Actin filament assembly , and how it points to intelligent design 1
Actin is a family of globular multi-functional proteins that form microfilaments. It is found in essentially all eukaryotic cells (the only known exception being nematode sperm), where it may be present at a concentration of over 100 μM. An actin protein's mass is roughly 42-kDa, with a diameter of 4 to 7 nm, and it is the monomeric subunit of two types of filaments in cells: microfilaments, one of the three major components of the cytoskeleton, and thin filaments, part of the contractile apparatus in muscle cells. It can be present as either a free monomer called G-actin (globular) or as part of a linear polymer microfilament called F-actin(filamentous), both of which are essential for such important cellular functions as the mobility and contraction of cells during cell division.
ACTIN AND ACTIN-BINDING PROTEINS
The actin cytoskeleton performs a wide range of functions in diverse cell types. Each actin subunit, sometimes called globular or G-actin, is a 375-amino-acid polypeptide carrying a tightly associated molecule of ATP or ADP (Figure16–11A).

Actin is extraordinarily well conserved among eukaryotes. The amino acid sequences of actins from different eukaryotic species are usually about 90% identical. Small variations in actin amino acid sequence can cause significant functional differences: In vertebrates, for example, there are three isoforms of actin, termed α, β, and γ, that differ slightly in their amino acid sequences and have distinct functions. α-Actin is expressed only in muscle cells, while β- and γ-actins are found together in almost all non-muscle cells.
Actin Subunits Assemble Head-to-Tail to Create Flexible, Polar Filaments
Actin subunits assemble head-to-tail to form a tight, right-handed helix, forming a structure about 8 nm wide called filamentous or F-actin (Figure 16–11B and C). Because the asymmetrical actin subunits of a filament all point in the same direction, filaments are polar and have structurally different ends: a slower-growing minus end and a faster-growing plus end. The minus end is also referred to as the “pointed end” and the plus end as the “barbed end,” because of the “arrowhead”appearance of the complex formed between actin filaments and the motor protein myosin (Figure 16–12).
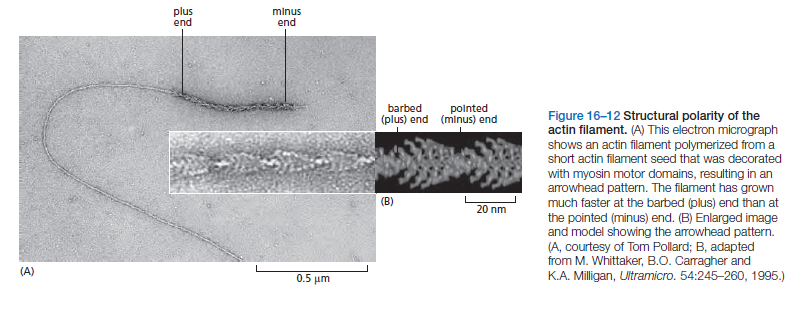
Within the filament, the subunits are positioned with their nucleotide-binding cleft directed toward the minus end. Individual actin filaments are quite flexible. The stiffness of a filament can be characterized by its persistence length, the minimum filament length at which random thermal fluctuations are likely to cause it to bend. The persistence length
of an actin filament is only a few tens of micrometers. In a living cell, however, accessory proteins cross-link and bundle the filaments together, making largescale actin structures that are much more rigid than an individual actin filament.
Nucleation Is the Rate-Limiting Step in the Formation of Actin Filaments
The regulation of actin filament formation is an important mechanism by which cells control their shape and movement. Small oligomers of actin subunits can assemble spontaneously, but they are unstable and disassemble readily because each monomer is bound to only one or two other monomers. For a new actin filament to form, subunits must assemble into an initial aggregate, or nucleus, that is stabilized by multiple subunit–subunit contacts and can then elongate rapidly by addition of more subunits. This process is called filament nucleation. Many features of actin nucleation and polymerization have been studied with purified actin in a test tube (Figure 16–13).
