https://reasonandscience.catsboard.com/t2430-molybdenum-essential-for-life
Molybdenum is bioavailable as molybdate (MoO42−) 20 Once molybdate enters the cell, it is subsequently incorporated by complex biosynthetic machineries into metal cofactors. Molybdenum cofactor Molybdenum utilization is very likely an ancient trait present in LUCA because (i) it is utilized by almost all phyla of Archaea and Bacteria and (ii) a number of molybdo-enzymes, including the arsenite oxidase, the formate dehydrogenase, the nitrate reductase and the polysulfide reductase, have been predicted to have existed before the Archaea/Bacteria divergence The synthesis of the molybdo-pterin cofactor is a conserved multi-step pathway that is present in molybdenum-utilizing organisms in all three domains of life (see Ref. [52] for review and references therein) and therefore might have been present in LUCA. 19 Molybdenum cofactor (Moco) is a metal-containing prosthetic group common to nearly all molybdoenzymes and is ubiquitous to all kingdoms of life. Moco-dependent enzymes play central roles in many biologically important processes such as purine and sulfur catabolism in mammals, anaerobic respiration in bacteria, and nitrate assimilation in plants. 16 Molybdoenzymes are widespread in eukaryotic and prokaryotic organisms where they play crucial functions in detoxification reactions in the metabolism of humans and bacteria, in nitrate assimilation in plants and in anaerobic respiration in bacteria. To be fully active, these enzymes require complex molybdenum-containing cofactors, which are inserted into the apoenzymes after folding. For almost all the bacterial molybdoenzymes, molybdenum cofactor insertion requires the involvement of specific chaperones. 17 Metals play unique and critical roles in biology, promoting structures and chemistries that would not otherwise be available to proteins alone. 8 Molybdenum is a naturally occurring element which is found in soil, water and in our bodies. In humans, molybdenum is needed to produce enzymes which play a vital role in maintaining our bodily functions. In fact, it’s essential for all human, animal and plant life. 14 The transition element molybdenum (Mo) is of essential importance for (nearly) all biological systems as it is required by enzymes catalyzing diverse key reactions in the global carbon, sulfur and nitrogen metabolism. 15 Molybdenum metabolism is strictly dependent on iron metabolism at different levels. FeMo-co biosynthesis and nitrogenase maturation are based on the synthesis of complex Fe–S clusters. 10
At the book A priviledged Planet, page 201, Gonzales writes:
The strong nuclear force is responsible for holding protons and neutrons together in the nuclei of atoms. In such close quarters, it is strong enough to overcome the electromagnetic force and bind the otherwise repulsive, positively charged protons together. It is as short-range as it is strong, extending no farther than atomic nuclei. But despite its short range, changing the strong nuclear force would have many wide-ranging consequences, most of them detrimental to life. The periodic table of the elements would look different with a changed strong nuclear force. If it were weaker, there would be fewer stable chemical elements. The more complex organisms require about twenty-seven chemical elements, iodine being the heaviest (with an atomic number of 53). Instead of ninety-two naturally occurring elements, a universe with a strong force weaker by 50 percent would have contained only about twenty to thirty. This would eliminate the life-essential elements iron and molybdenum.
Minerals containing the elements boron and molybdenum are key in assembling atoms into life-forming molecules. It is thought that the boron minerals needed to form RNA from pre-biotic soups were not available on early Earth in sufficient quantity, and the molybdenum minerals were not available in the correct chemical form. 11 The researcher points out that boron minerals help carbohydrate rings to form from pre-biotic chemicals, and then molybdenum takes that intermediate molecule and rearranges it to form ribose, and hence RNA. This raises problems for how life began on Earth, since the early Earth is thought to have been unsuitable for the formation of the necessary boron and molybdenum minerals. It is thought that the boron minerals needed to form RNA from pre-biotic soups were not available on early Earth in sufficient quantity, and the molybdenum minerals were not available in the correct chemical form. "It’s only when molybdenum becomes highly oxidised that it is able to influence how early life formed. "This form of molybdenum couldn’t have been available on Earth at the time life first began, because three billion years ago, the surface of the Earth had very little oxygen.
Hundreds of other minerals that incorporate relatively rare elements such as lithium, beryllium, and molybdenum appear to have taken a billion years or more to first appear because it is difficult to concentrate these elements sufficiently to form new minerals. So those slow-forming minerals are also excluded from the time of life’s origins. 12
That made Leslie Orgel propose the esoteric proposal of directed panspermia, as he wrote:
The chemical composition of living organisms must reflect to some extent the composition of the environment in which they evolved. Thus the presence in living organisms of elements that are extremely rare on the Earth might indicate that life is extraterrestrial in origin. Molybdenum is an essential trace element that plays an important role in many enzymatic reactions, while chromium and nickel are relatively unimportant in biochemistry. The abundance of chromium, nickel, and molybdenum on the Earth are 0.20, 3.16, and 0.02%, respectively. We cannot conclude anything from this single example, since molybdenum may be irreplaceable in some essential reaction nitrogen fixation, for example. However, if it could be shown that the elements represented in terrestrial living organisms correlate closely with those that are abundant in some class of star molybdenum stars, for example we might look more sympathetically at "infective” theories. 18
Chromium, molybdenum, selenium, and vanadium, for example, are essential for building proteins, and proteins serve as life’s molecular “factories.” 13
The atmosphere is the primary source of nitrogen, and the continents are the primary source of several mineral nutrients, including molybdenum. This suggests that planetary environments lacking a nitrogen-rich atmosphere and continents may not be able to support a robust biosphere.
