https://reasonandscience.catsboard.com/t2140-nadh-dehydrogenase-complex-i-in-mitochondria
NADH dehydrogenase ( Complex I ) in mitochondria 1
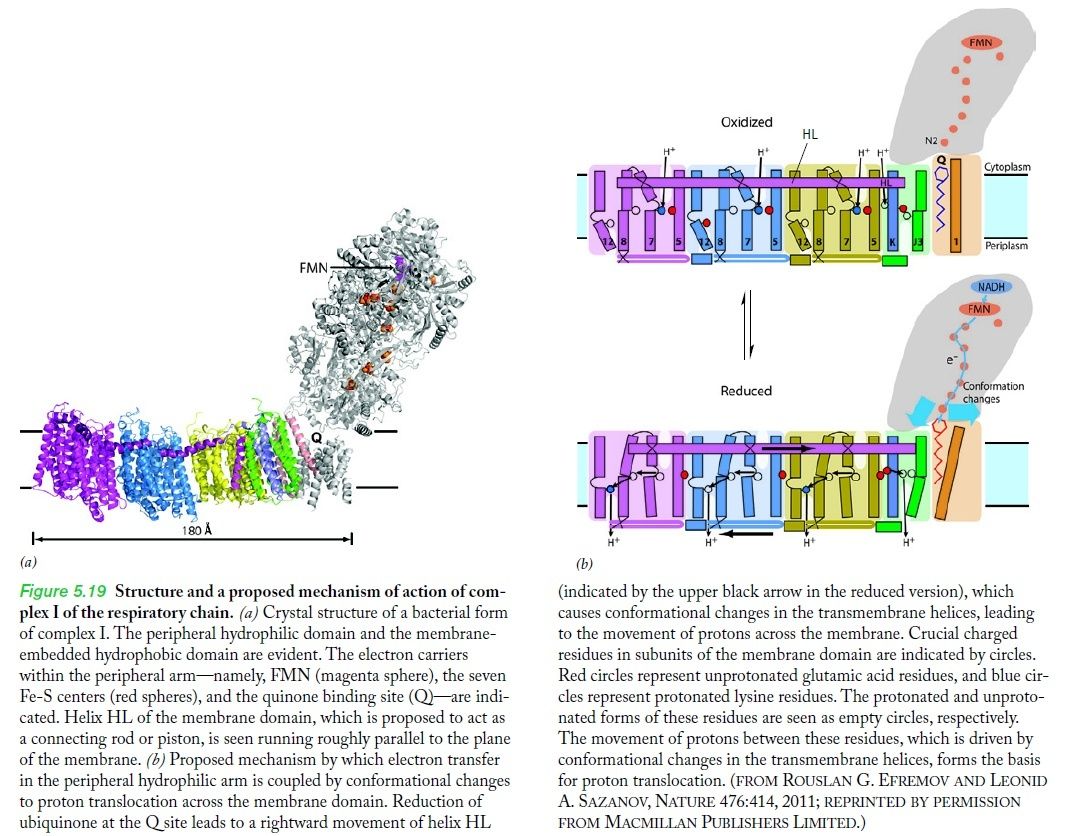
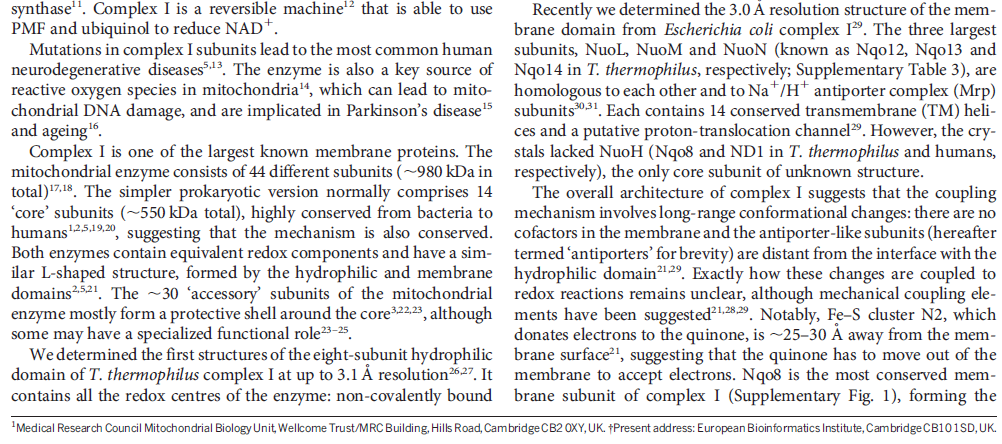
The molecular biological achievements of the last two decades culminated in 2010 with the deciphering of the crystal structure of another respiratory complex, the enormous (for a protein) complex I, by Walker's Cambridge colleague Leonid Sazanov (Efremov et al. 2010). Again, the structure betrays the mechanism — in this case not a rotary motor but, even more surprisingly, a lever mechanism not unlike the piston of a steam engine 6
ATP synthase operates at the end of a sequence of machines in the respiratory chain that generates chemical energy (in the form of ATP) from the food we eat (or from sunlight, in the case of plants). The enzyme runs on proton motive force – a flow of protons that drive its carousel-like rotor. But how does the proton gradient get established? That’s the job of Respiratory Complex I, the first machine (enzyme) in the chain. Complex I takes electrons from food, stored in NADH molecules, and transfers them down a chain of electron receptors to parts of the machine that pump protons across the mitochondrial membrane into the periplasm setting up a proton gradient. It now becomes evident that Complex I includes parts that move like pistons. Complex I was reported in a Science Express paper as having a railroad-like coupling rod.
It now appears that the we have trillions of mechanical devices similar like those coupling rods. They serve to transmit the energy in the food we eat into mechanical energy, driving a proton pump inside the mitochondrion. It’s all part of an amazing series of electromechanical machines in the powerhouses of the cell.