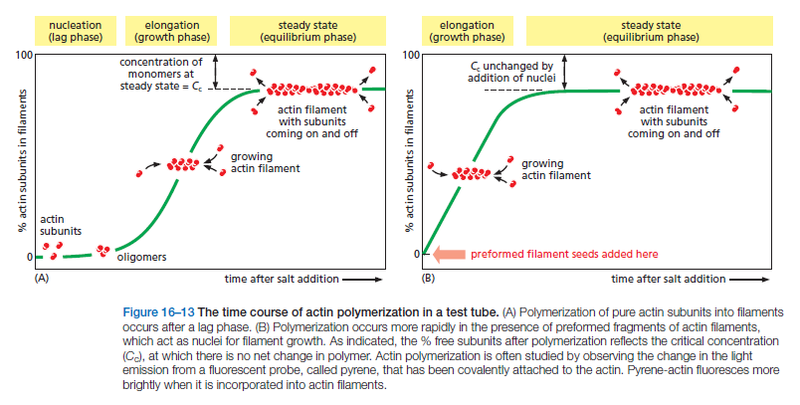
The instability of smaller actin aggregates creates a kinetic barrier to nucleation. When polymerization is initiated, this barrier results in a lag phase during which no filaments are observed. During this lag phase, however, a few of the small, unstable aggregates succeed in making the transition to a more stable form that resembles an actin filament. This leads to a phase of rapid filament elongation during which subunits are added quickly to the ends of the nucleated filaments (Figure 16–13A). Finally, as the concentration of actin monomers declines, the system approaches a steady state at which the rate of addition of new subunits to the filament ends exactly balances the rate of subunit dissociation. The concentration of free subunits left in solution at this point is called the critical concentration, Cc. As explained in Panel 16–2, the value of the critical concentration is equal to the rate constant for subunit loss divided by the rate constant for subunit addition—that is, Cc = koff/kon, which is equal to the dissociation constant, Kd, and the inverse of the equilibrium constant, K
Actin Filaments Have Two Distinct Ends That Grow at Different Rates
Due to the uniform orientation of asymmetric actin subunits in the filament, the structures at its two ends are different. This orientation makes the two ends of each polymer different in ways that have a profound effect on filament growth rates. The kinetic rate constants for actin subunit association and dissociation—kon and koff, respectively—are much greater at the plus end than the minus end. This can be seen when an excess of purified actin monomers is allowed to assemble onto polarity-marked filaments—the plus end of the filament elongates up to ten times faster (see Figure 16–12). If filaments are rapidly diluted so that the free subunit concentration drops below the critical concentration, the plus end also depolymerizes faster.
It is important to note, however, that the two ends of an actin filament have the same net affinity for actin subunits if all of the subunits are in the same nucleotide state. The addition of a subunit to either end of a filament of n subunits results in a filament of n + 1 subunits. Thus, the free-energy difference, and therefore the equilibrium constant (and the critical concentration), must be the same for the addition of subunits at either end of the polymer. In this case, the ratio of the rate constants, koff/kon, must be identical at the two ends, even though the absolute values of these rate constants are very different at each end. The cell takes advantage of actin filament dynamics and polarity to do mechanical work. Filament elongation proceeds spontaneously when the free-energy change (ΔG) for the addition of the soluble subunit is less than zero. This is the case when the concentration of subunits in solution exceeds the critical concentration. A cell can couple an energetically unfavorable process to this spontaneous process; thus, the cell can use free energy released during spontaneous filament polymerization to move an attached load. For example, by orienting the fast-growing plus ends of actin filaments toward its leading-edge, a motile cell can push its plasma membrane forward.
