All cellular functions are irreducibly complex 3
https://reasonandscience.catsboard.com/t2179-the-cell-is-a-interdependent-irreducible-complex-system
chemist Wilhelm Huck, professor at Radboud University Nijmegen
http://www.ru.nl/english/research/radboud/themes/genetics-cellular/vm/news-genetics-cell/@893712/protocells-formed/
A working cell is more than the sum of its parts. "A functioning cell must be entirely correct at once, in all its complexity,"
Paul Davies, the fifth miracle page 53:
Pluck the DNA from a living cell and it would be stranded, unable to carry out its familiar role. Only within the context of a highly specific molecular milieu will a given molecule play its role in life. To function properly, DNA must be part of a large team, with each molecule executing its assigned task alongside the others in a cooperative manner. Acknowledging the interdependability of the component molecules within a living organism immediately presents us with a stark philosophical puzzle. If everything needs everything else, how did the community of molecules ever arise in the first place? Since most large molecules needed for life are produced only by living organisms, and are not found outside the cell, how did they come to exist originally, without the help of a meddling scientist? Could we seriously expect a Miller-Urey type of soup to make them all at once, given the hit-and-miss nature of its chemistry?
Being part of a large team,cooperative manner,interdependability,everything needs everything else, are just other words for irreducibility and interdependence.
For a nonliving system, questions about irreducible complexity are even more challenging for a totally natural non-design scenario, because natural selection — which is the main mechanism of Darwinian evolution — cannot exist until a system can reproduce. For an origin of life we can think about the minimal complexity that would be required for reproduction and other basic life-functions. Most scientists think this would require hundreds of biomolecular parts, not just the five parts in a simple mousetrap or in my imaginary LMNOP system. And current science has no plausible theories to explain how the minimal complexity required for life (and the beginning of biological natural selection) could have been produced by natural process before the beginning of biological natural selection.
Determination of the Core of a Minimal Bacterial Gene Set
https://reasonandscience.catsboard.com/t2110-what-might-be-a-protocells-minimal-requirement-of-parts#3797
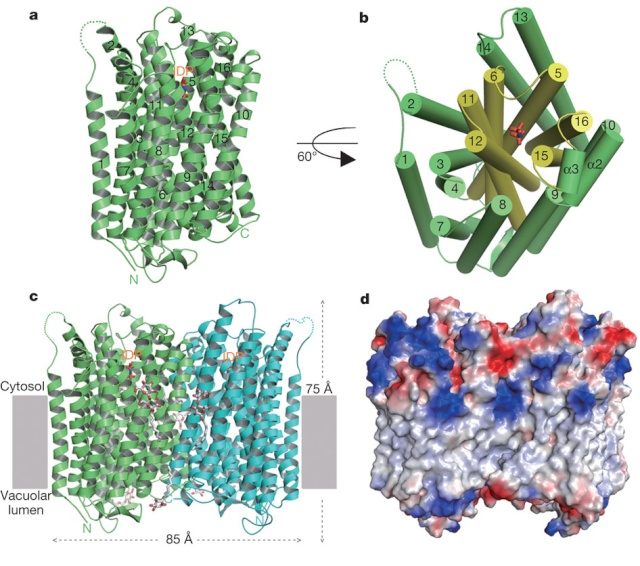
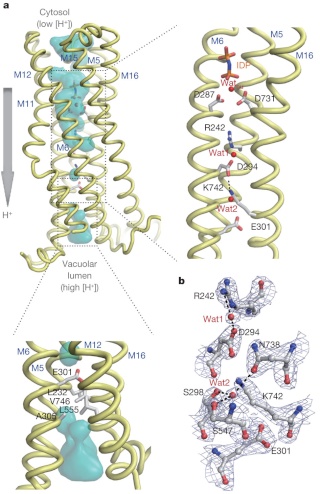
Prokaryotes are thought to differ from eukaryotes in that they lack membrane-bounded organelles. However, it has been demonstrated that there are bacterias which have membrane bound organelles named acidocalcisomes, and that V-H+PPase proton pumps are present in their surrounding membranes. Acidocalcisomes have been found in organisms as diverse as bacteria and humans. Volutin granules which are equivalent of acidocalcisomes also occur in Archaea and are, therefore, present in the three superkingdoms of life (Archaea, Bacteria and Eukarya). These volutin granule organelles occur in organisms spanning an enormous range of phylogenetic complexity from Bacteria and Archaea to unicellular eukaryotes to algae to plants to insects to humans. According to neo-darwinian thinking, the universal distribution of the V-H+PPase domain suggests the domain and the enzyme were already present in the Last Universal Common Ancestor (LUCA).
