Forces Stabilizing Proteins - essential for their correct folding
https://reasonandscience.catsboard.com/t2692-forces-stabilizing-proteins-essential-for-their-correct-folding
Proteins are the most complex molecules in life and are involved in basically all biochemical processes. Human cells contain 25000 genes, which can produce upon splicing up to over 100,000 different proteins.
The awe inspiringing spliceosome, the most complex macromolecular machine known, and pre-mRNA processing in eukaryotic cells
https://reasonandscience.catsboard.com/t2180-the-spliceosome-the-splicing-code-and-pre-mrna-processing-in-eukaryotic-cells
Proteins are the end products of the process of transcription and translation that starts with the information stored in the genome.
The function of proteins depends on the right sequence of amino acids, lined up into polypeptide chains of various sizes, ranging from short sequences, like extracellular hemoglobin with 140 amino acids, to monstrously large proteins, like tintin which is used in sarcomeres, responsible for passive elasticity of striated mu3scles. with lengths that vary from ~27,000 to ~33,000 amino acids (depending on the splice isoform), the largest known protein.
Titin the largest proteins known and titin-telethonin complex - the strongest protein bond found so far in nature
https://reasonandscience.catsboard.com/t2671-titin-the-largest-proteins-known-and-the-titin-telethonin-complex-the-strongest-protein-bond-found-so-far-in-nature
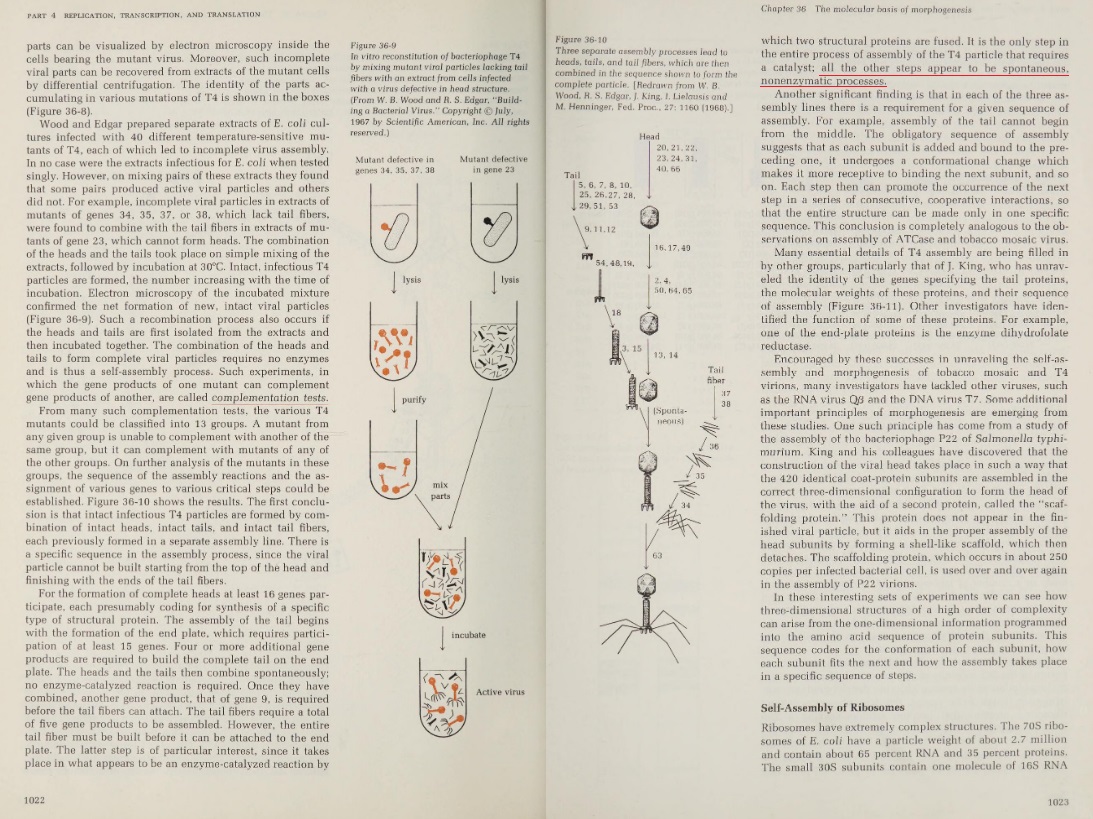
Proteins, in order to become functional, must fold into very specific 3D shapes, which happens right when they come out of the Ribosome, where they are synthesized. Specific protein shape and conformation depends on the interactions between its amino acid side chains. For a protein to function it must fold into a resting state which is a complex three-dimensional structure. If a protein fails to fold into its functional structure then it is not only without function but it cab become toxic to the cell. As proteins fold, they test a variety of conformations before reaching their final form, which is unique and compact. Folded proteins are stabilized by thousands of noncovalent bonds between amino acids. A relatively small protein of only 100 amino acids can take some 10^100 different configurations. If it tried these shapes at the rate of 100 billion a second, it would take longer than the age of the universe to find the correct one. Just how these molecules do the job in nanoseconds, nobody knows. 3
https://www.technologyreview.com/s/423087/physicists-discover-quantum-law-of-protein-folding/
Nobody knows, but since there was no evolution, to produce the first proteome for the first living cells, there are two options : blind, unguided, random lucky events on early earth, or the creative act of a super intelligent agent.
