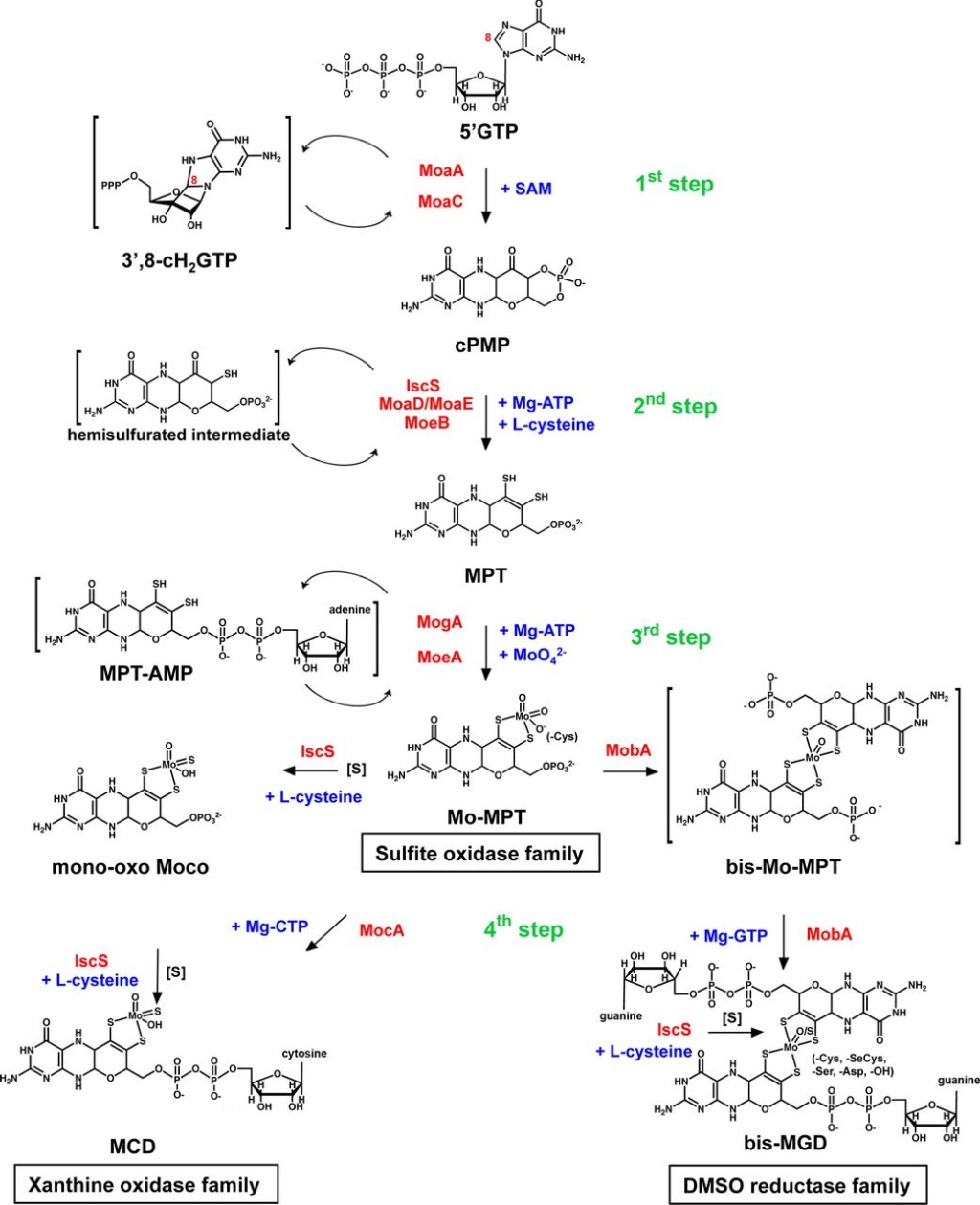
With the exception of nitrogenase containing the iron–molybdenum cofactor, all other molybdoenzymes possess in their active site a molybdenum atom coordinated to a dithiolene group on the 6-alkyl side chain of a pterin called molybdopterin (MPT) . 74% of bacteria representing almost all phyla utilize the molybdenum cofactor.