If the proton pumps of Volutin granules were present in LUCA, they had to emerge prior to self replication, which induces serious constraints to propose evolution as driving factor. But if evolution was not the mechanism, what else was ? There is not much left, namely chance, random chemical reactions, or physical necessity.
But lets for a instance accept the "fact of evolution", and suppose it was the driving force to make V-H+PPase proton pumps. In some period prior to the verge of non-life to life, natural selection or an other evolutionary mechanism would have had to start polymerisation of the right amino acid sequence to produce V-H+PPase proton pumps by addition of one amino acid monomer to the other. First, the whole extraordinarly production line of staggering complexity starting with DNA would have to be in place, that is :
The cell sends activator proteins to the site of the gene that needs to be switched on, which then jump-starts the RNA polymerase machine by removing a plug which blocks the DNA's entrance to the machine. The DNA strands do shift position so that the DNA lines up with the entrance to the RNA polymerase. Once these two movements have occurred and the DNA strands are in position, the RNA polymerase machine gets to work melting them out, so that the information they contain can be processed to produce mRNA 2 The process follows then after INITIATION OF TRANSCRIPTION through RNA polymerase enzyme complexes, the mRNA is capped through Post-transcriptional modifications by several different enzymes , ELONGATION provides the main transcription process from DNA to mRNA, furthermore SPLICING and CLEAVAGE , polyadenylation where a long string of repeated adenosine nucleotides is added, AND TERMINATION through over a dozen different enzymes, EXPORT FROM THE NUCLEUS TO THE CYTOSOL ( must be actively transported through the Nuclear Pore Complex channel in a controlled process that is selective and energy dependent ) INITIATION OF PROTEIN SYNTHESIS (TRANSLATION) in the Ribosome in a enormously complex process, COMPLETION OF PROTEIN SYNTHESIS AND PROTEIN FOLDING through chaperone enzymes. From there the proteins are transported by specialized proteins to the end destination. Most of these processes require ATP, the energy fuel inside the cell.
http://reasonandscience.heavenforum.org/t2067-there-is-no-selective-advantage-until-you-get-the-final-function?highlight=function
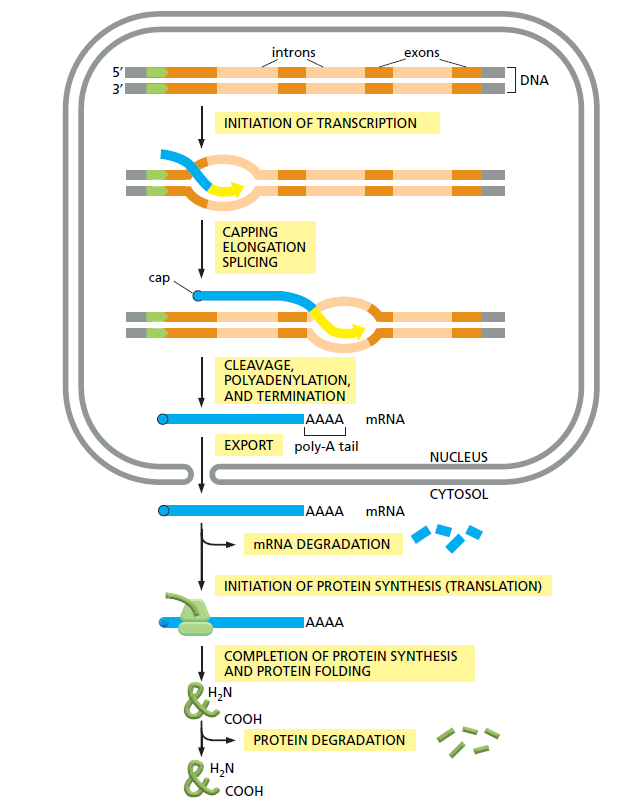
The genetic code to make the right ~600 amino acid sequence would have to be achieved not by mutation , since that would require a pre-existing amino acid sequence, but adding randomly a new amino acid, and select the advantageous sequence. Instead as by evolution, which evolves pre-existing proteins, in a origin of life scenario proteins would have all to be produced denovo. The problem in this stage is, when there is no selective advantage until you get the final function, the final function doesn't evolve. In other words, a chain of around 600 amino acids is required to make a funcional V-H+PPase proton pump, but there is no function, until polymerisation of all 600 monomers is completed and the right sequence achieved.