The 3D shape depends on the chemical forces, the chains are folded into a uniquely defined configuration, in which they are held by hydrogen bonds between the peptide nitrogen and oxygen atoms, which make a large contribution to protein stability, but other forces play an essential role as well, like hydrophobic interactions, disulfide bonds, Charge-charge interactions on the surface of proteins which are electrostatic interactions, pure ionic interactions, van der Waals forces, but also the surroundings of proteins, and their milieu. Chemical forces between a protein and its immediate environment contribute to protein shape and stability.
A special note deserve Van der Waals forces and interactions, which origins are ultimately a quantum mechanical. (At least the induced dipole portions: how the electrons move with respect to each other and an external electrical field is driven by quantum mechanics.) 2 Remarkably, quantum transition model fits the folding curves of 15 different proteins and even explains the difference in folding and unfolding rates of the same proteins. That means, the shape could change by quantum transition, meaning that the protein could ‘jump’ from one shape to another without necessarily forming the shapes in between. Impressive stuff !! 3
The 20 amino acids used in life are classified in Acidic amino acids, Basic amino acids with net positive charge, polar but uncharged, and nonpolar or hydrophobic amino acids. This variety contributes critically and is important for the processes that drive protein chains to “fold,” that is to form their natural (and functional) structures.
see more:
Amino Acids and peptide bonding
https://reasonandscience.catsboard.com/t2590-origins-what-cause-explains-best-our-existence-and-why#5845
But how could natural processes have foresight, which seems to be absolutely required, to "know" which amino acid sequences would provoke which forces, and how they would fold the protein structure to get functional for specific purposes within the cell ? Let's consider, that in order to have a minimal functional living cell, at least 561 proteins and protein complexes would have to be fully setup, working, and interacting together to confer a functional whole with all life-essential functions :
The last universal common ancestor represents the primordial cellular organism from which diversified life was derived
https://reasonandscience.catsboard.com/t2176-lucathe-last-universal-common-ancestor
A minimal estimate for the gene content of the last universal common ancestor
19 December 2005
A truly minimal estimate of the gene content of the last universal common ancestor, obtained by three different tree construction methods and the inclusion or not of eukaryotes (in total, there are 669 ortholog families distributed in 561 functional annotation descriptions, including 52 which remain uncharacterized)
So there had to be at least 561 proteins of various sizes, set up correctly in order to fold into functional 3D shapes, governed by chemical bond forces interacting between the individual amino acids. So not only is the right amino acid sequence essential but also as a side-effect the right dosage of the bond forces in between the amino acids.