ATP Hydrolysis Within Actin Filaments Leads to Treadmilling at Steady State
Actin can catalyze the hydrolysis of the nucleoside triphosphate ATP. For free actin subunits, this hydrolysis proceeds very slowly; however, it is accelerated when the subunits are incorporated into filaments. Shortly after ATP hydrolysis occurs, the free phosphate group is released from each subunit, but the ADP remains trapped in the filament structure. Thus, two different types of filament structures can exist, one with the “T form” of the nucleotide bound (ATP), and one with the “D form” bound (ADP). When the nucleotide is hydrolyzed, much of the free energy released by cleavage of the phosphate–phosphate bond is stored in the polymer. This makes the free-energy change for dissociation of a subunit from the D-form polymer more negative than the free-energy change for dissociation of a subunit from the T-form polymer. Consequently, the ratio of koff/kon for the D-form polymer, which is numerically equal to its critical concentration [Cc(D)], is larger than the corresponding ratio for the T-form polymer. Thus, Cc(D) is greater than Cc(T). At certain concentrations of free subunits, D-form polymers will therefore shrink while T-form polymers grow. In living cells, most soluble actin subunits are in the T form, as the free concentration of ATP is about tenfold higher than that of ADP. However, the longer the time that subunits have been in the actin filament, the more likely they are to have hydrolyzed their ATP. Whether the subunit at each end of a filament is in the T or the D form depends on the rate of this hydrolysis compared with the rate of subunit addition. If the concentration of actin monomers is greater than the critical concentration for both the T-form and D-form polymer, then subunits will add to the polymer at both ends before the nucleotides in the previously added subunits are hydrolyzed; as a result, the tips of the actin filament will remain in the T form. On the other hand, if the subunit concentration is less than the critical concentrations for both the T-form and D-form polymer, then hydrolysis may occur before the next subunit is added and both ends of the filament will be in the D form and will shrink. At intermediate concentrations of actin subunits, it is possible for the rate of subunit addition to be faster than nucleotide hydrolysis at the plus end, but slower than nucleotide hydrolysis at the minus end. In this case, the plus end of the filament remains in the T conformation, while the minus end adopts the D conformation. The filament then undergoes a net addition of subunits at the plus end, while simultaneously losing subunits from the minus end. This leads to the remarkable property of filament treadmilling.
Monomer Availability Controls Actin Filament Assembly
In most non-muscle vertebrate cells, approximately 50% of the actin is in filaments and 50% is soluble—and yet the soluble monomer concentration is 50–200 μM, well above the critical concentration. Why does so little of the actin
polymerize into filaments? The reason is that the cell contains proteins that bind to the actin monomers and make polymerization much less favorable. A small protein called thymosin is the most abundant of these proteins. Actin monomers bound to thymosin are in a locked state, where they cannot associate with either the plus or minus ends of actin filaments and can neither hydrolyze nor exchange their bound nucleotide. How do cells recruit actin monomers from this buffered storage pool and use them for polymerization? The answer depends on another monomer-binding protein called profilin. Profilin binds to the face of the actin monomer opposite the ATP-binding cleft, blocking the side of the monomer that would normally associate with the filament minus end, while leaving exposed the site on the monomer that binds to the plus end (Figure 16–15).
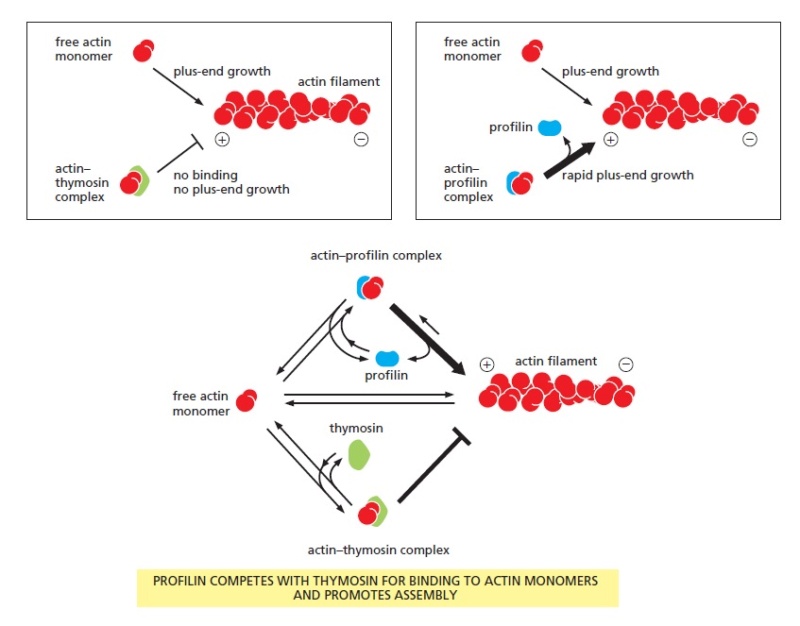
Effects of thymosin and profilin on actin polymerization.