Metals are essential in microbial cells. 4 Cells require a number of elements that could potentially provide biosignatures, including bioactive trace metals. Biological use of copper and molybdenum has developed along with bioavailability.
The majority of the presented subfamilies and, as a consequence, the Molybdo-enzyme superfamily as a whole, appear to have existed in LUCA. 5
Molybdoenzymes have been classified into three families:
1.xanthine oxidase (XO) family
2.sulfite oxidase (SO) family
3.the dimethyl sulfoxide (DMSO) reductase family
Molybdenum in Biology - An Essential Trace Element 1
Molybdenum is an essential trace element for several enzymes important to animal and plant metabolism, in special nitrate reductase and nitrogenase. Molybdenum functions as an electron carrier in those enzymes that catalyse the reduction of nitrogen and nitrate. Molybdenum is essential to plants and humans. Molybdenum is needed for at least three enzymes. Sulfite oxidase catalyses the oxidation of sulfite to sulfate, necessary for metabolism of sulfur amino acids. Sulfite oxidase deficiency or absence leads to neurological symptoms and early death. Xanthine oxidase catalyses oxidative hydroxylation of purines and pyridines including conversion of hypoxanthine to xanthine and xanthine to uric acid. Aldehyde oxidase oxidises purines, pyrimidines, pteridines and is involved in nicotinic acid metabolism. Low dietary molybdenum leads to low urinary and serum uric acid concentrations and excessive xanthine excretion.
Molybdenum (Mo) are trace elements that catalyze, upon binding to the appropriate cofactors, diverse and important redox reactions in the global carbon, nitrogen, and sulfur cycles. 7 Mo is found in two forms of oxygen-labile metal cofactors, a pterin-based and a Fe – S-cluster-based scaffold. Both oxyanions enter the cell via an ABC-type high affinity uptake system and are subsequently processed by a multistep biosynthetic machinery forming either Mo and W-pterin cofactors (Moco or Wco) in a large variety of Mo- and W-containing enzymes or the FeMo cofactor (FeMo-co) in nitrogenase-catalyzed nitrogen fixation. The functional diversity of pterin-based Mo and W cofactors is reflected by a large number of enzymes such as nitrate reductase, dimethyl sulfoxide reductase, formate dehydrogenase, aldehyde oxidoreductase and CO dehydrogenase. In these enzymes Mo and W are bound via thiolates to one or two unique tricyclic pterin moieties, commonly referred to as molybdopterin but the term “metal binding pterin” (MPT) is more appropriate due to its association with both, Mo and W. It is commonly believed, but still not demonstrated, that Moco and Wco are synthesized by a similar and highly conserved pathway. Synthesis of the Moco can be divided into four major steps, according to the biosynthetic intermediates cyclic pyranopterin monophosphate, MPT, and adenylated MPT. In contrast, FeMo-co biosynthesis is less understood in terms of reaction intermediate and mechanisms of different reactions catalyzed by the involved proteins. It starts with the formation of Fe – S cluster core structures that are assembled and arranged to a topology similar to mature FeMo-co. In the next steps, Mo and homocitrate are transferred before the mature cofactor is inserted into nitrogenase. Finally, a brief overview about Mo- and W-pterin enzymes as well as FeMo- and FeW-nitrogenases is given.