The different Moco-containing enzymes
One type of cofactor is the iron–sulfur-cluster-based iron-Mo cofactor, which is found only once in nature, namely in bacterial nitrogenase. The other type of cofactor is the pterin-based Mo-cofactor (Moco) that, in different variants, forms part of the active centers of all Mo-enyzmes in living organisms. Mo has a versatile redox chemistry that is used by the enzymes to catalyze diverse redox reactions. This redox chemistry is controlled both by the different ligands at the Mo atom and by the enzyme environment. Mo is very abundant in the oceans in the form of the molybdate anion.
four families based on the nature of the cofactor and other ligands bound in their active centres in the oxidized Mo(VI) forms

Molybdenum- and tungsten-containing cofactors and enzymes
(A) Chemical structures of molybdenum cofactor (Moco), Mo/W-bis pyranopterin guanosine dinucleotide (PGD) and W-bis pyranopterin cofactor.
(B) Domain structures of eukaryotic Mo enzymes of the xanthine oxidase (XO) and sulfite oxidase (SO) families. D, dimerization domain.
The first group is termed the xanthine oxidase (XO) family and features a single pterin cofactor and terminal sulfido and oxido-ligands.
The second group comprises the sulfite oxidase (SO) family and carries a single cofactor, a cysteine ligand and two oxido-ligands.
The third group is the dimethyl sulfoxide (DMSO) reductase family and harbours a bis-pyranopterin guanine dinucleotide cofactor and two oxido-ligands plus, and depending on the particular enzyme, an extra cysteine, seleno-cysteine, serine or aspartate ligand.
The fourth group comprises the archaeal aldehyde oxidoreductase family that harbours a bis-pyranopterin cofactor preferentially bound to a W atom.
Five molybdo-enzymes are found in eukaryotes and belong to only two of the families (Figure B):
nitrate reductase (NO), SO and the amidoximereducing component (mARC) are members of the SO family, while
XO/xanthine dehydrogenase (XDH) and aldehyde oxidase (AO) form part of the XO family.
Molybdo-enzymes (mainly bacterial) are involved in key processes in the global carbon, nitrogen and sulfur cycles, such as nitrate reduction, sulfite detoxification and purine catabolism.
Nitrate Reductase
Eukaryotic NR (EC 1.6.6.1) is a cytoplasmic, water-soluble enzyme involved in the reduction of nitrate to nitrite as the first step of assimilation of nitrogen in plants, algae and fungi.
Sulfite Oxidase
SO (EC 1.8.3.1) is essential in sulfur catabolism. In vertebrates, it catalyses the two-electron oxidation of sulfite to sulfate coupled to the reduction of two molecules of cytochrome c.37 Sulfite oxidation is the terminal step in the
oxidative degradation of cysteine.
Xanthine Dehydrogenase and Oxidase
Eukaryotic XDH/XO (EC 1.17.1.4) systems participate in the degradation of purines by oxidation of hypoxanthine to xanthine and xanthine to uric acid. The enzyme can function either as a dehydrogenase using NAD1 as electron
acceptor or, upon reversible cysteine oxidation, as an oxidase using dioxygen as terminal electron acceptor.
Aldehyde Oxidase
AO enzymes (EC 1.2.3.1) originate from a duplication of the xdh gene in eukaryotes before the origin of multicellularity.49 Consequently, both enzymes contain the same cofactor-binding domains (Fe-S clusters, FAD and
Moco) as well as a dimerization domain (Figure B).