The problem becomes even more severe when we consider that many, if not most proteins, are governed and build up by primary, secondary, tertiary, and quaternary structures. A quaternary structure refers to two or more polypeptide chains held together by intermolecular interactions to form a multi-subunit complex. And there are proteins, which require co-factors, often made of trace metals, like Iron, molybdenum etc. These cofactors require a pocket of the right size, and binding residues at the right place, and a tunnel where these cofactors can pass through during biosynthesis, to be inserted at the precise location inside the protein.
And there is more:
Molecular Chaperones Help Guide the Folding of Most Proteins
https://reasonandscience.catsboard.com/t1437-chaperones
Most proteins probably do not fold correctly during their synthesis and require a special class of proteins called molecular chaperones to do so. Molecular chaperones are useful for cells because there are many different folding paths available to an unfolded or partially folded protein. Without chaperones, some of these pathways would not lead to the correctly folded (and most stable) form because the protein would become “kinetically trapped” in structures that are off-pathway. Some of these off-pathway configurations would aggregate and be left as irreversible dead ends of nonfunctional (and potentially dangerous) structures. Proteins also require chaperones constantly, after correct folding, to maintain their functional states, which is called proteostasis.
That raises interesting questions: How should and could natural nonintelligent mechanisms forsee the necessity of chaperones in order to get a specific goal and result, that is functional proteins to make living organisms? Nonliving matter has no natural " drive " or purpose or goal to become living. The make of proteins to create life, however, is a multistep process of many parallel acting complex metabolic pathways and production-line like processes to make proteins and other life essential products like lipids, carbohydrates etc. The right folding of proteins is just one of several other essential processes in order to get a functional protein. But a functional protein by its own has no function unless correctly embedded through the right order of assembly at the right place.
Evolution of the correct protein foldings
To explain the origin of correct protein folds is paramount to explain the origin of life, and biodiversity. Its therefore of significant interest to see, how secular science papers explain its origin. Lets have a look at following:
Protein folding as an evolutionary process 1
7 December 2008
To provide a description that is consistent with other natural processes, protein folding is formulated from the principle of increasing entropy. It then becomes evident that protein folding is an evolutionary process among many others. During the course of folding protein structural hierarchy builds up in succession by diminishing energy density gradients in the quest for a stationary state determined by surrounding density-in-energy. Evolution toward more probable states, eventually attaining the stationary state, naturally selects steeply ascending paths on the entropy landscape that correspond to steeply descending paths on the free energy landscape.
The problem with this explanation is massive, and actually, untenable, naive, superficial and demanding. This explanation does not take into consideration that the right energy density at the right place must be precisely fine-tuned. Not any kind of energy density, anywhere within the protein structure will do, but on each place on the polypeptide " ladder ", there must be an emanating force from given amino acid, which will interact with the right strenght with an adjacent or nearby amino acid, which emanating force sums up with other forces to confer the right fold. Trial and error, or natural selection, are too unspecific and random to get a result which is functional
Structure Directing Amino acids (SDA)and the Protein Folding Code
The native ( amino acid) structure does not result from the participation of each amino acid inside the PPC but rather depends on some amino acids well positioned inside the sequence. Polypeptide chains presenting high sequence identity (up to 95%) can adopt different folds. Also, the growing number of molecules in PDB reveals that different polypeptide chains with sequence identity as low as 16% (e.g. 3RGK and 4O4T) can adopt very similar 3D-Structure. These reports clearly expose that there are well-defined amino acids inside the primary structure which dictate the form of the protein backbone. We call these amino acids SDA and have developed an alignment algorithm dedicated for their identification and classification.