Molybdoenzymes emerged as a superfamily of respiratory oxidoreductases that require a catalytic molybdenum/tungsten-based cofactor to catalyze redox reactions. 5 These enzymes are further classified into three families based on the active site structure that coordinates the molybdenum atom. A key feature that separates the dimethyl sulfoxide (DMSO) reductase family members from xanthine oxidase and sulfite oxidase families is that it has two pyranopterin groups coordinating the Mo atom, whereas the others have only one. Members of each family have similar structural folds around the catalytic cofactor, and a recent study demonstrated that the protein fold is directly correlated to the pyranopterin conformation

The complex iron sulfur containing molybdoenzyme DMSO reductase. (a) Structure of the molybdo-bis(pyranopterin guanine dinucleotide) (MobisPGD) catalytic cofactor and [4Fe-4S] clusters that makeup the electron transfer chain within DMSO reductase. (b) Overall architecture and composition of DMSO reductase from Escherichia coli, demonstrating the catalytic DmsA, electron conduit DmsB, and membrane anchor DmsC subunits
The distribution of life on earth is constrained also by the distribution of 20 bio-essential nutrients such as Calcium (Ca), Chloride (Cl−), Chromium (Cr), Cobalt (Co), Copper (Cu), Magnesium (Mg), Manganese (Mn), Nickel (Ni), Iodine (I), Iron (Fe), Molybdenum (Mo), Phosphorus (P), Potassium (K), Selenium (Se), Sodium (Na), Sulfur (S), Tugsten (W), Vanadium (V), and Zinc (Zn), which are relatively rare, but that are key components of DNA, RNA, enzymes, and other biomolecules. 2
Molybdenum in pre-biotic chemistry-the nitrogen cycle
The nitrogen cycle provides essential nutrients to the biosphere, but its antiquity in modern form is unclear. In a drill core though homogeneous organic- rich shale in the 2.5- billion- year- old Mount McRae Shale, Australia, nitrogen isotope values vary from +1.0 to +7.5 per mil (parts per thousand) and back to +2.5 parts per thousand over similar to 30 meters. These changes evidently record a transient departure from a largely anaerobic to an aerobic nitrogen cycle complete with nitrification and denitrification. Complementary molybdenum abundance and sulfur isotopic values suggest that nitrification occurred in response to a small increase in surface- ocean oxygenation. These data imply that nitrifying and denitrifying microbes had already evolved by the late Archean and were present before oxygen first began to accumulate in the atmosphere
Molybdenum and the origin of life
An evolutionary tree of key enzymes from the Complex-Iron-Sulfur-Molybdoenzyme (CISM) superfamily distinguishes "ancient" members, i.e. enzymes in the last universal common ancestor (LUCA) of prokaryotes, from more recently evolved subfamilies. The molybdo-enzyme superfamily existed in LUCA. The results are discussed with respect to the nature of bioenergetic substrates available to early life and to problems arising from the low solubility of molybdenum under conditions of the primordial Earth.
Whereas the details of iron's and copper's involvement in numerous biological reactions have been studied for more than a century, the precise role of other metals, although recognized as vital trace elements in enzyme catalysis, became elucidated only recently. Molybdenum (Mo) has during the last 2 decades been shown to constitute an essential cofactor in at least 3 distinct enzyme superfamilies, the most widespread of which is the so-called Complex Iron-Sulfur Molybdoenzyme (CISM) superfamily of molybdo-pterin containing enzymes. Incidentally, this denomination ignores the fact that a few members of the family use tungsten (W) instead of molybdenum in their active sites. In the periodic table of elements, tungsten lies directly below molybdenum in the d-block and is thus expected to feature chemical properties related to those of Mo.
The ineluctable requirement for the trans-iron elements molybdenum and/or tungsten in the origin of life 3
The majority of the presented subfamilies and, as a consequence, the Molybdo-enzyme superfamily as a whole, appear to have existed in LUCA.
A vast number of enzymes rely on metal cofactors for catalysis and/or redox conversions. The structural unit of the CISM protein thus appears to have served multiple purposes for life, especially in energy harvesting, right from its very beginnings. CISM enzymes in LUCA likely performed energy conversion through the reduction of carbon dioxide, polysulfide or nitrate as well as from the oxidation of arsenite. Reduction of CO2 and sulfur with H2 as electron donor would be viable bioenergetic pathways in the geochemical setting of the early Archaean and have indeed been put forward as ancestral bioenergetic mechanisms. The presence of the CISM superfamily in LUCA implies a vital role of its metal cofactors in early life.
What then of the availability of these two transition metals?
W occurs in both acid and alkaline solutions and was thus available to emerging life, whereas Mo is relatively insoluble in reduced and neutral waters, but does occur in mixed valence sulfide and selenide and/or oxide complexes in alkaline solutions. Mo's insolubility at neutral pH values, exacerbated by an anoxic atmosphere, suggested a low bioavailability of this element for early life. Mo-isotope analyses on samples from the Archaean era indeed show substantially lower levels than during Phanerozoic times. Two scenarios can reconcile the results of molecular phylogeny and paleogeochemistry. (i) The ancestral CISM enzyme exclusively used W (tungsten) which was later replaced by Mo. (ii) CISM-catalyzed reactions in early life used Mo supplied by alkaline hydrothermal vents, proposed as cradles for life.
What we see here, are the usual "ad-hoc" explanations.