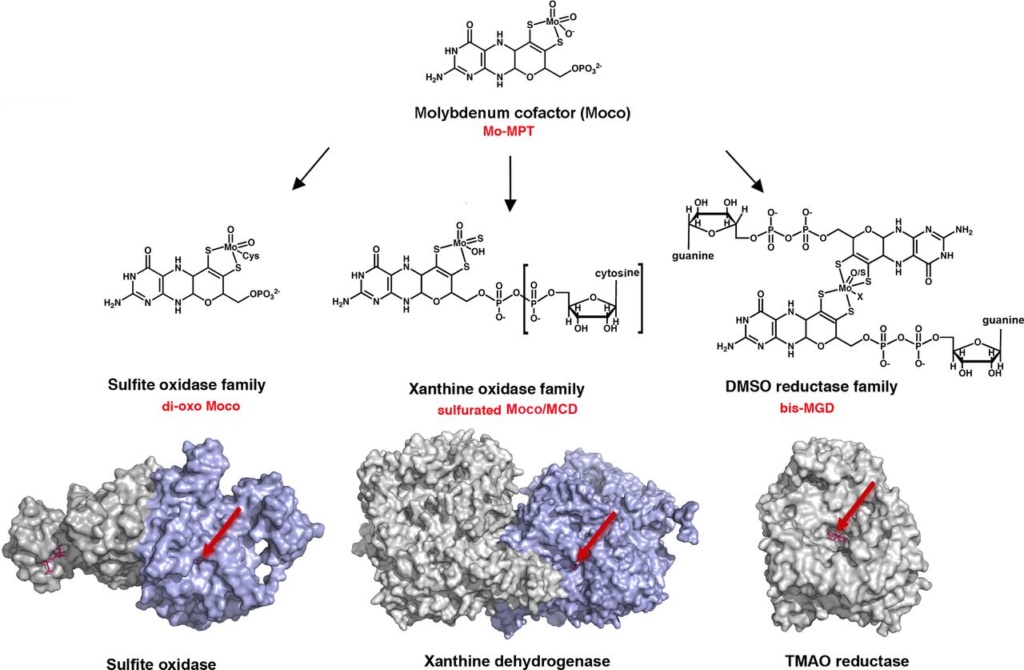
The families of molybdoenzymes.
The cofactor is not located on the surface of the protein, but is buried deeply within the interior of the enzyme and a tunnel-like structure makes it accessible to the cognate substrates.
The basic form of Moco is a 5,6,7,8-tetrahydropyranopterin, named Mo-MPT, which coordinates the molybdenum atom by the characteristic dithiolene group at the C1′ and C2′ positions of the pyranopterin ring. Mo-MPT (shown in the tri-oxo structure ) can be further modified and three different molybdenum-containing enzyme families are classified according to their coordination at the molybdenum atom: the XO, SO and DMSO reductase families. The SO family is characterized by a MPT-MoVIO2Cys ligand sphere. The XO family contains a MPT-MoVIOS(OH) core. Here, the MPT core can be modified by an additional CMP nucleotide at the phosphate group, forming MCD. The DMSO reductase family contains a MGD2-MoVIXY core with X being either a sulfur or an oxygen ligand and Y either being a hydroxo or amino acid ligand (Ser, Cys, Sec and Asp ligands were identified so far). Shown are structures of enzymes representative enzymes from each family: chicken sulfite oxidase (pdb 1SOX), bovine xanthine dehydrogenase (pdb 1FIQ) and S. massilia TMAO reductase (pdb 1TMO). The surface representations show that the Moco in each enzyme is deeply buried at the end of a funnel-like passage, giving access only to the substrate molecules (entrance site is shown by the arrow).
In eukaryotes, the most prominent Mo-enzymes are
(1) sulfite oxidase, which catalyzes the final step in the degradation of sulfur-containing amino acids and is involved in detoxifying excess sulfite,
(2) xanthine dehydrogenase, which is involved in purine catabolism and reactive oxygen production,
(3) aldehyde oxidase, which oxidizes a variety of aldehydes and is essential for the biosynthesis of the phytohormone abscisic acid, and in autotrophic organisms also
(4) nitrate reductase, which catalyzes the key step in inorganic nitrogen assimilation.