The problem for those who accept the truth of evolution is, they cannot accept the idea that any biological structure with a beneficial function, however complex, is very far removed from the next closest functional system or subsystem within the potential of "sequence space" that might be beneficial if it were ever found by random mutations of any kind. In our case the situation is even more drastic, since DENOVO genetic sequence and subsequently amino acid chain for a new formation of a new amino acids strand is required. A further constraint is the fact that 100% of amino acids used and needed for life are left handed, while DNA and RNA requires D-sugars. Until today, science has not sorted out how nature is able to select the right chiral handedness. The problem is its believed there to be a warm soup consisting of racemic mixtures of amino acid enantiomers (and sugars). How did this homogenous phase separate into chirally pure components? How did an asymmetry (assumed to be small to start with) arise in the population of both enantiomers? How did the preference of one chiral form over the other, propagate so that all living systems are made of 100 percent optically pure components?
What is sequence space ? 1
Imagine 20 amino acids mixed up in a pool, randomly mixed , one adjacent to the other. The pool with all the random amino acids is the sequence space. This space can be two dimentional, tridimensional, or multidimensional. In evolutionary biology, sequence space is a way of representing all possible sequences (for a protein, gene or genome). Most sequences in sequence space have no function, leaving relatively small regions that are populated by naturally occurring genes. Each protein sequence is adjacent to all other sequences that can be reached through a single mutation. Evolution can be visualised as the process of sampling nearby sequences in sequence space and moving to any with improved fitness over the current one.
Functional sequences in sequence space
Despite the diversity of protein superfamilies, sequence space is extremely sparsely populated by functional proteins. That is, amongst all the possible amino acid sequences, only a few permit the make of functional proteins. Most random protein sequences have no fold or function. To exemplify: In order to write METHINKS IT IS LIKE A WEASEL , there are 10^40 possible random combinations possible to get the right sequence. But only one is correct.
Enzyme superfamilies, therefore, exist as tiny clusters of active proteins in a vast empty space of non-functional sequence.The density of functional proteins in sequence space, and the proximity of different functions to one another is a key determinant in understanding evolvability. Protein sequence space has been compared to the Library of Babel a theoretical library containing all possible books that are 410 pages long. In the Library of Babel, finding any book that made sense was impossible due to the sheer number and lack of order.
How would a bacterium evolve a function like a single protein enzyme? - like a V-H+PPase proton pump? The requirement is about 600 specified residues at minimum. A useful V-H+PPase cannot be made with significantly lower minimum size and specificity requirements. These minimum requirements create a kind of threshold beyond which the V-H+PPase function simply cannot be built up gradually where very small one or two residues changes at a time result in a useful change in the degree of the proton pump function. Therefore, such functions cannot have evolved in a gradual, step by step manner. There simply is no template or gradual pathway from just any starting point to the minimum threshold requirement. Only after this threshold has been reached can evolution take over and make further refinements - but not until then.
All Functions are "Irreducibly Complex" 4
The fact is that all cellular functions are irreducibly complex in that all of them require a minimum number of parts in a particular order or orientation. I go beyond what Behe proposes and make the suggestion that even single-protein enzymes are irreducibly complex. A minimum number of parts in the form of amino acid residues are required for them to have their particular functions. The proton pump function cannot be realized in even the smallest degree with a string of only 5 or 10 or even 500 residues of any arrangement. Also, not only is a minimum number of parts required for the proton pump function to be realized, but the parts themselves, once they are available in the proper number, must be assembled in the proper order and three-dimensional orientation. Brought together randomly, the residues, if left to themselves, do not know how to self-assemble themselves to form a much of anything as far as a functional system that even comes close to the level of complexity of a even a relatively simple function like a proton pump. And yet, their specified assembly and ultimate order is vital to function.