Certainly tungsten and most likely molybdenum ought to be added to the list of metals vital already to earliest life on Earth

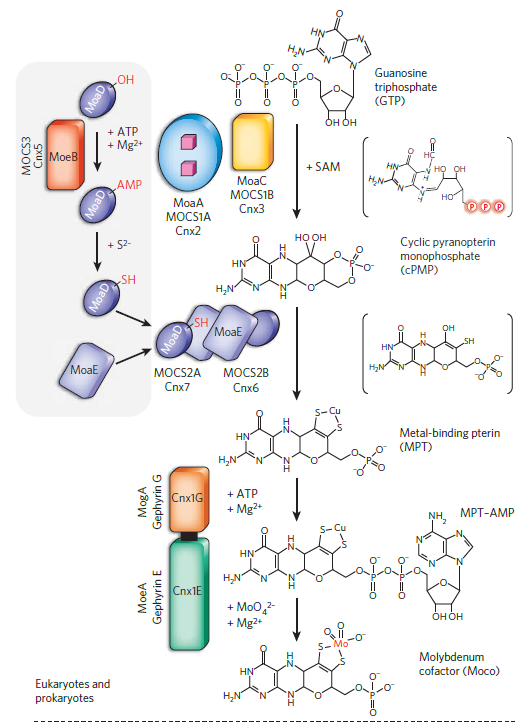
The biosynthesis of the molybdenum cofactor 17
In E. coli, nine proteins with known function are directly involved in Moco biosynthesis (MoaA, MoaC, MobA, MocA, MoaD, MoaE, MoeA, MoeB, MogA) 17 In all prokaryotes, Moco is synthesized by a conserved pathway which can be divided into four general steps:
(i) the synthesis of cyclic pyranopterin monophosphate (cPMP) from 5′GTP ,
(ii) insertion of two sulfur atoms into cPMP and formation of MPT,
(iii) formation of Moco by insertion of molybdate to the sulfur atoms of MPT,
(iv) further modification of Moco by the attachment of GMP or CMP to the phosphate group of MPT, forming the MGD cofactor or MCD cofactor (Fig. 1). The four steps of Moco biosynthesis are described below:
First step: The biosynthesis of Moco starts from 5′-GTP. The first stable intermediate of Moco was isolated in 1993 and later identified to be a 6-alkyl pterin with a cyclic phosphate group at the C2′ and C4′ atoms, named cPMP . The reaction of cPMP formation from 5′GTP is catalyzed by the two proteins MoaA and MoaC in bacteria. MoaA belongs to the superfamily of S-adenosyl methionine (SAM)-dependent radical enzymes. The protein contains two [4Fe4S] clusters, with the N-terminal [4Fe4S] cluster binding SAM and generating the 5′-deoxyadenosyl radical, and the C-terminal [4Fe4S] cluster binding the 5′GTP. While the individual catalytic functions of MoaA and MoaC had long been unknown, recent studies showed that MoaA catalyzes the conversion of 5′GTP to (8S)-3′,8-cyclo-7,8-dihydroguanosine 5′triphosphate (3′,8-cH2GTP), and MoaC catalyzes the conversion of 3′,8-cH2GTP to cPMP . In this reaction, the C8 of GTP is inserted between the C2′ and C3′ carbons of the ribose. MoaC further converts 3′,8-cH2GTP to cPMP, a reaction which involves pyrophosphate cleavage in addition to the formation of the cyclic phosphate group.
Second step: In the next step of Moco biosynthesis, cPMP is converted to MPT by the insertion of two sulfur atoms to the C1′ and C2′ positions of cPMP. This reaction is catalyzed by MPT synthase, a (αβ)2 heterotetrameric complex composed of two MoaD and two MoaE subunits. The sulfur atoms required for this reaction are present at the C-terminus of MoaD in form of a thiocarboxylate group. Studies on the reaction mechanism lead to a model in which two MoaD molecules are required for the sulfur insertion and cPMP is bound to MoaE during the reaction. Both subunits of the MoaE dimer core act independently, so that the two MoaD proteins are exchanged on each side of the dimer during the reaction. The first sulfur is added to the C2′ position cPMP by one MoaD-SH molecule, resulting in a hemisulfurated intermediate. This reaction is coupled to the hydrolysis of the cPMP cyclic phosphate group. In the hemisulfurated intermediate, the MoaD C-terminus is covalently linked via a thioester linkage. In the next reaction, the thioester is hydrolyzed by a water molecule, the first MoaD subunit dissociates from the MPT synthase complex and a new MoaD-SH molecule associates with the complex. After the opening of the cyclic phosphate in the first sulfur transfer step to the C2′ position, the hemisulfurated intermediate is proposed to shift its location within MoaE protein, so that the C1′ position becomes more accessible to the attack by the second MoaD-SH. In the second sulfur transfer step, a covalent intermediate is formed with the new MoaD-SH protein. Further, MPT is formed by hydrolysis of this MoaD-thioester intermediate and release of MoaD.
For a new round of catalysis, a new thiocarboxylate group needs to be formed on MoaD. The activation of MoaD is catalyzed by the MoeB protein, which forms a (MoaD-MoeB)2 complex with MoaD. During the reaction, the C-terminus of MoaD is activated by the formation of an acyl-adenylate group at its terminal glycine. In this complex, MoaD-AMP receives the sulfur from a sulfurtransferase and MoaD-SH is formed. The sulfur is directly transferred to MoaD in the (MoaD-MoeB)2 complex, additionally releasing (MoeB)2 and AMP. MoaD-SH dissociates from the complex, and reassociates with MoaE to form the active MPT synthase heterotetramer (Fig. 1) . It was shown that in E. coli, L-cysteine serves as the origin of the MPT dithiolene sulfurs. In the sulfurtransfer reaction to MoaD, the proteins IscS and TusA are involved in E. coli, forming a sulfur relay system in which a persulfide sulfur is transferred from one protein to another. It is believed that under specific conditions TusA can be replaced by other sulfurtransferases in the cell, like the rhodanese-like protein YnjE or the L-cysteine desulfurase SufS. It has been proposed that the sulfur of one of these persulfide-containing proteins interacts with the (MoaD-MoeB)2 complex and attacks the MoaD-AMP bond, releasing AMP and creating a transient MoaD perthiocarboxylate intermediate with the sulfurtransferase, which is further reductively cleaved, finally releasing MoaD-SH.