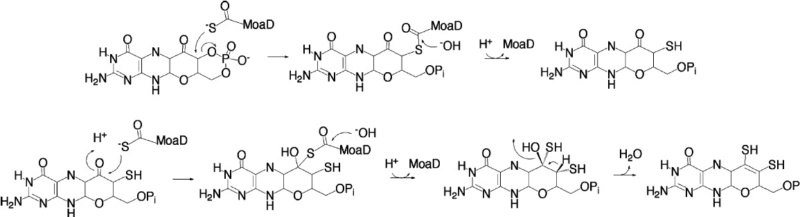
All Mo-enzymes, except plant sulfite oxidase, need at least one more redox active center, many of them involving iron in electron transfer. The biosynthesis of Moco involves the complex interaction of six proteins and is a process of four steps, which also includes iron as well as copper in an indespensable way. Moco as released after synthesis is likely to be distributed to the apoproteins of Mo-enzymes by putative Moco-carrier proteins. Xanthine dehydrogenase and aldehyde oxidase, but not sulfite oxidase and nitrate reductase, require the postranslational sulfuration of their Mo-site for becoming active. This final maturation step is catalyzed by a Moco-sulfurase enzyme, which mobilizes sulfur from l-cysteine in a pyridoxal phosphate-dependent manner as typical for cysteine desulfurases. 11
How does it work, what does it do
Mo-enzymes generally catalyze the transfer of an oxygen atom, ultimately derived from or incorporated into water, to or from a substrate. Each reaction, either reduction or oxidation, involves the transfer of two electrons, thereby causing a change in the oxidation state of the Mo atom in the substrate-binding site from IV to VI or vice versa.
The task of the cofactor is to position the catalytic metal Mo correctly within the active center, to control its redox behaviour and to participate with its pterin ring system in the electron transfer to or from the Mo atom. 11 The pterin with its several possible reduction states as well as different structural conformations might also be important in channeling electrons from or to other prosthetic groups
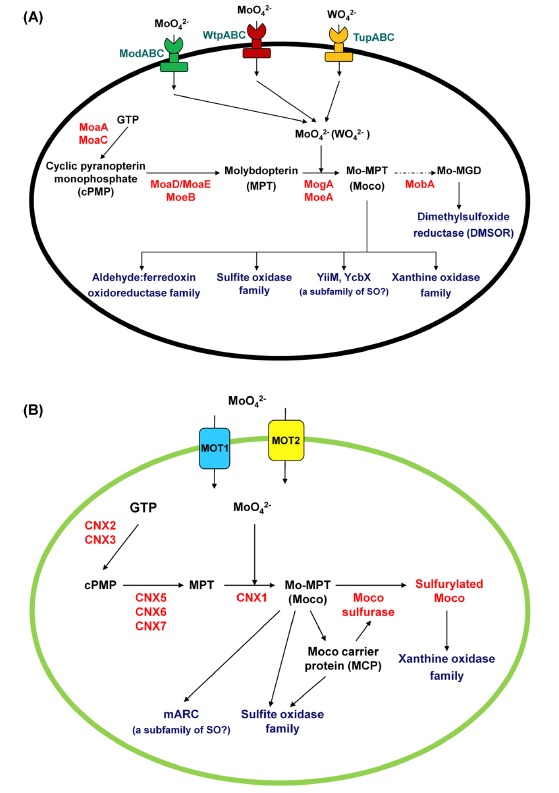
Assembly pathway of a bacterial complex iron sulfur molybdoenzyme
The path goes from molybdenum uptake into the cell, via formation of the molybdenum cofactor and its storage, to the final modification of the molybdenum cofactor and its insertion into apo-metalloenzymes 12 In soils, the molybdate anion is the only form of Mo that is available for plants and bacteria. Moco is a tricyclic pterin that co-ordinates the metal via a dithiolene group at the third (pyrano) ring.