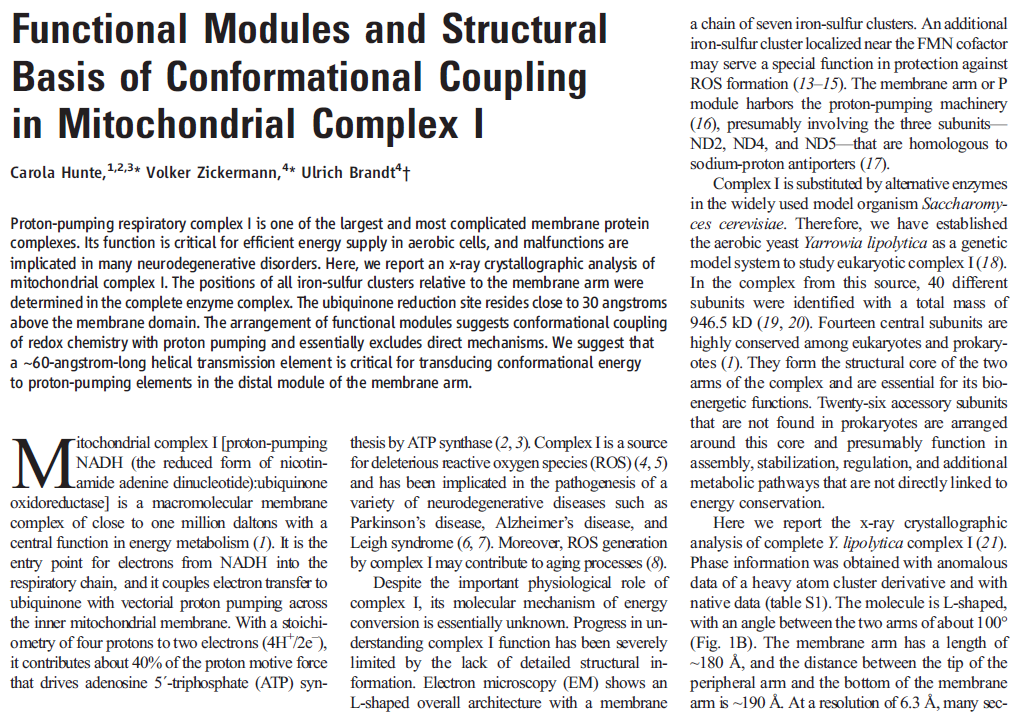
NADH dehydrogenase, also called Mitochondrial Complex I, is an essential part of the respiration process (also called oxidative phosphorylation) that passes electrons, protons and oxygen through a sophisticated energy transport chain so that energy can be stored in ATP.Complex I, composed of four major parts and shaped somewhat like a hockey stick, produces 40% of the proton motive force used by ATP synthase to produce ATP. Its job is to derive protons from NADH and hand them off to additional cofactors and enzymes in the transport chain that will pump the protons outside the mitochondrial membrane. The electrical potential thus created across the membrane drives the ATP synthase rotary engine at the end of the chain.
In discussing the paper published in Science Express 3, Science Daily contained some amazing facts about the machinery of respiration and how it delicately handles explosive ingredients:
In a laboratory experiment, hydrogen and oxygen gas would react in an explosion and the energy contained would be released as heat. In biological oxidation, the energy will be released by the membrane bound protein complexes of the respiratory chain in a controlled manner in small packages. Comparable to a fuel cell, this process generates an electrical membrane potential, which is the driving force of ATP synthesis. The total surface of all mitochondrial membranes in a human body covers about 14 square meters. This accounts for a daily production of about 65 kg of ATP.
That 65 kg, by the way, is near a typical human body weight. That’s how much ATP the human body synthesizes each day – even during sleep. At any one time, though, our body only contains the ATP equivalent of a AA battery . The electrical potential generated across that 14 square meters of mitochondrial membrane drives the ATP synthesis that keeps us – and every living thing alive.
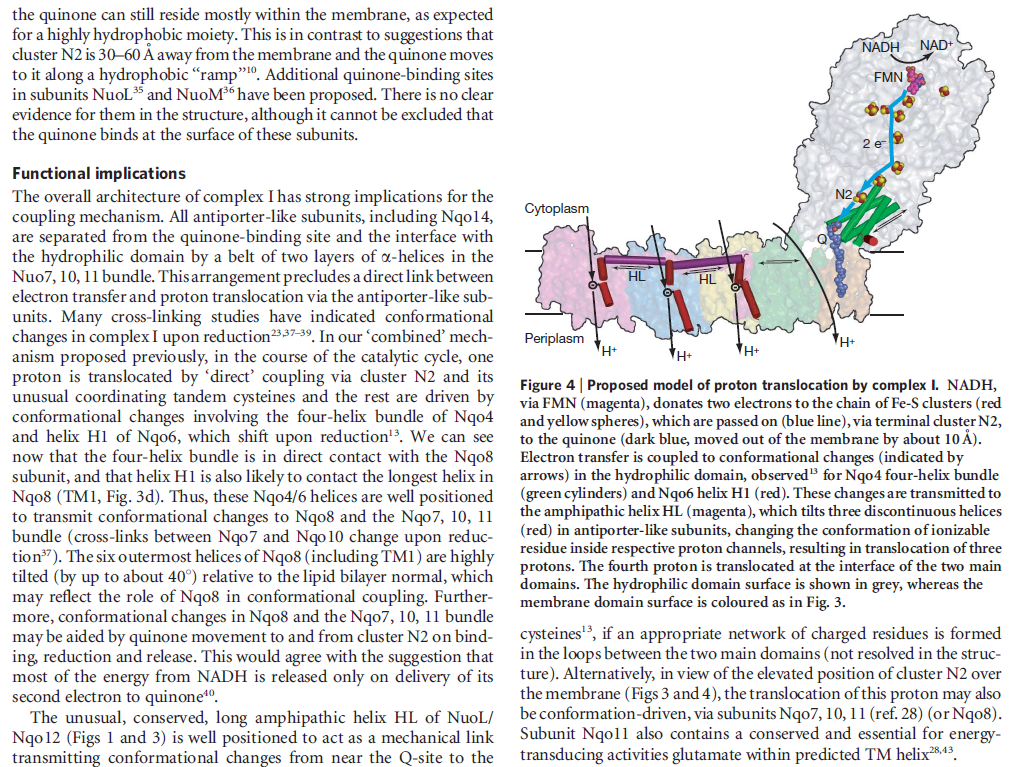
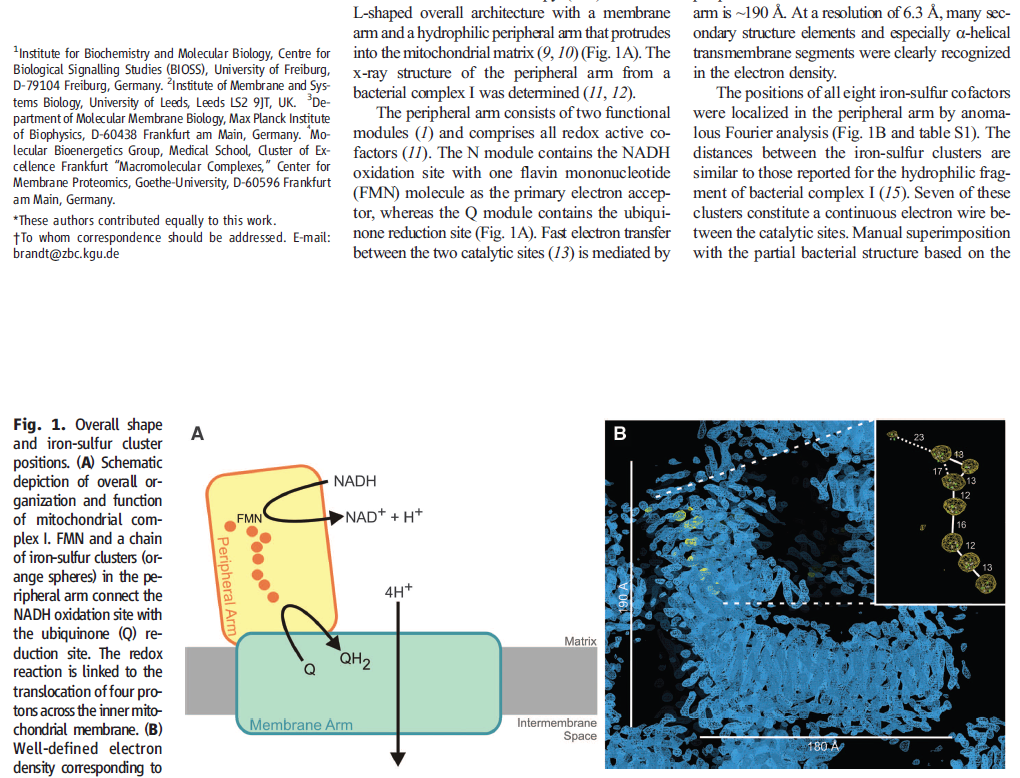