AlphaFold: a solution to a 50-year-old grand challenge in biology November 30, 2020
What a protein does largely depends on its unique 3D structure. Figuring out what shapes proteins fold into is known as the “protein folding problem”, and has stood as a grand challenge in biology for the past 50 years. In a major scientific advance, the latest version of our AI system AlphaFold has been recognized as a solution to this grand challenge by the organizers of the biennial Critical Assessment of protein Structure Prediction (CASP).
https://www.deepmind.com/blog/alphafold-a-solution-to-a-50-year-old-grand-challenge-in-biology
Highly accurate protein structure prediction with AlphaFold
Xiaoliang Ma Physical Folding Codes for Proteins 2019
Exploring and understanding the protein-folding problem has been a long-standing challenge in molecular biology. Here, using molecular dynamics simulation, we reveal how parallel distributed adjacent planar peptide groups of unfolded proteins fold reproducibly following explicit physical folding codes in aqueous environments due to electrostatic attractions. Superfast folding of protein is found to be powered by the contribution of the formation of hydrogen bonds. Temperature-induced torsional waves propagating along unfolded proteins break the parallel distributed state of specific amino acids, inferred as the beginning of folding. Electric charge and rotational resistance differences among neighboring side-chains are used to decipher the physical folding codes by means of which precise secondary structures develop. We present a powerful method of decoding amino acid sequences to predict native structures of proteins. The method is verified by comparing the results available from experiments in the literature
Protein products are the basis of life on Earth and serve nearly all functions in the essential biochemistry of life science. The intrinsic biological functions of a protein are expressed and determined by its native three-dimensional (3D) structure derived from protein folding, which should be regarded as a central dogma of molecular biology. Protein folding performs the phenomenon of functionalizing polymer-like polypeptides into activated proteins and bringing millions of kinds of protein into existence. Protein folding can be considered the most important mechanism, principle, and motivation of biological existence, functionalization, diversity, and evolution. Based on the complexity of protein folding, the protein-folding problem has been summarized in three unanswered questions1 :
(i) What is the physical folding code in the amino acid sequence that determines the particular native 3D structure rather than any other of the unfathomable number of possible conformations?
(ii) What is the folding mechanism that enables proteins to fold so quickly?
(iii) Is it possible to devise a computer algorithm to effectively predict a protein’s native structure from its amino acid sequence? Moreover, protein folding requires aqueous environments and specific temperature ranges.
Here, a new algorithm, the microcanonical (NVE) ensemble relaxation engine (NVERE) is employed to reveal the mysteries of the four issues. Protein folding is considered a spontaneous free energy minimizing process or a relaxation process that is guided mainly by the following physical forces:
(i) formation of intramolecular hydrogen bonds,
(ii) van der Waals interactions,
(iii) electrostatic interactions,
(iv) hydrophobic interactions,
(v) chain entropy of protein.
Because molecular dynamics (MD) is capable of simultaneously describing all these physical forces and providing atomic-level resolution of protein models, MD has grown in popularity in protein-folding research since the 1980s . Using MD to answer the three main questions of protein folding has been an enduring goal. Laser temperature-jump studies have experimentally determined that alpha-helices and beta-sheets form very quickly at microsecond timescales. These findings all indicate that there may be an explicit folding code dominating the folding process or pathway of a given protein.
The existence, functionalization, diversity, and evolution of millions of kinds of protein on Earth must have been derived from the delicate coding strategies that fully utilize the differences in torsional resistance and in the electrical charges of side-chains. 5
Jon Rumbley An amino acid code for protein folding January 2, 2001
Direct structural information obtained for many proteins supports the following conclusions. The amino acid sequences of proteins can stabilize not only the final native state but also a small set of discrete partially folded native-like intermediates. Intermediates are formed in steps that use as units the cooperative secondary structural elements of the native protein. Earlier intermediates guide the addition of subsequent units in a process of sequential stabilization mediated by native-like tertiary interactions. The resulting stepwise self-assembly process automatically constructs a folding pathway, whether linear or branched. These conclusions are drawn mainly from hydrogen exchange-based methods, which can depict the structure of infinitesimally populated folding intermediates at equilibrium and kinetic intermediates with subsecond lifetimes. Other kinetic studies show that the polypeptide chain enters the folding pathway after an initial free-energy-uphill conformational search. The search culminates by finding a native-like topology that can support forward (native-like) folding in a free-energy-downhill manner. This condition automatically defines an initial transition state, the search for which sets the maximum possible (two-state) folding rate. It also extends the sequential stabilization strategy, which depends on a native-like context, to the first step in the folding process. Thus the native structure naturally generates its own folding pathway. The same amino acid code that translates into the final equilibrium native structure—by virtue of propensities, patterning, secondary structural cueing, and tertiary context—also produces its kinetic accessibility.