The biosynthesis of the molybdenum cofactors.
Shown is a scheme of the biosynthetic pathway for Moco biosynthesis. The central part shows the three conserved steps of Moco biosynthesis present in all organisms, the formation of cPMP, MPT and Mo-MPT. Unstable intermediates formed during the reactions are shown in brackets: 3,8-cH2GTP, the hemisulfurated MPT intermediate, MPT-AMP and bis-Mo-MPT. Bacteria contain a fourth step of Moco modification in which Mo-MPT is further modified by the addition of nucleotides, GMP or CMP. Additionally, Moco can be further modified by the replacement of one oxo ligand by a sulfido ligand, forming the mono-oxo Moco present in the xanthine oxidase family of molybdoenzymes. The SO family contains the Mo-MPT cofactor with a proteinogenic cysteine ligand. The DMSO reductase family of molybdoenzymes present only in bacteria binds the bis-MGD cofactor in which the molybdenum atom contains an additional ligand, which can be a cysteine, a selenocysteine, a serine, an aspartate or a hydroxo-ligand. Here, also a Moco sulfuration step exists, in which an oxo-ligand at the bis-MGD cofactor is replaced by a sulfur ligand. The proteins involved in the reactions are colored in red, and additional cosubstrates required for the reactions are colored in blue.
Third step: For the formation of Moco, molybdate is bound to the dithiolene sulfurs of MPT. The specific insertion of molybdenum into MPT was shown to be catalyzed by the joined action of the MoeA and MogA proteins. During the reaction, MogA thereby forms an MPT-AMP intermediate under ATP consumption, and this intermediate is further transferred to MoeA, which mediates molybdenum ligation at low concentrations of MoO42-. The end product of the MoeA and MogA reaction is Mo-MPT in a tri-oxo form, the basic form of the molybdenum cofactor which can be further modified by nucleotide addition in the next step. Alternatively, the Mo-MPT cofactor can be directly inserted into enzymes of the SO family, where Moco is coordinated by a cysteine ligand which is provided by the polypeptide chain of the protein, forming an MPT-MoVIO2 core in its oxidized state (Fig. 2).

Figure 2. The families of molybdoenzymes.
The basic form of Moco is a 5,6,7,8-tetrahydropyranopterin, named Mo-MPT, which coordinates the molybdenum atom by the characteristic dithiolene group at the C1′ and C2′ positions of the pyranopterin ring. Mo-MPT (shown in the tri-oxo structure; Reschke et al.2013) can be further modified and three different molybdenum-containing enzyme families are classified according to their coordination at the molybdenum atom: the XO, SO and DMSO reductase families. The SO family is characterized by a MPT-MoVIO2Cys ligand sphere. The XO family contains a MPT-MoVIOS(OH) core. Here, the MPT core can be modified by an additional CMP nucleotide at the phosphate group, forming MCD. The DMSO reductase family contains a MGD2-MoVIXY core with X being either a sulfur or an oxygen ligand and Y either being a hydroxo or amino acid ligand (Ser, Cys, Sec and Asp ligands were identified so far). Shown are structures of enzymes representative enzymes from each family: chicken sulfite oxidase (pdb 1SOX), bovine xanthine dehydrogenase (pdb 1FIQ) and S. massilia TMAO reductase (pdb 1TMO). The surface representations show that the Moco in each enzyme is deeply buried at the end of a funnel-like passage, giving access only to the substrate molecules (entrance site is shown by the arrow).
Fourth step: In the fourth step of Moco biosynthesis in bacteria, Mo-MPT can be further modified by the addition of a GMP or CMP to the terminal phosphate group. (A) The proteins of the DMSO reductase family in bacteria contain the bis-MGD cofactor (Fig. 2). The synthesis of the bis-MGD was shown to occur in a two-step reaction which requires Mo-MPT, MobA and Mg-GTP (Reschke et al.2013). In the first reaction, the bis-Mo-MPT intermediate is formed on MobA with Mo-MPT as substrate. For this reaction, the ligation of molybdenum to MPT is essential but no further cofactors or molecules are required (Temple and Rajagopalan 2000). In the second reaction, two GMP moieties from GTP are added to the C4′ phosphate of bis-Mo-MPT, forming the bis-MGD cofactor (Palmer et al.1996; Lake et al.2000) (Fig. 1). After the attachment of two GMP molecules to the bis-Mo-MPT intermediate, the bis-MGD cofactor is formed and released from MobA. Since bis-MGD is not stable in its free form, it is immediately bound by Moco-binding chaperones, which insert the cofactor specifically into target enzymes of the DMSO reductase family (see below).