What’s new and exciting is that a coupling rod is part of the energy transport chain. Higher up in the enzyme, a series of iron-sulfur clusters works something like an electrical wire, transferring electrons from the first domain into the second domain. The transport is constructed to prevent the formation of dangerous reactive oxygen species (ROS). Between the second and third domains, it appears that the electrical energy is converted into mechanical energy via a coupling rod composed of a 6-nanometer alpha coil that is “critical for transducing conformational energy to proton-pumping elements in the distal module of the membrane arm.” In other words, it has moving parts.
Quips Pumping protons: a complex problem 7
Cells use a lot of ATP. An E.coli cell can use a billion ATP molecules per minute and humans use their own weight in ATP every day. Because of this, cells need an efficient mechanism to recombine ADP with phosphate to regenerate ATP, and much of the energy in our food is used in this process. ATP synthase, the enzyme that makes ATP, is located in the mitochondrial inner membrane (or the bacterial cell membrane), and is powered by a concentration difference (gradient) of protons across the membrane.
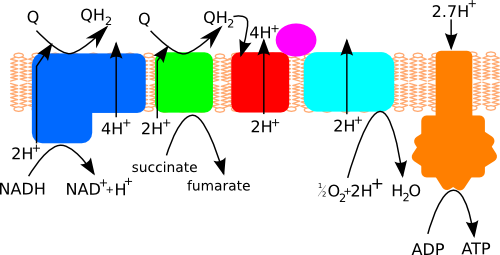
Figure 1. Schematic overview of the respiratory chain showing how complex I (blue ), complex II (green), complex III (red) and complex IV (light blue) produce the proton gradient used by ATP synthase (orange) to generate ATP. Cytochrome c is shown in magenta and shuttles electrons between complexes III and IV. Q stands for quinone.
Four different protein complexes make up the respiratory chain that pumps protons across the cell membrane: ubiquinone oxidoreductase (complex I), succinate dehydrogenase (complex II), cytochrome bc1 complex (complex III) and cytochrome c oxidase (complex IV). These generate the proton gradient that drives ATP synthase (Figure 1). Complex I oxidises NADH (formed in glycolysis and the Krebs cycle) to pump four protons across the membrane. Complex I also uses the reductive power of NADH to reduce quinone, which is then used as a substrate by complex III to pump two more protons across the membrane.
One ‘ell of a structure.