Of course, such relatively simply systems, though truly irreducibly complex, have evolved. This is because the sequence space at such relatively low levels of functional complexity is fairly dense. It is fairly easy to come across new beneficial sequences if the density of potentially beneficial sequences in sequence space is relatively high. This density does in fact get higher and higher at lower and lower levels of functional complexity - in an exponential manner. Darwinian evolution can work fine when one small step (e.g., a single point mutation) along an evolutionary pathway gives an advantage. The theory of intelligent design has no problem with this.
It is much like moving between 3-letter words in the English language system. Since the ratio of meaningful vs. meaningless 3-letter words in the English language is somewhere around 1:18, one can randomly find a new meaningful and even beneficial 3-letter word via single random letter changes/mutations in relatively short order. This is not true for those ideas/functions/meanings that require more and more letters. For example, the ratio of meaningful vs. meaningless 7-letter words and combinations of smaller words equaling 7-letters is far far lower at about 1 in 250,000. It is therefore just a bit harder to evolve between 7-letter words, one mutation at a time, than it was to evolve between 3-letter words owing to the exponential decline in the ratio of meaningful vs. meaningless sequences.
The same thing is true for the evolution of codes, information systems, and systems of function in living things as it is for non-living things (i.e., computer systems etc). The parts of these codes and systems of function, if brought together randomly, simply do not have enough meaningful information to do much of anything. So, how are they brought together in living things to form such high level functional order?
1) https://en.wikipedia.org/wiki/Sequence_space_(evolution)
2) http://creationwiki.org/Weasel_program
3) http://www.detectingdesign.com/irreduciblemousetrap.html
4) http://www.sciencedaily.com/releases/2013/07/130702100115.htm
5) http://www.ru.nl/english/research/radboud/themes/genetics-cellular/vm/news-genetics-cell/@893712/protocells-formed/
https://reasonandscience.catsboard.com/t2179-the-cell-is-a-interdependent-irreducible-complex-system
chemist Wilhelm Huck, professor at Radboud University Nijmegen
http://www.ru.nl/english/research/radboud/themes/genetics-cellular/vm/news-genetics-cell/@893712/protocells-formed/
A working cell is more than the sum of its parts. "A functioning cell must be entirely correct at once, in all its complexity,"
Paul Davies, the fifth miracle page 53:
Pluck the DNA from a living cell and it would be stranded, unable to carry out its familiar role. Only within the context of a highly specific molecular milieu will a given molecule play its role in life. To function properly, DNA must be part of a large team, with each molecule executing its assigned task alongside the others in a cooperative manner. Acknowledging the interdependability of the component molecules within a living organism immediately presents us with a stark philosophical puzzle. If everything needs everything else, how did the community of molecules ever arise in the first place? Since most large molecules needed for life are produced only by living organisms, and are not found outside the cell, how did they come to exist originally, without the help of a meddling scientist? Could we seriously expect a Miller-Urey type of soup to make them all at once, given the hit-and-miss nature of its chemistry?
Being part of a large team,cooperative manner,interdependability,everything needs everything else, are just other words for irreducibility and interdependence.
For a nonliving system, questions about irreducible complexity are even more challenging for a totally natural non-design scenario, because natural selection — which is the main mechanism of Darwinian evolution — cannot exist until a system can reproduce. For an origin of life we can think about the minimal complexity that would be required for reproduction and other basic life-functions. Most scientists think this would require hundreds of biomolecular parts, not just the five parts in a simple mousetrap or in my imaginary LMNOP system. And current science has no plausible theories to explain how the minimal complexity required for life (and the beginning of biological natural selection) could have been produced by natural process before the beginning of biological natural selection.
Determination of the Core of a Minimal Bacterial Gene Set
https://reasonandscience.catsboard.com/t2110-what-might-be-a-protocells-minimal-requirement-of-parts#3797
Prokaryotes are thought to differ from eukaryotes in that they lack membrane-bounded organelles. However, it has been demonstrated that there are bacterias which have membrane bound organelles named acidocalcisomes, and that V-H+PPase proton pumps are present in their surrounding membranes. Acidocalcisomes have been found in organisms as diverse as bacteria and humans. Volutin granules which are equivalent of acidocalcisomes also occur in Archaea and are, therefore, present in the three superkingdoms of life (Archaea, Bacteria and Eukarya). These volutin granule organelles occur in organisms spanning an enormous range of phylogenetic complexity from Bacteria and Archaea to unicellular eukaryotes to algae to plants to insects to humans. According to neo-darwinian thinking, the universal distribution of the V-H+PPase domain suggests the domain and the enzyme were already present in the Last Universal Common Ancestor (LUCA).