(B) Enzymes of the XO family in some bacteria like E. coli contain the MCD form of Moco (Fig. 2). MCD formation is catalyzed by MocA, a protein that shares a high amino acid sequence identities to MobA. The overall catalytic reaction of MocA is similar to the second part of the reaction of MobA, in that it acts as a MPT CTP transferase and covalently links MPT and CMP with the concomitant release of the α- and γ-phosphates of CTP as pyrophosphate (Fig. 1). However, in this reaction MCD is the end product and the bis-Mo-MPT form is not formed. Instead, the MCD cofactor for all enzymes of the XO family is further modified and contains an equatorial sulfido ligand at its active site, which is essential for enzyme activity.
Molybdenum cofactor synthesis is linked to copper: a copper ion is bound to molybdopterin dithiolate sulfurs as an intermediate in the biosynthetic pathway (Kuper et al. 2004). 7 Its synthesis involves the insertion of molybdenum into molybdopterin by the Cnx1 G-domain. The identification of copper bound to the molybdopterin dithiolate sulphurs in Cnx1G, coupled with the observation that copper inhibited Cnx1G activity, suggests a link between molybdenum and copper metabolism, which would require cytoplasmic copper (Schwarz and Mendel 2006).
MoCo-binding Chaperones
The crystal structures of several molybdoenzymes revealed that Moco is deeply buried inside the proteins, at the end of a funnel-shaped passage giving access only to the substrate (Fig. 2). These structures suggested that chaperones might be required to facilitate the insertion of Moco into the target enzyme. Additionally, it was suggested that only after the insertion of Moco, the apo-enzymes adopt their final structure. It was also shown that Moco insertion is usually the final step of molybdoenzyme maturation, which occurs after protein folding, subunit assembly and the insertion of additional redox cofactors such as cytochromes, FeS clusters or flavin mono/dinucleotides. The Moco insertion step is catalyzed by Moco-binding molecular chaperones, which bind the respective Moco variant and insert it into the specific target molybdoenzyme (Fig. 3). It is suggested that most molybdoenzymes in bacteria, especially enzymes of the DMSO reductase family, have a specific chaperone for Moco insertion. Deletion of the chaperone gene generally leads to the loss of activity of the molybdoenzyme due to the lack of Moco insertion into the enzyme.

Figure 3. Chaperone-assisted Moco insertion into molybdoenzymes.
On the left side, the TorD/TorA system for bis-MGD insertion is shown: TorD binds bis-MGD and inserts the cofactor into apo-TorA. Shown are the structures of dimeric TorD from S. massilia (pdb 1N1C) and monomeric TorA from S. massilia (1TMO).
In the middle, a model of the FdsC/FdsA system for insertion of sulfurated bis-MGD from R. capsulatus is shown. Rhodobacter capsulatus FdsC binds bis-MGD and further transfers it to the FdsA subunit of R. capsulatus FDH, which is composed of the (FdsGBA)2 heterotrimer. It is proposed that bis-MGD is further modified by sulfuration. For the homologous E. coli system, it was shown that IscS is involved in sulfurtransfer to bis-MGD. In R. capsulatus, the NifS4 protein performs a similar role for the XdhC/XdhB system. Here it is suggested that FdsC binds bis-MGD and an L-cysteine desulfurase (IscS/NifS4) transfers the sulfur to the Mo atom by exchanging an oxo-group and adding the sulfur ligand. Afterwards, sulfurated bis-MGD is inserted into FdsA, which is already assembled as a (FdsGBA)2 heterotrimer containing various FeS clusters and FMN. The crystal structure for the FdhD-homologous protein from Desulfotalea psychrophila is shown (pdb 2PW9).
On the right-hand side, the XdhC/XdhB system for insertion of sulfurated Mo-MPT from R. capsulatus is shown. It was shown that XdhC binds Mo-MPT. The equatorial Mo=S ligand of Mo-MPT is inserted into Moco while bound to XdhC by the sulfurtransferase function of the NifS4 protein. After the formation of sulfurated Mo-MPT, XdhC interacts with XDH (here the XdhB subunit of R. capsulatus XDH is shown, pdb 1JRO) for final Moco insertion. The crystal structure for the XdhC-homologous protein from Bacillus halodurans is depicted (pdb 3ON5).
One well-studied example for the action of a molecular chaperone with its apo-enzyme is the TorD/TorA system for TMAO reduction in E. coli. TorD was shown to be the specific chaperone of TorA and plays a direct role in the insertion of Moco into apoTorA. In E. coli, specific chaperones were also identified for nitrate reductase, DMSO reductase and FDH and their specific function was analyzed in detail: NarJ is the chaperone for nitrate reductase A NarGHI, NarW is the chaperone for nitrate reductase Z NarZYV, DmsD is the chaperone for DmsABC and YnfE/F , NapD is the chaperone for periplasmic nitrate reductase NapA and FdhD is the chaperone for FdhF . However, in the same host, no defined specific chaperone has been identified so far for the cytoplasmic molybdoenzyme BisC or the periplasmic molybdoenzyme TorZ. Crystal structures of several molybdoenzyme-specific chaperones were solved, and TorD, DmsD, NarJ and NapD chaperones were fully described. These dedicated chaperones are sometimes also called REMPs for redox enzyme maturation proteins.
Molecular chaperones were also identified for the XO family of molybdoenzymes. Here, the best characterized chaperone from this family is the Rhodobacter capsulatus XdhC protein which is essential for the maturation of R. capsulatus XDH (Neumann and Leimkühler 2011) (Fig. 3, Table 1).