An actin monomer bound to thymosin is sterically prevented from binding to and elongating the plus end of an actin filament (left). An actin monomer bound to profilin, on the other hand, is capable of elongating a filament (right). Thymosin and profilin cannot both bind to a single actin monomer at the same time. In a cell in which most of the actin monomer is bound to thymosin, the activation of a small amount of profilin can produce rapid filament assembly. As indicated (bottom), profilin binds to actin monomers that are transiently released from the thymosin-bound monomer pool, shuttles them onto the plus ends of actin filaments, and is then released and recycled for
further rounds of filament elongation.
When the profilin–actin complex binds a free plus end, a conformational change in actin reduces its affinity for profilin and the profilin falls off, leaving the actin filament one subunit longer. Profilin competes with thymosin for binding to individual actin monomers. Thus, by regulating the local activity of profilin, cells can control the movement of actin subunits from the sequestered thymosin-bound pool onto filament plus ends. Several mechanisms regulate profilin activity, including profilin phosphorylation and profilin binding to inositol phospholipids. These mechanisms can define the sites where profilin acts. For example, profilin is required for filament assembly at the plasma membrane, where it is recruited by an interaction with acidic membrane phospholipids. At this location, extracellular signals can activate profilin to produce local actin polymerization and the extension of actin-rich motile structures such as filopodia and lamellipodia.
This is clearly an irreducible process, where all three players have to be in place, thymosin, profilin, and the actin subunits, in order to make the elongation process possible.
Actin is a family of globular multi-functional proteins that form microfilaments. It is found in essentially all eukaryotic cells (the only known exception being nematode sperm), where it may be present at a concentration of over 100 μM. An actin protein's mass is roughly 42-kDa, with a diameter of 4 to 7 nm, and it is the monomeric subunit of two types of filaments in cells: microfilaments, one of the three major components of the cytoskeleton, and thin filaments, part of the contractile apparatus in muscle cells. It can be present as either a free monomer called G-actin (globular) or as part of a linear polymer microfilament called F-actin(filamentous), both of which are essential for such important cellular functions as the mobility and contraction of cells during cell division.
ACTIN AND ACTIN-BINDING PROTEINS
The actin cytoskeleton performs a wide range of functions in diverse cell types. Each actin subunit, sometimes called globular or G-actin, is a 375-amino-acid polypeptide carrying a tightly associated molecule of ATP or ADP (Figure16–11A).

Actin is extraordinarily well conserved among eukaryotes. The amino acid sequences of actins from different eukaryotic species are usually about 90% identical. Small variations in actin amino acid sequence can cause significant functional differences: In vertebrates, for example, there are three isoforms of actin, termed α, β, and γ, that differ slightly in their amino acid sequences and have distinct functions. α-Actin is expressed only in muscle cells, while β- and γ-actins are found together in almost all non-muscle cells.
Actin Subunits Assemble Head-to-Tail to Create Flexible, Polar Filaments
Actin subunits assemble head-to-tail to form a tight, right-handed helix, forming a structure about 8 nm wide called filamentous or F-actin (Figure 16–11B and C). Because the asymmetrical actin subunits of a filament all point in the same direction, filaments are polar and have structurally different ends: a slower-growing minus end and a faster-growing plus end. The minus end is also referred to as the “pointed end” and the plus end as the “barbed end,” because of the “arrowhead”appearance of the complex formed between actin filaments and the motor protein myosin (Figure 16–12).

Within the filament, the subunits are positioned with their nucleotide-binding cleft directed toward the minus end. Individual actin filaments are quite flexible. The stiffness of a filament can be characterized by its persistence length, the minimum filament length at which random thermal fluctuations are likely to cause it to bend. The persistence length
of an actin filament is only a few tens of micrometers. In a living cell, however, accessory proteins cross-link and bundle the filaments together, making largescale actin structures that are much more rigid than an individual actin filament.