The task of the cofactor is to position the catalytic metal Mo correctly within the active centre, to control its redox behavior, and to participate with its pterin ring system in the electron transfer to or from the Mo atom. The pterin, with its several possible reduction states as well as different structural conformations, might also be important in channeling electrons from or to other prosthetic groups
Once entered the cell, molybdate has to be complexed by a unique scaffold in order to gain biological activity. This compound is a unique tricyclic pterin called molybdopterin or metalcontaining pterin (MPT)
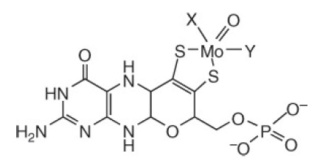
The molybdenum cofactor as found in eukaryotic molybdenum enzymes. In enzymes of the sulfite oxidase family, X is represented by a single-bonded sulfur provided by a cysteine residue of the respective protein, while Y corresponds to a double-bonded oxygen. In enzymes of the xanthine oxidase family, X is represented by a double-bonded inorganic sulfur and Y by a hydroxyl group.
Protein folding and assembly into macromolecule complexes within the living cell are complex processes requiring intimate coordination. The biogenesis of complex iron sulfur molybdoenzymes (CISM) requires use of a system specific chaperone – a redox enzyme maturation protein (REMP) – to help mediate final folding and assembly. The CISM dimethyl sulfoxide (DMSO) reductase is a bacterial oxidoreductase that utilizes DMSO as a final electron acceptor for anaerobic respiration. The REMP DmsD strongly interacts with DMSO reductase to facilitate folding, cofactor-insertion, subunit assembly and targeting of the multi-subunit enzyme prior to membrane translocation and final assembly and maturation into a bioenergetic catalytic unit.
There must be correct folding and targeting coordinated with cofactor insertion. Proteins must be targeted to their correct subcellular locations, and there has to be precise control of the assembly of large multimeric complexes.
Targeting, coordination, correct insertion, precise control of assembly are usually actions either directly performed by intelligence, or preprogrammed by intelligence to be exsercised in a robot-like manner.
Maturation of a CISM into a functional holoenzyme requires numerous stages that involve initial translational ribosome integrated folding, cofactor insertion and coordination, subsequent folding and assembly with other subunits that may have had similarly complex folding pathway. The many steps comprising the cytoplasmic biogenesis processes are highly complex and must be intricately coordinated by numerous assistant proteins to produce a functional CISM.
Of special interest to CISM maturation are system specific chaperones that help mediate the complete final folding and assembly. Such chaperones were termed redox enzyme maturation proteins (REMPs) and are essential for the proper assembly of CISMs albeit absent in the final assembled holoenzyme.
REMPs ‘escort’ their CISM substrates though the entire maturation process. Accordingly, many potential roles for REMPs have been proposed, including functioning as:
1. foldases to ensure correct secondary and tertiary structure;
2. unfoldases to correct folding mistakes;
3. avoidance chaperones to prevent incorrect membrane targeting during folding and assembly;
4. cofactor-assembly chaperones to maintain apoenzymes in a cofactor-binding competent conformation;
5. cofactor-binding proteins, which bind the cofactor prior to its transfer to the apoenzyme;
6. targeting proteins directing substrates to specific cellular locations;
7. escort chaperones to promote transmembrane transport of enzyme complexes;
8. proofreading chaperones to suppress transport until essential prior steps in the assembly process are complete;
9. protease protection chaperones to prevent degradation during assembly
The crystal structures of several molybdoenzymes revealed that Moco is deeply buried inside the proteins, at the end of a funnel-shaped passage giving access only to the substrate. Chaperones are required to facilitate the insertion of Moco into the target enzyme. Only after the insertion of Moco, the apo-enzymes adopt their final structure. Moco insertion is usually the final step of molybdoenzyme maturation, which occurs after protein folding, subunit assembly and the insertion of additional redox cofactors such as cytochromes, FeS clusters or flavin mono/dinucleotides. The Moco insertion step is catalyzed by Moco-binding molecular chaperones, which bind the respective Moco variant and insert it into the specific target molybdoenzyme. Most molybdoenzymes in bacteria, especially enzymes of the DMSO reductase family, have a specific chaperone for Moco insertion.