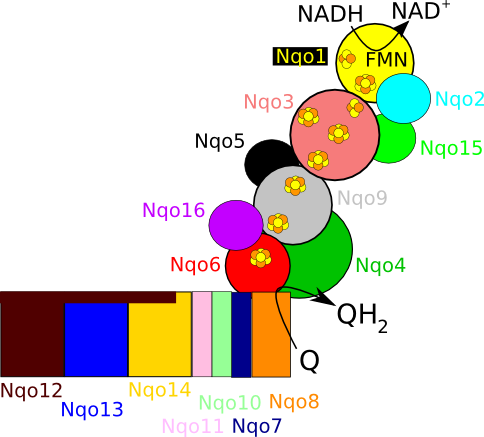
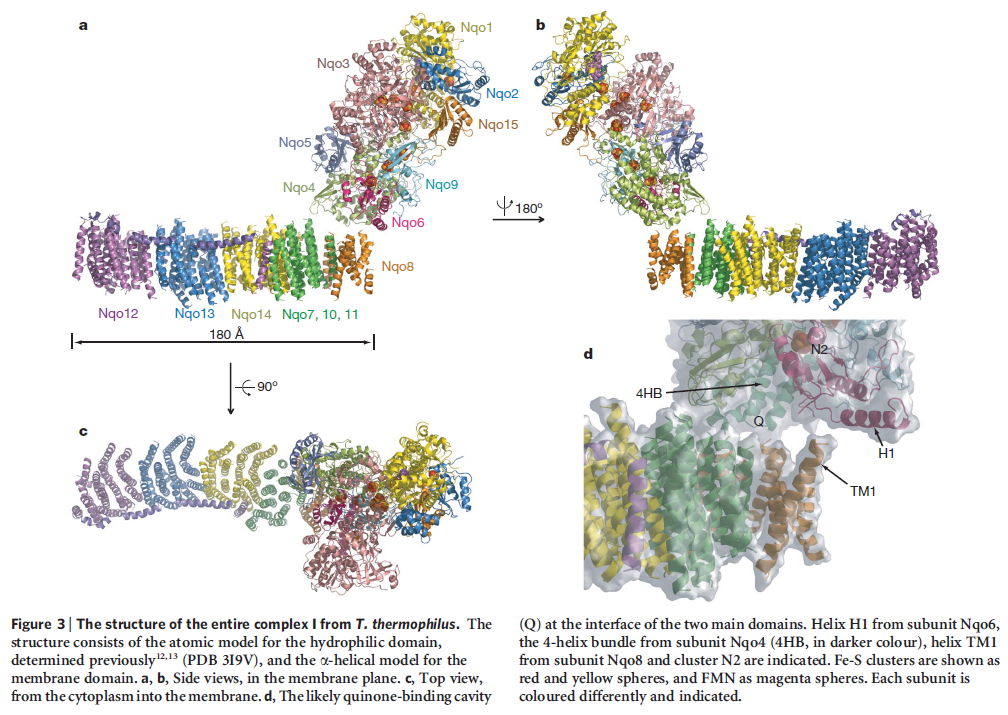
Figure 2. Schematic overview of T. thermophilus complex I, highlighting the 16 subunits, the iron sulfur cluster chain and the NADH and quinone (Q) binding sites.
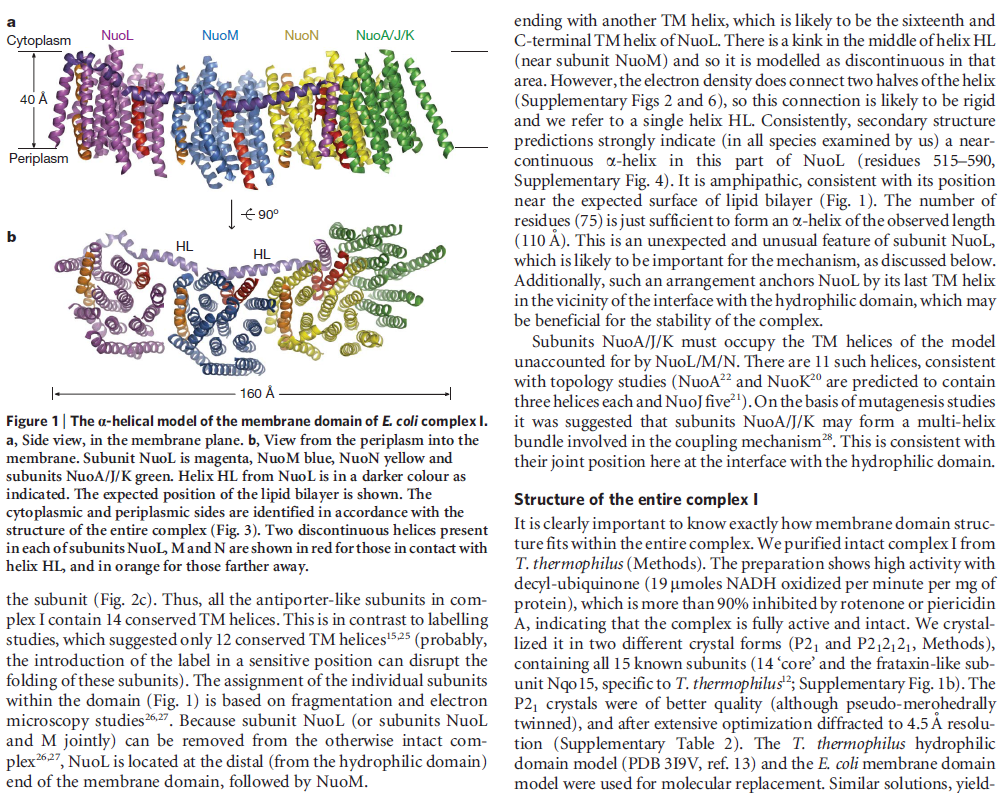
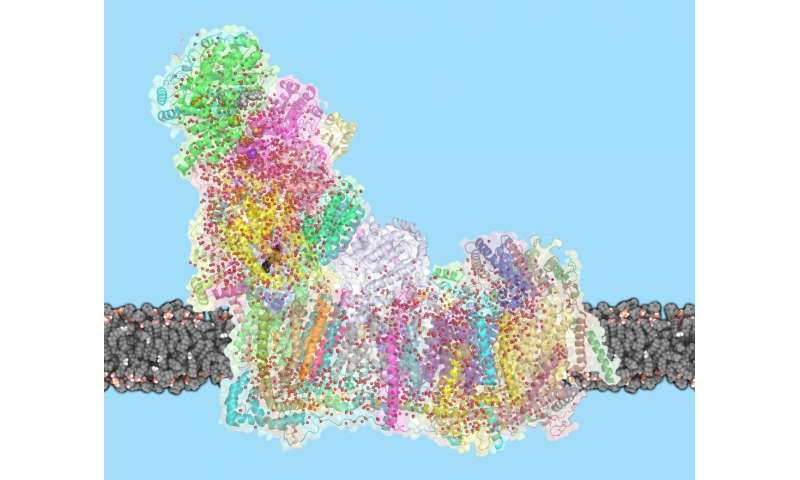
In mammals, complex I consists of 44 different subunits and electron microscopy studies have revealed that it has an L shape. One arm of the L is inserted in the membrane (the membrane arm) and the other arm (the hydrophilic arm) projects some 100 Å into the mitochondrial matrix. Bacteria also possess a complex I, but it is a ‘cut down’ version of its eukaryotic counterpart, containing at least 14 of the subunits found in the mitochondrial enzyme.