Nucleation Is the Rate-Limiting Step in the Formation of Actin Filaments
The regulation of actin filament formation is an important mechanism by which cells control their shape and movement. Small oligomers of actin subunits can assemble spontaneously, but they are unstable and disassemble readily because each monomer is bound to only one or two other monomers. For a new actin filament to form, subunits must assemble into an initial aggregate, or nucleus, that is stabilized by multiple subunit–subunit contacts and can then elongate rapidly by addition of more subunits. This process is called filament nucleation. Many features of actin nucleation and polymerization have been studied with purified actin in a test tube (Figure 16–13).

The instability of smaller actin aggregates creates a kinetic barrier to nucleation. When polymerization is initiated, this barrier results in a lag phase during which no filaments are observed. During this lag phase, however, a few of the small, unstable aggregates succeed in making the transition to a more stable form that resembles an actin filament. This leads to a phase of rapid filament elongation during which subunits are added quickly to the ends of the nucleated filaments (Figure 16–13A). Finally, as the concentration of actin monomers declines, the system approaches a steady state at which the rate of addition of new subunits to the filament ends exactly balances the rate of subunit dissociation. The concentration of free subunits left in solution at this point is called the critical concentration, Cc. As explained in Panel 16–2, the value of the critical concentration is equal to the rate constant for subunit loss divided by the rate constant for subunit addition—that is, Cc = koff/kon, which is equal to the dissociation constant, Kd, and the inverse of the equilibrium constant, K
Actin Filaments Have Two Distinct Ends That Grow at Different Rates
Due to the uniform orientation of asymmetric actin subunits in the filament, the structures at its two ends are different. This orientation makes the two ends of each polymer different in ways that have a profound effect on filament growth rates. The kinetic rate constants for actin subunit association and dissociation—kon and koff, respectively—are much greater at the plus end than the minus end. This can be seen when an excess of purified actin monomers is allowed to assemble onto polarity-marked filaments—the plus end of the filament elongates up to ten times faster (see Figure 16–12). If filaments are rapidly diluted so that the free subunit concentration drops below the critical concentration, the plus end also depolymerizes faster.
It is important to note, however, that the two ends of an actin filament have the same net affinity for actin subunits if all of the subunits are in the same nucleotide state. The addition of a subunit to either end of a filament of n subunits results in a filament of n + 1 subunits. Thus, the free-energy difference, and therefore the equilibrium constant (and the critical concentration), must be the same for the addition of subunits at either end of the polymer. In this case, the ratio of the rate constants, koff/kon, must be identical at the two ends, even though the absolute values of these rate constants are very different at each end. The cell takes advantage of actin filament dynamics and polarity to do mechanical work. Filament elongation proceeds spontaneously when the free-energy change (ΔG) for the addition of the soluble subunit is less than zero. This is the case when the concentration of subunits in solution exceeds the critical concentration. A cell can couple an energetically unfavorable process to this spontaneous process; thus, the cell can use free energy released during spontaneous filament polymerization to move an attached load. For example, by orienting the fast-growing plus ends of actin filaments toward its leading-edge, a motile cell can push its plasma membrane forward.