Conclusions
How does the amino acid sequence code for kinetic folding? Most simply stated, it appears to do so according to the same design principles that code for the native state. The structure-based information reviewed here consistently affirms that the folding process is dominated by native-like structure and interactions throughout. The supporting native-like structural context is laid down at the very first on-pathway step. Native-like tertiary interactions promote the progressive association of cooperative secondary structural units to form discrete folding intermediates. These factors determine a limited set of possible folding pathways. The effective kinetic barriers represent an initial large-scale conformational search for a native-like topology that ultimately limits the folding rate, subsequent smaller search-dependent barriers for putting sequential intermediates into place, and, when necessary, an additionally inserted thermal search to reverse nonnative misfolding errors. These same principles appear to explain the folding behavior of many proteins. 4

1. http://www.helsinki.fi/~aannila/arto/folding.pdf
2. https://chemistry.stackexchange.com/questions/53843/does-quantum-mechanics-play-a-role-in-protein-folding
3. https://www.technologyreview.com/s/423087/physicists-discover-quantum-law-of-protein-folding/
4. https://www.pnas.org/doi/10.1073/pnas.98.1.105
5. https://arxiv.org/ftp/arxiv/papers/1901/1901.00991.pdf
https://reasonandscience.catsboard.com/t2692-forces-stabilizing-proteins-essential-for-their-correct-folding
Proteins are the most complex molecules in life and are involved in basically all biochemical processes. Human cells contain 25000 genes, which can produce upon splicing up to over 100,000 different proteins.
The awe inspiringing spliceosome, the most complex macromolecular machine known, and pre-mRNA processing in eukaryotic cells
https://reasonandscience.catsboard.com/t2180-the-spliceosome-the-splicing-code-and-pre-mrna-processing-in-eukaryotic-cells
Proteins are the end products of the process of transcription and translation that starts with the information stored in the genome.
The function of proteins depends on the right sequence of amino acids, lined up into polypeptide chains of various sizes, ranging from short sequences, like extracellular hemoglobin with 140 amino acids, to monstrously large proteins, like tintin which is used in sarcomeres, responsible for passive elasticity of striated mu3scles. with lengths that vary from ~27,000 to ~33,000 amino acids (depending on the splice isoform), the largest known protein.
Titin the largest proteins known and titin-telethonin complex - the strongest protein bond found so far in nature
https://reasonandscience.catsboard.com/t2671-titin-the-largest-proteins-known-and-the-titin-telethonin-complex-the-strongest-protein-bond-found-so-far-in-nature
Proteins, in order to become functional, must fold into very specific 3D shapes, which happens right when they come out of the Ribosome, where they are synthesized. Specific protein shape and conformation depends on the interactions between its amino acid side chains. For a protein to function it must fold into a resting state which is a complex three-dimensional structure. If a protein fails to fold into its functional structure then it is not only without function but it cab become toxic to the cell. As proteins fold, they test a variety of conformations before reaching their final form, which is unique and compact. Folded proteins are stabilized by thousands of noncovalent bonds between amino acids. A relatively small protein of only 100 amino acids can take some 10^100 different configurations. If it tried these shapes at the rate of 100 billion a second, it would take longer than the age of the universe to find the correct one. Just how these molecules do the job in nanoseconds, nobody knows. 3
https://www.technologyreview.com/s/423087/physicists-discover-quantum-law-of-protein-folding/
Nobody knows, but since there was no evolution, to produce the first proteome for the first living cells, there are two options : blind, unguided, random lucky events on early earth, or the creative act of a super intelligent agent.