The crystal structure of complex I from the bacterium Thermus thermophilus (PDB entry 4hea) shows that it also has the characteristic L shape(view-1) with the hydrophilic arm projecting into the cytoplasm.
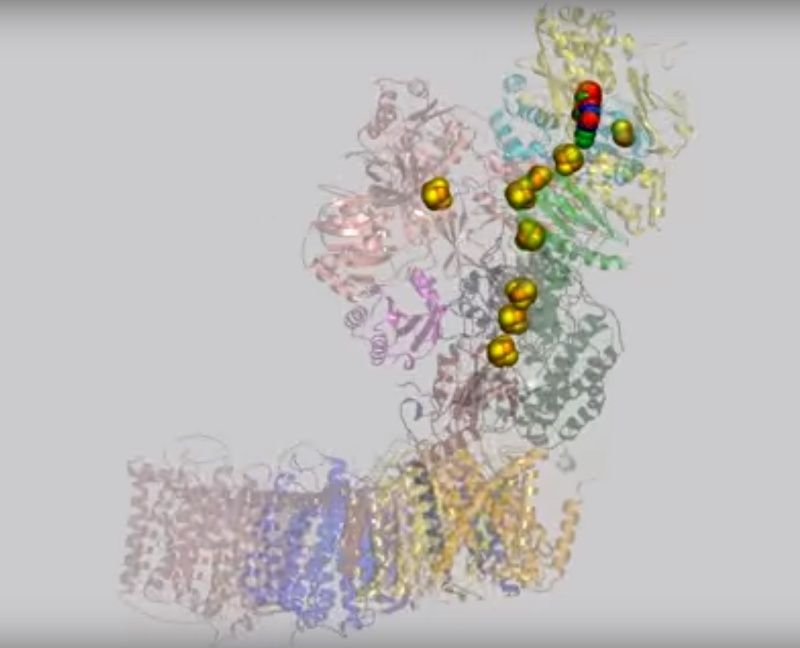
(view-1)View 1: Overall architecture of complex I.The 16 proteins making up complex I are shown in cartoon representation. Initially, the hydrophilic (red) and membrane (cyan) arms are highlighted; subsequently, each chain is coloured separately using the same colour scheme as in Figure 2. The iron-sulfur clusters (yellow and orange) and FMN (withgreen
The complex consists of 16 protein subunits with a combined molecular weight of 536 . The complex contains seven Fe4S4 and two Fe2S2 iron-sulfur clusters and a bound flavin mononucleotide (FMN) as co-factors (Figure 2). Seven of the subunits span the membrane and contain 64 transmembrane helices in total.
Where does NADH enter complex I?
The NADH substrate-binding site is at the distal end of the hydrophilic arm in subunit Nqo1 (view-2). The site is formed from a modified Rossmann fold which also incorporates a FMN-binding site. The structure of the hydrophilic arm of T. thermophilus complex I with bound NADH (PDB entry 3iam) shed light on the interactions between FMN and NADH. The adenine ring of NADH is held in place by stacking against three phenylalanine residues. This allows the nicotinamide ring of NADH to stack against the isolloxazine ring of the bound FMN (view-2). Electron transfer then occurs between the C4N atom of the nicotinamide ring of NADH and N5 of FMN. The architecture of the binding site positions these atoms within 3.2 Å of each other for efficient electron transfer. A glutamate residue in the binding site appears to help position the two rings close together.

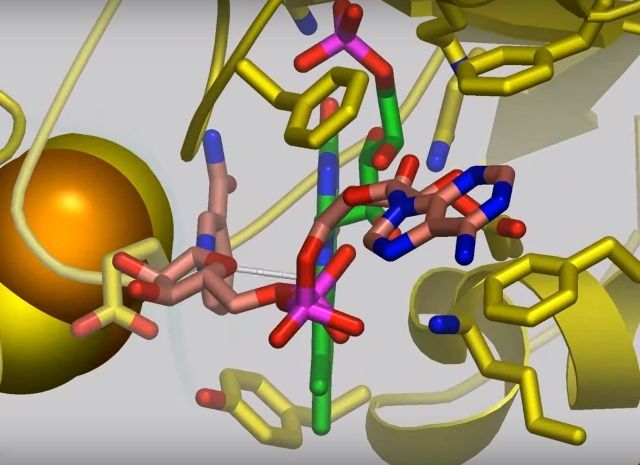
View 2: NADH binding and electron transfer. NADH (salmon carbons) in Nqo1 is shown in stick representation at the top of complex I, adjacent to the bound FMN molecule (green carbons). A dashed line between FMN atom N5 and C4N of NADH indicates the electron transfer path. Nqo1 residues that interact with the FMN or NADH are shown as sticks. Iron-sulfur clusters are shown as large yellow and orangespheres.
Reducing the problem.
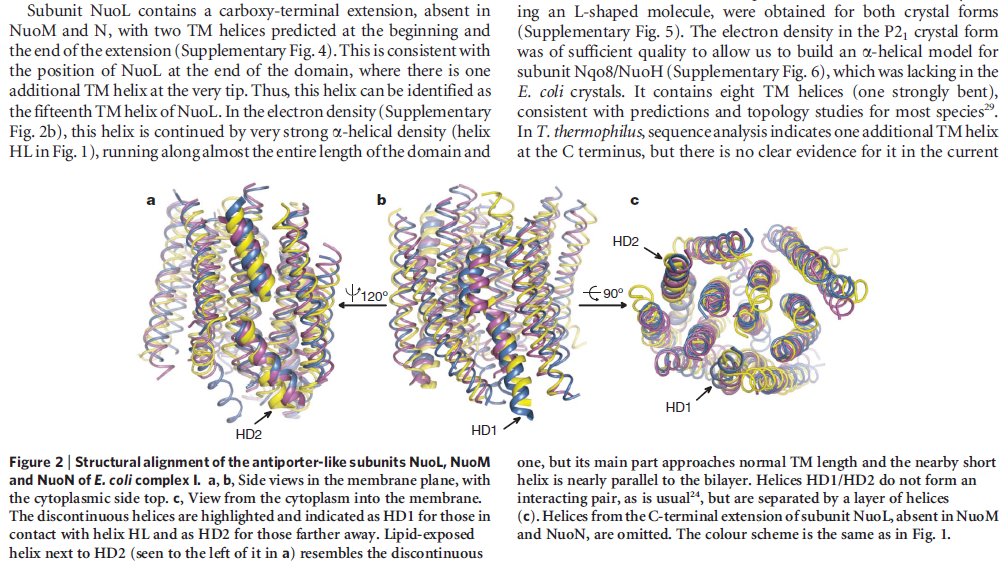
Subunits Nqo1, Nqo3, Nqo6 and Nqo9 contain seven of the iron-sulfur clusters in the complex. These form a “wire” some 90 Å long which carries electrons from the FMN, through the hydrophilic arm to the quinone-binding site formed by Nqo4, Nqo6 and Nqo8 (view-3). The iron-sulfur clusters are at most 14 Å apart (edge to edge distance), allowing efficient electron transfer between the clusters(view-3). Nqo2 and Nqo3 contain additional iron-sulfur clusters, but the function of these clusters is unknown.