ATP Hydrolysis Within Actin Filaments Leads to Treadmilling at Steady State
Actin can catalyze the hydrolysis of the nucleoside triphosphate ATP. For free actin subunits, this hydrolysis proceeds very slowly; however, it is accelerated when the subunits are incorporated into filaments. Shortly after ATP hydrolysis occurs, the free phosphate group is released from each subunit, but the ADP remains trapped in the filament structure. Thus, two different types of filament structures can exist, one with the “T form” of the nucleotide bound (ATP), and one with the “D form” bound (ADP). When the nucleotide is hydrolyzed, much of the free energy released by cleavage of the phosphate–phosphate bond is stored in the polymer. This makes the free-energy change for dissociation of a subunit from the D-form polymer more negative than the free-energy change for dissociation of a subunit from the T-form polymer. Consequently, the ratio of koff/kon for the D-form polymer, which is numerically equal to its critical concentration [Cc(D)], is larger than the corresponding ratio for the T-form polymer. Thus, Cc(D) is greater than Cc(T). At certain concentrations of free subunits, D-form polymers will therefore shrink while T-form polymers grow. In living cells, most soluble actin subunits are in the T form, as the free concentration of ATP is about tenfold higher than that of ADP. However, the longer the time that subunits have been in the actin filament, the more likely they are to have hydrolyzed their ATP. Whether the subunit at each end of a filament is in the T or the D form depends on the rate of this hydrolysis compared with the rate of subunit addition. If the concentration of actin monomers is greater than the critical concentration for both the T-form and D-form polymer, then subunits will add to the polymer at both ends before the nucleotides in the previously added subunits are hydrolyzed; as a result, the tips of the actin filament will remain in the T form. On the other hand, if the subunit concentration is less than the critical concentrations for both the T-form and D-form polymer, then hydrolysis may occur before the next subunit is added and both ends of the filament will be in the D form and will shrink. At intermediate concentrations of actin subunits, it is possible for the rate of subunit addition to be faster than nucleotide hydrolysis at the plus end, but slower than nucleotide hydrolysis at the minus end. In this case, the plus end of the filament remains in the T conformation, while the minus end adopts the D conformation. The filament then undergoes a net addition of subunits at the plus end, while simultaneously losing subunits from the minus end. This leads to the remarkable property of filament treadmilling.
Monomer Availability Controls Actin Filament Assembly
In most non-muscle vertebrate cells, approximately 50% of the actin is in filaments and 50% is soluble—and yet the soluble monomer concentration is 50–200 μM, well above the critical concentration. Why does so little of the actin
polymerize into filaments? The reason is that the cell contains proteins that bind to the actin monomers and make polymerization much less favorable. A small protein called thymosin is the most abundant of these proteins. Actin monomers bound to thymosin are in a locked state, where they cannot associate with either the plus or minus ends of actin filaments and can neither hydrolyze nor exchange their bound nucleotide. How do cells recruit actin monomers from this buffered storage pool and use them for polymerization? The answer depends on another monomer-binding protein called profilin. Profilin binds to the face of the actin monomer opposite the ATP-binding cleft, blocking the side of the monomer that would normally associate with the filament minus end, while leaving exposed the site on the monomer that binds to the plus end (Figure 16–15).

Effects of thymosin and profilin on actin polymerization.
An actin monomer bound to thymosin is sterically prevented from binding to and elongating the plus end of an actin filament (left). An actin monomer bound to profilin, on the other hand, is capable of elongating a filament (right). Thymosin and profilin cannot both bind to a single actin monomer at the same time. In a cell in which most of the actin monomer is bound to thymosin, the activation of a small amount of profilin can produce rapid filament assembly. As indicated (bottom), profilin binds to actin monomers that are transiently released from the thymosin-bound monomer pool, shuttles them onto the plus ends of actin filaments, and is then released and recycled for
further rounds of filament elongation.
When the profilin–actin complex binds a free plus end, a conformational change in actin reduces its affinity for profilin and the profilin falls off, leaving the actin filament one subunit longer. Profilin competes with thymosin for binding to individual actin monomers. Thus, by regulating the local activity of profilin, cells can control the movement of actin subunits from the sequestered thymosin-bound pool onto filament plus ends. Several mechanisms regulate profilin activity, including profilin phosphorylation and profilin binding to inositol phospholipids. These mechanisms can define the sites where profilin acts. For example, profilin is required for filament assembly at the plasma membrane, where it is recruited by an interaction with acidic membrane phospholipids. At this location, extracellular signals can activate profilin to produce local actin polymerization and the extension of actin-rich motile structures such as filopodia and lamellipodia.
This is clearly an irreducible process, where all three players have to be in place, thymosin, profilin, and the actin subunits, in order to make the elongation process possible.