If the proton pumps of Volutin granules were present in LUCA, they had to emerge prior to self replication, which induces serious constraints to propose evolution as driving factor. But if evolution was not the mechanism, what else was ? There is not much left, namely chance, random chemical reactions, or physical necessity.
But lets for a instance accept the "fact of evolution", and suppose it was the driving force to make V-H+PPase proton pumps. In some period prior to the verge of non-life to life, natural selection or an other evolutionary mechanism would have had to start polymerisation of the right amino acid sequence to produce V-H+PPase proton pumps by addition of one amino acid monomer to the other. First, the whole extraordinarly production line of staggering complexity starting with DNA would have to be in place, that is :
The cell sends activator proteins to the site of the gene that needs to be switched on, which then jump-starts the RNA polymerase machine by removing a plug which blocks the DNA's entrance to the machine. The DNA strands do shift position so that the DNA lines up with the entrance to the RNA polymerase. Once these two movements have occurred and the DNA strands are in position, the RNA polymerase machine gets to work melting them out, so that the information they contain can be processed to produce mRNA 2 The process follows then after INITIATION OF TRANSCRIPTION through RNA polymerase enzyme complexes, the mRNA is capped through Post-transcriptional modifications by several different enzymes , ELONGATION provides the main transcription process from DNA to mRNA, furthermore SPLICING and CLEAVAGE , polyadenylation where a long string of repeated adenosine nucleotides is added, AND TERMINATION through over a dozen different enzymes, EXPORT FROM THE NUCLEUS TO THE CYTOSOL ( must be actively transported through the Nuclear Pore Complex channel in a controlled process that is selective and energy dependent ) INITIATION OF PROTEIN SYNTHESIS (TRANSLATION) in the Ribosome in a enormously complex process, COMPLETION OF PROTEIN SYNTHESIS AND PROTEIN FOLDING through chaperone enzymes. From there the proteins are transported by specialized proteins to the end destination. Most of these processes require ATP, the energy fuel inside the cell.
http://reasonandscience.heavenforum.org/t2067-there-is-no-selective-advantage-until-you-get-the-final-function?highlight=function

The genetic code to make the right ~600 amino acid sequence would have to be achieved not by mutation , since that would require a pre-existing amino acid sequence, but adding randomly a new amino acid, and select the advantageous sequence. Instead as by evolution, which evolves pre-existing proteins, in a origin of life scenario proteins would have all to be produced denovo. The problem in this stage is, when there is no selective advantage until you get the final function, the final function doesn't evolve. In other words, a chain of around 600 amino acids is required to make a funcional V-H+PPase proton pump, but there is no function, until polymerisation of all 600 monomers is completed and the right sequence achieved.
The problem for those who accept the truth of evolution is, they cannot accept the idea that any biological structure with a beneficial function, however complex, is very far removed from the next closest functional system or subsystem within the potential of "sequence space" that might be beneficial if it were ever found by random mutations of any kind. In our case the situation is even more drastic, since DENOVO genetic sequence and subsequently amino acid chain for a new formation of a new amino acids strand is required. A further constraint is the fact that 100% of amino acids used and needed for life are left handed, while DNA and RNA requires D-sugars. Until today, science has not sorted out how nature is able to select the right chiral handedness. The problem is its believed there to be a warm soup consisting of racemic mixtures of amino acid enantiomers (and sugars). How did this homogenous phase separate into chirally pure components? How did an asymmetry (assumed to be small to start with) arise in the population of both enantiomers? How did the preference of one chiral form over the other, propagate so that all living systems are made of 100 percent optically pure components?
What is sequence space ? 1
Imagine 20 amino acids mixed up in a pool, randomly mixed , one adjacent to the other. The pool with all the random amino acids is the sequence space. This space can be two dimentional, tridimensional, or multidimensional. In evolutionary biology, sequence space is a way of representing all possible sequences (for a protein, gene or genome). Most sequences in sequence space have no function, leaving relatively small regions that are populated by naturally occurring genes. Each protein sequence is adjacent to all other sequences that can be reached through a single mutation. Evolution can be visualised as the process of sampling nearby sequences in sequence space and moving to any with improved fitness over the current one.