Figure 3. Menaquinone 8 used by T. thermophilus complex I.
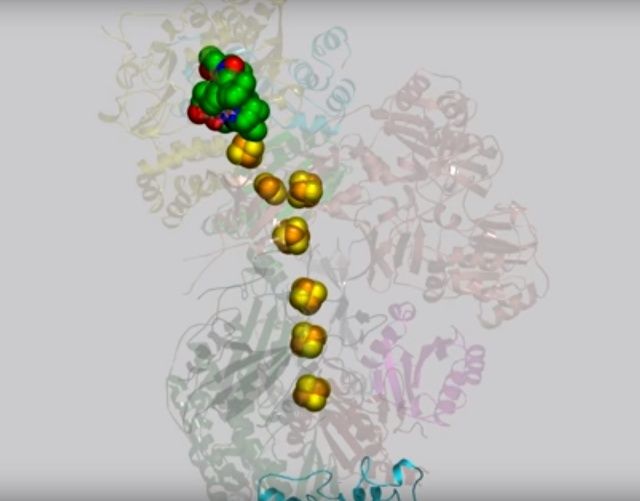
View 3: Electrons travel down the "wire" of iron-sulfur clusters. The chain of iron-sulfur clusters down which electrons travel from NADH to the quinone-binding site is shown using spheres. Their path is highlighted in the animation by enlarging each cluster in turn.
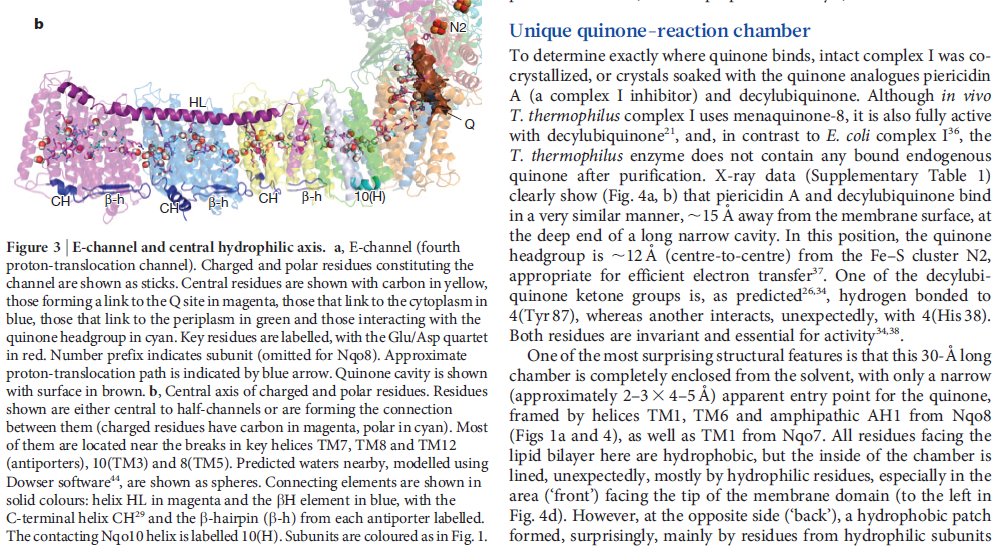
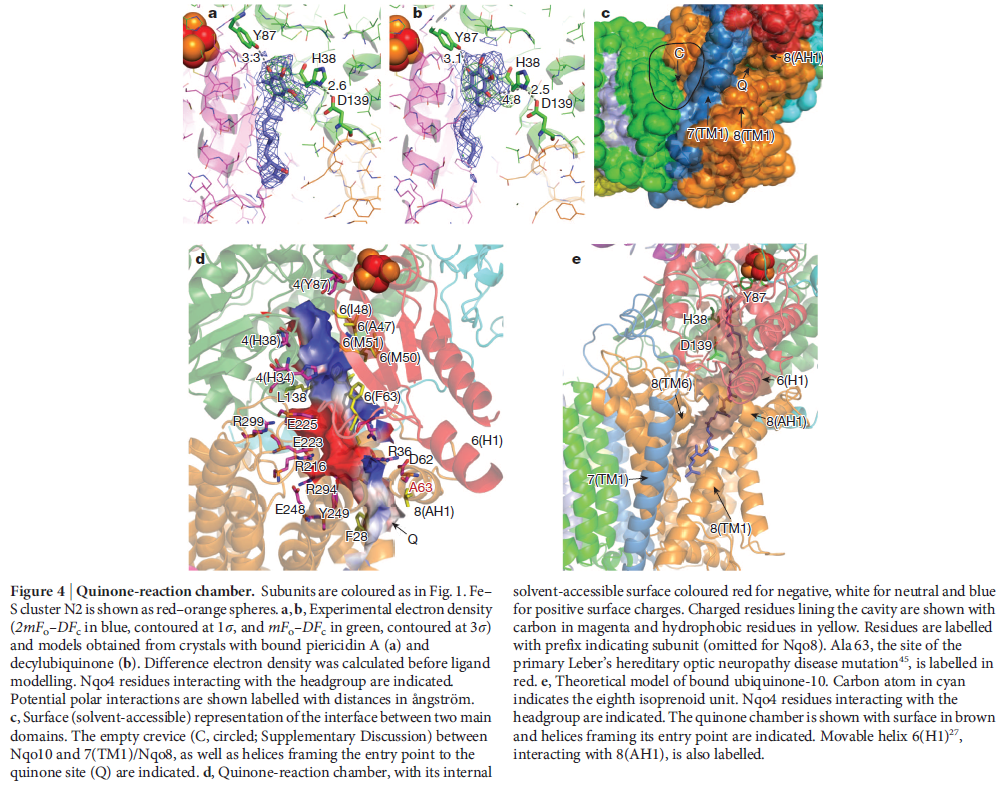
NADH donates two electrons to the iron-sulfur cluster “wire” which are transferred one at a time along the clusters and used to reduce the bound quinone. Different organisms use different quinones, and T. thermophilus usesmenaquinone-8 (Figure 3). Menaquinone-8 binds in a 30 Å long cavity formed by subunits Nqo4, Nqo6 and Nqo8 at the interface between the two arms. The entrance to the cavity, through which the quinone has to pass, is surprisingly narrow, around 2.3 x 4.5 Å (view-4), suggesting movement in this region to allow quinone binding. The quinone-binding cavity contains hydrophilic residues along its length, which may interact with the hydrophilic head group of the quinone as it enters the cavity. Residues His38, Tyr87 and Asp139 in Nqo4, at the distal end of the cavity, probably position the quinone close to the last iron-sulfur cluster in the “wire” to allow for efficient reduction of the quinone. After being reduced, the quinone leaves complex I and is subsequently used as a substrate by complex III.