The 3D shape depends on the chemical forces, the chains are folded into a uniquely defined configuration, in which they are held by hydrogen bonds between the peptide nitrogen and oxygen atoms, which make a large contribution to protein stability, but other forces play an essential role as well, like hydrophobic interactions, disulfide bonds, Charge-charge interactions on the surface of proteins which are electrostatic interactions, pure ionic interactions, van der Waals forces, but also the surroundings of proteins, and their milieu. Chemical forces between a protein and its immediate environment contribute to protein shape and stability.

A special note deserve Van der Waals forces and interactions, which origins are ultimately a quantum mechanical. (At least the induced dipole portions: how the electrons move with respect to each other and an external electrical field is driven by quantum mechanics.) 2 Remarkably, quantum transition model fits the folding curves of 15 different proteins and even explains the difference in folding and unfolding rates of the same proteins. That means, the shape could change by quantum transition, meaning that the protein could ‘jump’ from one shape to another without necessarily forming the shapes in between. Impressive stuff !! 3
The 20 amino acids used in life are classified in Acidic amino acids, Basic amino acids with net positive charge, polar but uncharged, and nonpolar or hydrophobic amino acids. This variety contributes critically and is important for the processes that drive protein chains to “fold,” that is to form their natural (and functional) structures.
see more:
Amino Acids and peptide bonding
https://reasonandscience.catsboard.com/t2590-origins-what-cause-explains-best-our-existence-and-why#5845
But how could natural processes have foresight, which seems to be absolutely required, to "know" which amino acid sequences would provoke which forces, and how they would fold the protein structure to get functional for specific purposes within the cell ? Let's consider, that in order to have a minimal functional living cell, at least 561 proteins and protein complexes would have to be fully setup, working, and interacting together to confer a functional whole with all life-essential functions :
The last universal common ancestor represents the primordial cellular organism from which diversified life was derived
https://reasonandscience.catsboard.com/t2176-lucathe-last-universal-common-ancestor
A minimal estimate for the gene content of the last universal common ancestor
19 December 2005
A truly minimal estimate of the gene content of the last universal common ancestor, obtained by three different tree construction methods and the inclusion or not of eukaryotes (in total, there are 669 ortholog families distributed in 561 functional annotation descriptions, including 52 which remain uncharacterized)
So there had to be at least 561 proteins of various sizes, set up correctly in order to fold into functional 3D shapes, governed by chemical bond forces interacting between the individual amino acids. So not only is the right amino acid sequence essential but also as a side-effect the right dosage of the bond forces in between the amino acids.
The problem becomes even more severe when we consider that many, if not most proteins, are governed and build up by primary, secondary, tertiary, and quaternary structures. A quaternary structure refers to two or more polypeptide chains held together by intermolecular interactions to form a multi-subunit complex. And there are proteins, which require co-factors, often made of trace metals, like Iron, molybdenum etc. These cofactors require a pocket of the right size, and binding residues at the right place, and a tunnel where these cofactors can pass through during biosynthesis, to be inserted at the precise location inside the protein.
And there is more:
Molecular Chaperones Help Guide the Folding of Most Proteins
https://reasonandscience.catsboard.com/t1437-chaperones
Most proteins probably do not fold correctly during their synthesis and require a special class of proteins called molecular chaperones to do so. Molecular chaperones are useful for cells because there are many different folding paths available to an unfolded or partially folded protein. Without chaperones, some of these pathways would not lead to the correctly folded (and most stable) form because the protein would become “kinetically trapped” in structures that are off-pathway. Some of these off-pathway configurations would aggregate and be left as irreversible dead ends of nonfunctional (and potentially dangerous) structures. Proteins also require chaperones constantly, after correct folding, to maintain their functional states, which is called proteostasis.
That raises interesting questions: How should and could natural nonintelligent mechanisms forsee the necessity of chaperones in order to get a specific goal and result, that is functional proteins to make living organisms? Nonliving matter has no natural " drive " or purpose or goal to become living. The make of proteins to create life, however, is a multistep process of many parallel acting complex metabolic pathways and production-line like processes to make proteins and other life essential products like lipids, carbohydrates etc. The right folding of proteins is just one of several other essential processes in order to get a functional protein. But a functional protein by its own has no function unless correctly embedded through the right order of assembly at the right place.