The TorD family of Moco-binding chaperones
The TorD family of molecular chaperones was shown to contain hundreds of members that are mainly bacterial but also a few archaeal cytoplasmic proteins. Except YcdY, which in E. coli is linked to a zinc protein, these chaperones carry out essential roles in the biogenesis of both periplasmic and cytoplasmic molybdoenzymes from the DMSO reductase family. Thus, it was shown that for all studied chaperone–molybdoenzyme couple, the absence of the chaperone affects the stability or the activity of their target protein. In general, these Moco-binding chaperones are highly specific for their target molybdoenzyme.
While the TorD-like proteins in general present a low level of sequence identity (20% or less), the members of this family are characterized by a common fold. Their 3D structures showed that they generally are organized in 10–12 α-helices that account for 60–70% of the protein residues. They contain in addition a long loop region separating the N- and C-terminal domains of the proteins. At first, a motif, ‘E(Q)PxDH’, held by the loop region was proposed to characterize members of the TorD family, but with the increase of available sequences it became clear that the sequence motif is not present in all species. It was established that these chaperones bind at least to one region of their target corresponding to the twin-arginine signal sequences of exported molybdoenzymes to the periplasm or N-terminal sequences of cytoplasmic ones. The maturation process of various bis-MGD containing enzymes has been reported. A common framework in the maturation mechanism has been suggested according to the complexity or the location of the molybdoenzymes; however, differences also appear.
Maturation of TorA, a simple periplasmic molybdoenzyme
The mechanism of TorA maturation by TorD has been studied in detail in the past. TorA is a bacterial respiratory enzyme catalyzing the reduction of TMAO into the volatile compound trimethylamine (TMA). It is a monomeric protein located in the periplasm and contains solely the bis-MGD cofactor as prosthetic group. Therefore, it is a well-defined model system to study the insertion of Moco without interferences linked to the assembly of subunits or the incorporation of other cofactors, e.g. iron–sulfur centers. TorA maturation is a cytoplasmic event which occurs before its translocation across the membrane. It is now established that TorD is involved in the stabilization and maturation of TorA (Fig. 4).

Figure 4. Model for TorA and NarGHI maturation in E. coli.
TorD-dependent TorA maturation: by interacting with the leader peptide (LP) and the core of apoTorA, TorD protects it against proteolytic attack of the Lon protease and maintain the apoenzyme in a competent conformation for Moco insertion. TorD recruits the components involved in the final step of bis-MGD synthesis and transfers the mature cofactor to the catalytic site of TorA. Translocation of TorA is dependent on the TAT machinery, role of TorD in TorA targeting to the TAT components is not defined. NarJ-dependent NarGHI maturation: NarJ binds the N-terminal part and the core of NarG. NarJ is required to insert the iron–sulfur cluster and Moco into the NarG subunit. NarJ interacts with enzymes involved in Moco synthesis but a direct binding to bis-MGD was not shown yet. When NarG maturation is complete, the NarGHI complex is anchored to the membrane.
Maturation of membrane-associated complex molybdoenzymes
NarGHI is a membranous nitrate reductase responsible for anaerobic respiration when nitrate is abundant. The catalytic subunit NarG is facing the cytoplasm and contains the bis-MGD cofactor and also an iron–sulfur cluster named FS0 , NarH is an electron transfer unit containing four iron–sulfur centers and NarI contains a b-type cytochrome and permits the anchoring of the complex to the cytoplasmic membrane (Fig. 4). The molecular chaperone for NarGHI is the soluble protein NarJ. NarJ is required for the stability and the location of the complex and the insertion of Moco. Like TorD, the role of NarJ is mediated by its interaction with two distinct regions on NarG, the N terminal part and the core of the protein . The 1–15 N-terminal amino acids of NarG adopt an α-helical conformation in solution, which is recognized by NarJ via hydrophobic interactions. Moreover, the NarJ conformation is modified when interacting with NarG. The absence of NarJ leads to a lack of the cofactor in NarG, a defect in the assembly of the NarGHI complex which results in a soluble NarGH complex and an alteration of the b-type heme content of the membranous NarI cytochrome. Finally, it was also demonstrated that the maturation of the FS0 center must precede bis-MGD insertion into NarG. Thus, NarJ ensures complete maturation and anchoring of the cytoplasmic NarGH complex to NarI.