Functional sequences in sequence space
Despite the diversity of protein superfamilies, sequence space is extremely sparsely populated by functional proteins. That is, amongst all the possible amino acid sequences, only a few permit the make of functional proteins. Most random protein sequences have no fold or function. To exemplify: In order to write METHINKS IT IS LIKE A WEASEL , there are 10^40 possible random combinations possible to get the right sequence. But only one is correct.
Enzyme superfamilies, therefore, exist as tiny clusters of active proteins in a vast empty space of non-functional sequence.The density of functional proteins in sequence space, and the proximity of different functions to one another is a key determinant in understanding evolvability. Protein sequence space has been compared to the Library of Babel a theoretical library containing all possible books that are 410 pages long. In the Library of Babel, finding any book that made sense was impossible due to the sheer number and lack of order.
How would a bacterium evolve a function like a single protein enzyme? - like a V-H+PPase proton pump? The requirement is about 600 specified residues at minimum. A useful V-H+PPase cannot be made with significantly lower minimum size and specificity requirements. These minimum requirements create a kind of threshold beyond which the V-H+PPase function simply cannot be built up gradually where very small one or two residues changes at a time result in a useful change in the degree of the proton pump function. Therefore, such functions cannot have evolved in a gradual, step by step manner. There simply is no template or gradual pathway from just any starting point to the minimum threshold requirement. Only after this threshold has been reached can evolution take over and make further refinements - but not until then.
All Functions are "Irreducibly Complex" 4
The fact is that all cellular functions are irreducibly complex in that all of them require a minimum number of parts in a particular order or orientation. I go beyond what Behe proposes and make the suggestion that even single-protein enzymes are irreducibly complex. A minimum number of parts in the form of amino acid residues are required for them to have their particular functions. The proton pump function cannot be realized in even the smallest degree with a string of only 5 or 10 or even 500 residues of any arrangement. Also, not only is a minimum number of parts required for the proton pump function to be realized, but the parts themselves, once they are available in the proper number, must be assembled in the proper order and three-dimensional orientation. Brought together randomly, the residues, if left to themselves, do not know how to self-assemble themselves to form a much of anything as far as a functional system that even comes close to the level of complexity of a even a relatively simple function like a proton pump. And yet, their specified assembly and ultimate order is vital to function.
Of course, such relatively simply systems, though truly irreducibly complex, have evolved. This is because the sequence space at such relatively low levels of functional complexity is fairly dense. It is fairly easy to come across new beneficial sequences if the density of potentially beneficial sequences in sequence space is relatively high. This density does in fact get higher and higher at lower and lower levels of functional complexity - in an exponential manner. Darwinian evolution can work fine when one small step (e.g., a single point mutation) along an evolutionary pathway gives an advantage. The theory of intelligent design has no problem with this.
It is much like moving between 3-letter words in the English language system. Since the ratio of meaningful vs. meaningless 3-letter words in the English language is somewhere around 1:18, one can randomly find a new meaningful and even beneficial 3-letter word via single random letter changes/mutations in relatively short order. This is not true for those ideas/functions/meanings that require more and more letters. For example, the ratio of meaningful vs. meaningless 7-letter words and combinations of smaller words equaling 7-letters is far far lower at about 1 in 250,000. It is therefore just a bit harder to evolve between 7-letter words, one mutation at a time, than it was to evolve between 3-letter words owing to the exponential decline in the ratio of meaningful vs. meaningless sequences.
The same thing is true for the evolution of codes, information systems, and systems of function in living things as it is for non-living things (i.e., computer systems etc). The parts of these codes and systems of function, if brought together randomly, simply do not have enough meaningful information to do much of anything. So, how are they brought together in living things to form such high level functional order?
1) https://en.wikipedia.org/wiki/Sequence_space_(evolution)
2) http://creationwiki.org/Weasel_program
3) http://www.detectingdesign.com/irreduciblemousetrap.html
4) http://www.sciencedaily.com/releases/2013/07/130702100115.htm
5) http://www.ru.nl/english/research/radboud/themes/genetics-cellular/vm/news-genetics-cell/@893712/protocells-formed/
Last edited by Admin on Sun May 13, 2018 8:18 am; edited 11 times in total