Evolution of the correct protein foldings
To explain the origin of correct protein folds is paramount to explain the origin of life, and biodiversity. Its therefore of significant interest to see, how secular science papers explain its origin. Lets have a look at following:
Protein folding as an evolutionary process 1
7 December 2008
To provide a description that is consistent with other natural processes, protein folding is formulated from the principle of increasing entropy. It then becomes evident that protein folding is an evolutionary process among many others. During the course of folding protein structural hierarchy builds up in succession by diminishing energy density gradients in the quest for a stationary state determined by surrounding density-in-energy. Evolution toward more probable states, eventually attaining the stationary state, naturally selects steeply ascending paths on the entropy landscape that correspond to steeply descending paths on the free energy landscape.
The problem with this explanation is massive, and actually, untenable, naive, superficial and demanding. This explanation does not take into consideration that the right energy density at the right place must be precisely fine-tuned. Not any kind of energy density, anywhere within the protein structure will do, but on each place on the polypeptide " ladder ", there must be an emanating force from given amino acid, which will interact with the right strenght with an adjacent or nearby amino acid, which emanating force sums up with other forces to confer the right fold. Trial and error, or natural selection, are too unspecific and random to get a result which is functional
Structure Directing Amino acids (SDA)and the Protein Folding Code
The native ( amino acid) structure does not result from the participation of each amino acid inside the PPC but rather depends on some amino acids well positioned inside the sequence. Polypeptide chains presenting high sequence identity (up to 95%) can adopt different folds. Also, the growing number of molecules in PDB reveals that different polypeptide chains with sequence identity as low as 16% (e.g. 3RGK and 4O4T) can adopt very similar 3D-Structure. These reports clearly expose that there are well-defined amino acids inside the primary structure which dictate the form of the protein backbone. We call these amino acids SDA and have developed an alignment algorithm dedicated for their identification and classification.
AlphaFold: a solution to a 50-year-old grand challenge in biology November 30, 2020
What a protein does largely depends on its unique 3D structure. Figuring out what shapes proteins fold into is known as the “protein folding problem”, and has stood as a grand challenge in biology for the past 50 years. In a major scientific advance, the latest version of our AI system AlphaFold has been recognized as a solution to this grand challenge by the organizers of the biennial Critical Assessment of protein Structure Prediction (CASP).
https://www.deepmind.com/blog/alphafold-a-solution-to-a-50-year-old-grand-challenge-in-biology
Highly accurate protein structure prediction with AlphaFold
Xiaoliang Ma Physical Folding Codes for Proteins 2019
Exploring and understanding the protein-folding problem has been a long-standing challenge in molecular biology. Here, using molecular dynamics simulation, we reveal how parallel distributed adjacent planar peptide groups of unfolded proteins fold reproducibly following explicit physical folding codes in aqueous environments due to electrostatic attractions. Superfast folding of protein is found to be powered by the contribution of the formation of hydrogen bonds. Temperature-induced torsional waves propagating along unfolded proteins break the parallel distributed state of specific amino acids, inferred as the beginning of folding. Electric charge and rotational resistance differences among neighboring side-chains are used to decipher the physical folding codes by means of which precise secondary structures develop. We present a powerful method of decoding amino acid sequences to predict native structures of proteins. The method is verified by comparing the results available from experiments in the literature
Protein products are the basis of life on Earth and serve nearly all functions in the essential biochemistry of life science. The intrinsic biological functions of a protein are expressed and determined by its native three-dimensional (3D) structure derived from protein folding, which should be regarded as a central dogma of molecular biology. Protein folding performs the phenomenon of functionalizing polymer-like polypeptides into activated proteins and bringing millions of kinds of protein into existence. Protein folding can be considered the most important mechanism, principle, and motivation of biological existence, functionalization, diversity, and evolution. Based on the complexity of protein folding, the protein-folding problem has been summarized in three unanswered questions1 :
(i) What is the physical folding code in the amino acid sequence that determines the particular native 3D structure rather than any other of the unfathomable number of possible conformations?