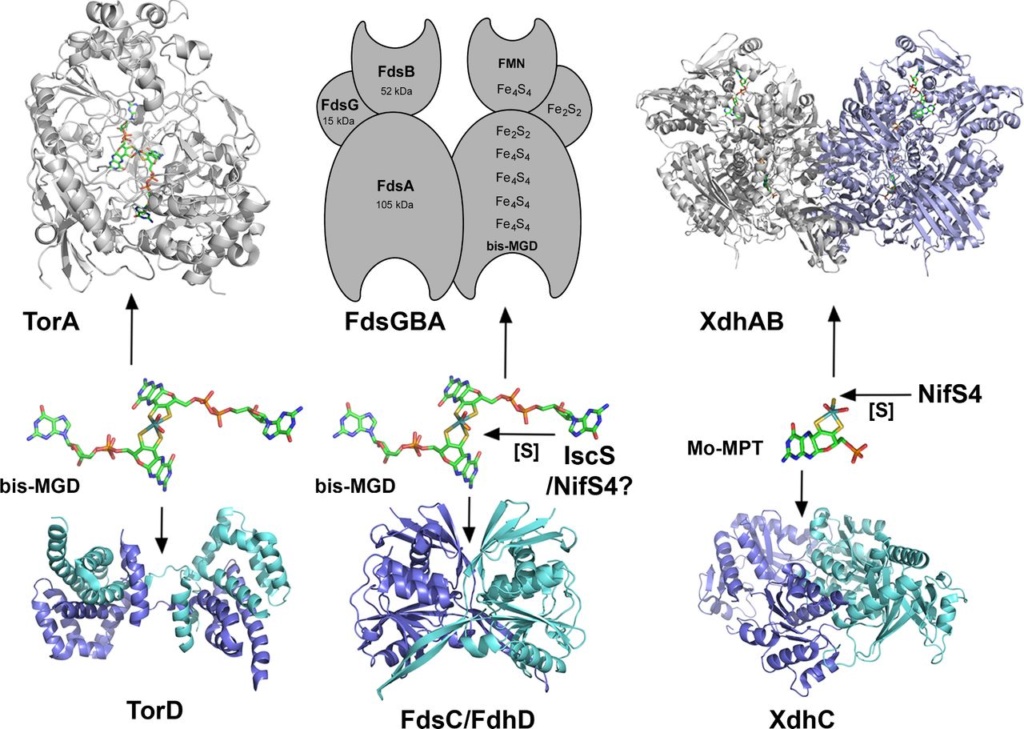
Chaperone-assisted Moco insertion into molybdoenzymes.
On the left side, the TorD/TorA system for bis-MGD insertion is shown: TorD binds bis-MGD and inserts the cofactor into apo-TorA. Shown are the structures of dimeric TorD from S. massilia and monomeric TorA from S. massilia .
In the middle, a model of the FdsC/FdsA system for insertion of sulfurated bis-MGD from R. capsulatus is shown. Rhodobacter capsulatus FdsC binds bis-MGD and further transfers it to the FdsA subunit of R. capsulatus FDH, which is composed of the (FdsGBA)2 heterotrimer. It is proposed that bis-MGD is further modified by sulfuration. For the homologous E. coli system, it was shown that IscS is involved in sulfurtransfer to bis-MGD. In R. capsulatus, the NifS4 protein performs a similar role for the XdhC/XdhB system. Here it is suggested that FdsC binds bis-MGD and an L-cysteine desulfurase (IscS/NifS4) transfers the sulfur to the Mo atom by exchanging an oxo-group and adding the sulfur ligand. Afterwards, sulfurated bis-MGD is inserted into FdsA, which is already assembled as a (FdsGBA)2 heterotrimer containing various FeS clusters and FMN. The crystal structure for the FdhD-homologous protein from Desulfotalea psychrophila is shown.