TorD interacts with two distinct regions of the Moco-free apoform of TorA, which encompasses the signal sequence at the N-terminal part of the protein and a binding site in the core of the apoprotein. This was confirmed by analysis of the TorA–TorD complex by SAXS analyses which showed a 1:1 binding stoichiometry of the two proteins (Dow et al.2013). Moreover, TorD is able to bind to both sites simultaneously. Recently, it was demonstrated that TorD binding to the core of apoTorA prevents proteolytic attack of the Lon protease and also the proteolysis of the N-terminal extremity by an additional, still unknown protease (Genest et al.2006a,b; Redelberger et al.2013). While the region of TorD involved in the recognition of the core of apoTorA was defined and corresponds to the fifth alpha helix in TorD structure, the region of TorD responsible for the protection of the apoTorA signal sequence is still controversial (Jack et al.2004; Genest et al.2008; Genest, Mejean and Iobbi-Nivol 2009). However, two residues (D124 and H125, E. coli numbering) located in the loop region were predicted to be involved in the binding of TorA leader sequence (Jack et al.2004; Hatzixanthis et al.2005). The TorD chaperone is also required for the insertion of the Moco into the catalytic site of TorA (Pommier et al.1998; Ilbert, Mejean and Iobbi-Nivol 2004) (Fig. 3). By binding to the core of the apoprotein, TorD induces a conformational change of apoTorA that becomes thus competent for Moco insertion. Further, TorD plays a role also in the last step of the Moco maturation by interacting with the MobA protein involved in bis-MGD formation (Genest et al.2008). Thus, the chaperone could act as a facilitator to insert the synthesized bis-MGD cofactor into the binding site of apoTorA and thereby render the mature enzyme protease resistant (Redelberger et al.2013). After bis-MGD insertion into apoTorA, mature TorA and release of TorD from the complex, TorA has to be targeted to the TAT machinery (Fig. 4). This step is facilitated by the fact that insertion of Moco into the catalytic site of TorA modifies the affinity between the interacting region of TorA and TorD allowing the release of TorD (Pommier et al.1998). This event can be considered as a proofreading mechanism directed by TorD or a passive competition between proteins of the TAT machinery and TorD, whose affinity for the signal sequence is thereby decreased (Jack et al.2004; Genest, Mejean and Iobbi-Nivol 2009). Consequently, the TAT leader peptide is exposed after TorD release and bis-MGD containing TorA can be targeted to the TAT machinery. Alternatively, a competition can occur between TorD and the TAT components leading to the release of TorD. These models remain to be proven.
Conclusions
Although their biosynthesis is a complex multistep process, molybdoproteins are ancient enzymes which are present in all kingdoms of living organisms. It has been shown that their maturation requires a precise pattern drawn almost specifically for each of them. This characteristic can surely be linked to both the variety of forms of the Moco that can be inserted and the diversity in the catalytic sites and localizations of these enzymes. The specific chaperones are a control point for the discrimination of the correct Moco to be inserted. Moreover, the way they handle the protection or the maturation process appears adapted to their proper target.
Importance of molybdoenzymes was already depicted in humans where the absence of Moco leads to a death in early childhood. In bacteria, it appears that the wide range of enzymatic activities catalyzed by these enzymes can be a great advantage for survival in many niches including polluted sites and an efficient adaptive mechanism. In an era submitted to ecological pressure to counteract anthropogenic discharges, bacterial molybdoenzymes can be surely adapted tools.
Parts in the cell required for the biosynthesis of Molybdenum cofactor synthesis
High-affinity ATP-binding cassette (ABC) transporter for molybdenum uptake
Copper
pterin
iron
ATP
Moco-binding proteins
MoaA and MoaC
MPT synthase, a (αβ)2 heterotetrameric complex
MoaD
MoeA
MogA
bis-MGD cofactor
TorD/TorA system
periplasmic molybdate-binding protein (ModA)
transmembrane channel (ModB)
cytoplasmic protein (ModC)
Transmembrane protein, ModB
Molybdenum metabolism is strictly dependent on iron metabolism at different levels. FeMo-co biosynthesis and nitrogenase maturation are based on the synthesis of complex Fe–S clusters
1. http://www.imoa.info/HSE/environmental_data/molybdenum_trace_element.php
2. Molybdenum Cofactors and Their role in the Evolution of Metabolic Pathways , page 10
3. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3278043/
4. http://www.ajsonline.org/content/305/6-8/467.abstract
5. http://www.nature.com/articles/srep00263
6. Prokaryotic Systems Biology page 215
7. Molecular microbiology of heavy metals, page 271
8. http://www.cell.com/current-biology/fulltext/S0960-9822(11)01079-7
9. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3650355/
10. http://www.nature.com/nature/journal/v460/n7257/full/nature08302.html
11. http://www.bbc.com/news/science-environment-23872765
12. https://www.sciencedaily.com/releases/2013/11/131125164814.htm
13. http://www.reasons.org/articles/vital-poisons
14. http://www.imoa.info/essentiality/molybdenum-essential-for-life.php
15. http://www.sciencedirect.com/science/article/pii/S0167488906001017
16. https://en.wikipedia.org/wiki/Molybdenum_cofactor
17. https://academic.oup.com/femsre/article-lookup/doi/10.1093/femsre/fuv043
18.http://www.checktheevidence.co.uk/Disclosure/PDF%20Documents/Directed%20Panspermia%20F.%20H.%20C.%20CRICK%20AND%20L.%20E.%20Orgel.pdf
19. http://www.sciencedirect.com.https.sci-hub.hk/science/article/pii/S0005272812010407
20. http://sci-hub.tw/https://www.nature.com/articles/nature08302
Further readings:
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0028170
Last edited by Admin on Thu Mar 22, 2018 9:56 am; edited 36 times in total