(ii) What is the folding mechanism that enables proteins to fold so quickly?
(iii) Is it possible to devise a computer algorithm to effectively predict a protein’s native structure from its amino acid sequence? Moreover, protein folding requires aqueous environments and specific temperature ranges.
Here, a new algorithm, the microcanonical (NVE) ensemble relaxation engine (NVERE) is employed to reveal the mysteries of the four issues. Protein folding is considered a spontaneous free energy minimizing process or a relaxation process that is guided mainly by the following physical forces:
(i) formation of intramolecular hydrogen bonds,
(ii) van der Waals interactions,
(iii) electrostatic interactions,
(iv) hydrophobic interactions,
(v) chain entropy of protein.
Because molecular dynamics (MD) is capable of simultaneously describing all these physical forces and providing atomic-level resolution of protein models, MD has grown in popularity in protein-folding research since the 1980s . Using MD to answer the three main questions of protein folding has been an enduring goal. Laser temperature-jump studies have experimentally determined that alpha-helices and beta-sheets form very quickly at microsecond timescales. These findings all indicate that there may be an explicit folding code dominating the folding process or pathway of a given protein.
The existence, functionalization, diversity, and evolution of millions of kinds of protein on Earth must have been derived from the delicate coding strategies that fully utilize the differences in torsional resistance and in the electrical charges of side-chains. 5
Jon Rumbley An amino acid code for protein folding January 2, 2001
Direct structural information obtained for many proteins supports the following conclusions. The amino acid sequences of proteins can stabilize not only the final native state but also a small set of discrete partially folded native-like intermediates. Intermediates are formed in steps that use as units the cooperative secondary structural elements of the native protein. Earlier intermediates guide the addition of subsequent units in a process of sequential stabilization mediated by native-like tertiary interactions. The resulting stepwise self-assembly process automatically constructs a folding pathway, whether linear or branched. These conclusions are drawn mainly from hydrogen exchange-based methods, which can depict the structure of infinitesimally populated folding intermediates at equilibrium and kinetic intermediates with subsecond lifetimes. Other kinetic studies show that the polypeptide chain enters the folding pathway after an initial free-energy-uphill conformational search. The search culminates by finding a native-like topology that can support forward (native-like) folding in a free-energy-downhill manner. This condition automatically defines an initial transition state, the search for which sets the maximum possible (two-state) folding rate. It also extends the sequential stabilization strategy, which depends on a native-like context, to the first step in the folding process. Thus the native structure naturally generates its own folding pathway. The same amino acid code that translates into the final equilibrium native structure—by virtue of propensities, patterning, secondary structural cueing, and tertiary context—also produces its kinetic accessibility.
Conclusions
How does the amino acid sequence code for kinetic folding? Most simply stated, it appears to do so according to the same design principles that code for the native state. The structure-based information reviewed here consistently affirms that the folding process is dominated by native-like structure and interactions throughout. The supporting native-like structural context is laid down at the very first on-pathway step. Native-like tertiary interactions promote the progressive association of cooperative secondary structural units to form discrete folding intermediates. These factors determine a limited set of possible folding pathways. The effective kinetic barriers represent an initial large-scale conformational search for a native-like topology that ultimately limits the folding rate, subsequent smaller search-dependent barriers for putting sequential intermediates into place, and, when necessary, an additionally inserted thermal search to reverse nonnative misfolding errors. These same principles appear to explain the folding behavior of many proteins. 4

1. http://www.helsinki.fi/~aannila/arto/folding.pdf
2. https://chemistry.stackexchange.com/questions/53843/does-quantum-mechanics-play-a-role-in-protein-folding
3. https://www.technologyreview.com/s/423087/physicists-discover-quantum-law-of-protein-folding/
4. https://www.pnas.org/doi/10.1073/pnas.98.1.105
5. https://arxiv.org/ftp/arxiv/papers/1901/1901.00991.pdf
Last edited by Otangelo on Wed Jun 22, 2022 11:15 am; edited 3 times in total