On the right-hand side, the XdhC/XdhB system for insertion of sulfurated Mo-MPT from R. capsulatus is shown. It was shown that XdhC binds Mo-MPT. The equatorial Mo=S ligand of Mo-MPT is inserted into Moco while bound to XdhC by the sulfurtransferase function of the NifS4 protein. After the formation of sulfurated Mo-MPT, XdhC interacts with XDH (here the XdhB subunit of R. capsulatus XDH is shown, pdb 1JRO) for final Moco insertion. The crystal structure for the XdhC-homologous protein from Bacillus halodurans is depicted (pdb 3ON5).
Chaperones in maturation of molybdoenzymes: Why specific is better than general? 25 Apr 2018
An interesting feature is that molybdoenzymes and their cognate chaperones, which are usually genetically related, seem to have coevolved. This latter point could be an explanation concerning the high level of specificity between the molybdoenzyme and the associated chaperone 7
This is a remarkable claim. Since these enzymes had to become functional at LUCA, they had to emerge without evolution as a causal mechanism. Secondly, chaperones have the only function when used in the maturation process of the enzyme. The high specificity on top of that seems far better to infer that a designer projected these enzymes and the requirement of "helper" proteins for maturation and synthesis of the protein complex.
a Hydroxylation is a chemical process that introduces a hydroxyl group (-OH) into an organic compound. In biochemistry, hydroxylation reactions are often facilitated by enzymes called hydroxylases. Hydroxylation is the first step in the oxidative degradation of organic compounds in air. It is extremely important in detoxification since hydroxylation converts lipophilic compounds into water-soluble (hydrophilic) products that are more readily removed by the kidneys or liver and excreted.
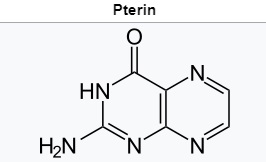
b.Pterin is a heterocyclic compound composed of a pteridine ring system, with a "keto group" (a lactam) and an amino group on positions 4 and 2 respectively. It is structurally related to the parent bicyclic heterocycle called pteridine. Pterins, as a group, are compounds related to pterin with additional substituents. Pterin itself is of no biological significance. 8
1. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5817353/
2. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3278043/
3. https://royalsocietypublishing.org/doi/10.1098/rstb.2012.0258
4. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3311541/
5. https://sci-hub.tw/https://www.sciencedirect.com/science/article/abs/pii/S0010854508000167
6. https://academic.oup.com/femsre/article/40/1/1/2467802
7. https://hal.archives-ouvertes.fr/hal-01413222/document
8. https://en.wikipedia.org/wiki/Pterin
9. https://sci-hub.tw/https://www.sciencedirect.com/science/article/abs/pii/S0010854511000671
10. https://link.springer.com/chapter/10.1007/1-4020-2179-8_10
11. https://www.sciencedirect.com/science/article/pii/S0167488906001017
12. https://academic.oup.com/jxb/article/58/9/2289/542465
13. https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1007518#pgen.1007518.ref051
14. https://www.nature.com/articles/srep00263
15. https://blogs.scientificamerican.com/thoughtomics/a-spoonful-of-molybdenum-some-ulysses-and-the-origin-of-life/
16. https://www.imoa.info/essentiality/molybdenum_trace_element.php
17. https://boa.unimib.it/retrieve/handle/10281/53873/82319/phd_unimib_057957.pdf
Last edited by Admin on Sat Mar 21, 2020 3:39 pm; edited 36 times in total