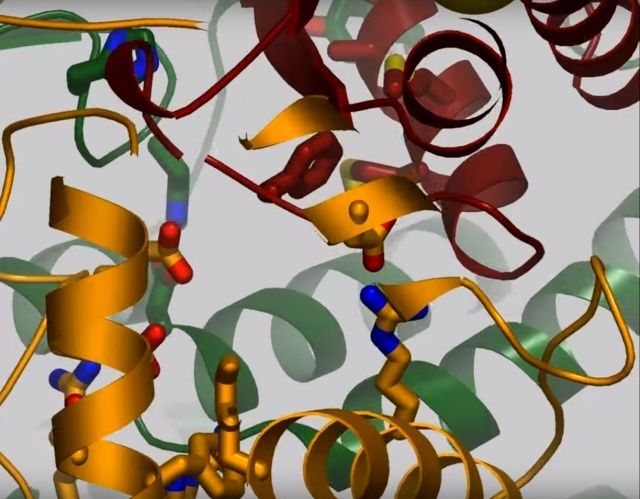
View 4: The quinone binding site. The residues that line the quinone binding site in Nqo4 (darkgreen), Nqo6 (darkred) and Nqo8 (orange) are shown in stick representation.
We all pump together.
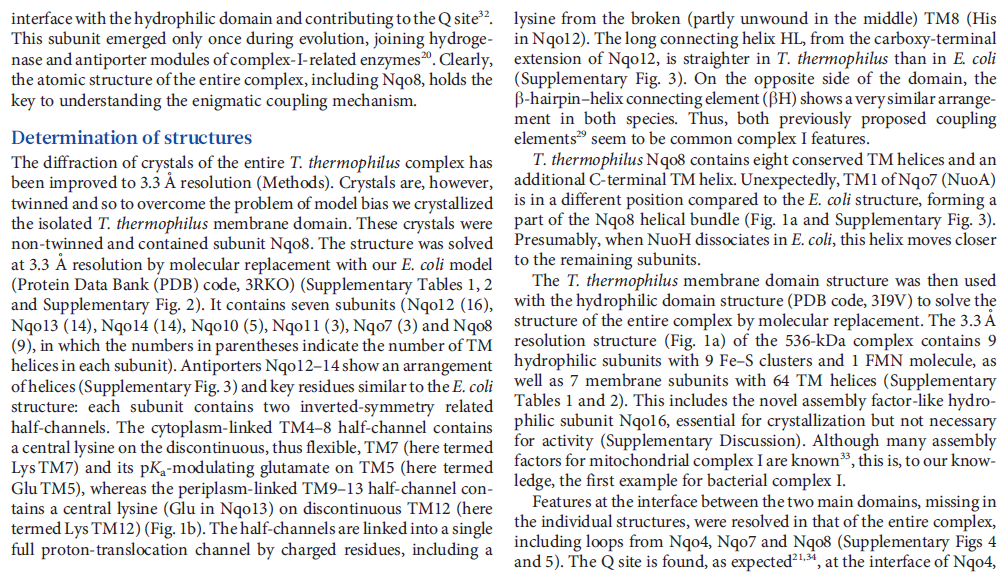
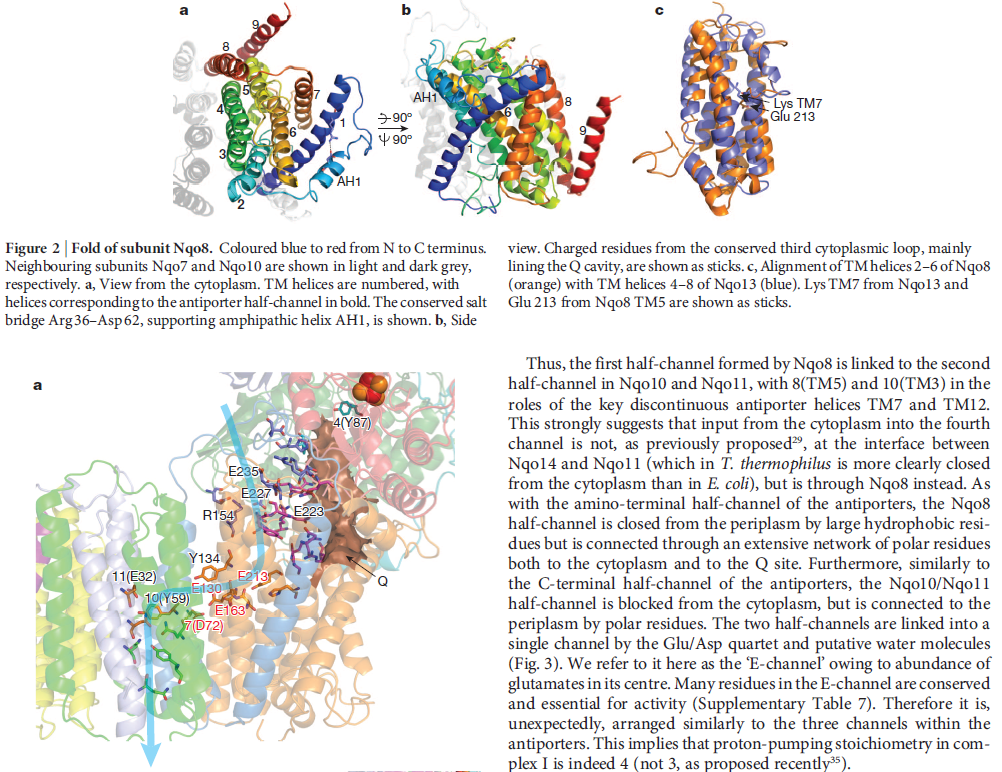
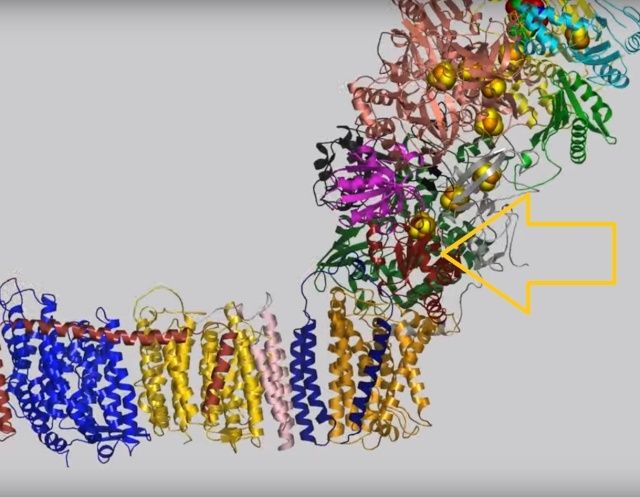
When complex I reduces the bound quinone it also pumps four protons across the membrane. Complex I contains four potential channels within the membrane which are likely to perform this role. Subunits Nqo12, 13 and 14 are homologous to each other and to sodium/proton antiporters, indicating that they most likely form the proton-pumping channels 1, 2 and 3, respectively. Subunits Nqo8, Nqo10 and Nqo11 also form a channel across the membrane, channel 4, which links the other channels to the hydrophilic domain (view-5).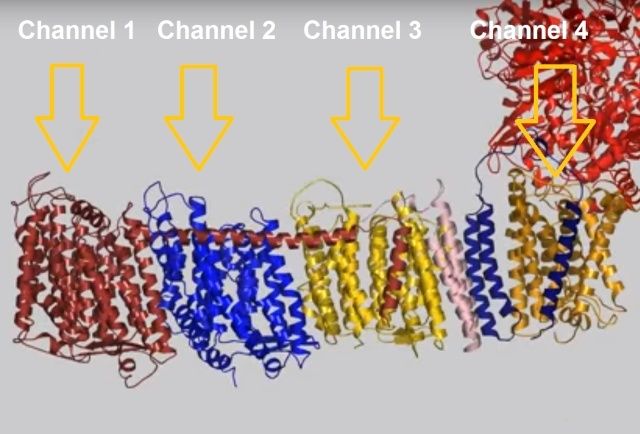
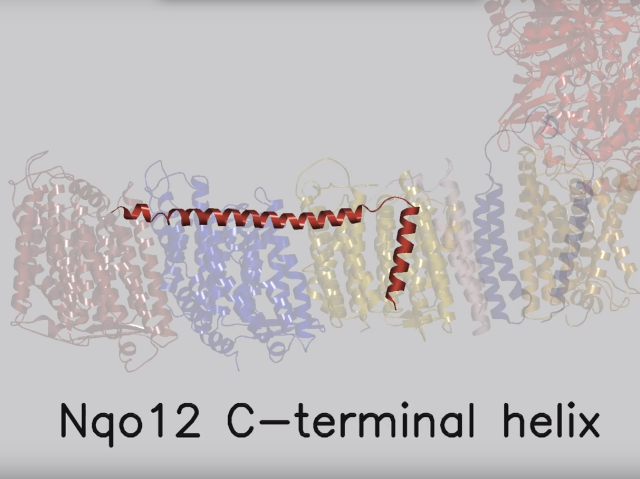
View 5: Proton-pumping channels. The subunits containing the four proton pumping channels are highlighted, followed by the C-terminal helix in Nqo12. Subsequently, charged residues are shown as sticks, highlighting a line of charged residues in the centre of the membrane arm.
So how does reduction of the quinone result in the pumping of four protons across the membrane? The most obvious assumption is that each channel pumps a single proton. But this begs the question how the channels are coordinated, especially as the distal channel is 130 Å from the quinone binding site. Each channel contains two transmembrane helices which have a break in the middle with a charged residue at this point (view-5). It is likely that these charged residues form a path for the proton as it makes its way through the channel. Other charged residues appear to connect the proton pumping channels together, possibly providing a mechanism of coordination between the channels. The C-terminus of Nqo12 (containing channel 1) consists of a 104 Å long α-helix which runs along the surface of the membrane before terminating adjacent to Nqo14. The role of this helix is unknown, although it interacts with one of the two broken transmembrane helices in each of the channels. Additional charged residues link the quinone binding site to the adjacent channel (channel 4), suggesting that they may initiate proton pumping upon reduction of the quinone (view-5). Further work is needed to investigate the mechanism of proton pumping.
A long-distance relationship that works.
NADH is used as a reducing agent by complex I, donating two electrons that reduce a quinone molecule and drive the pumping of four protons across the membrane in four channels. Amazingly, the NADH substrate is 85 Å away from the quinone and the quinone is 140 Å removed from the most distant channel! Complex I displays a truly remarkable form of long-distance communication.
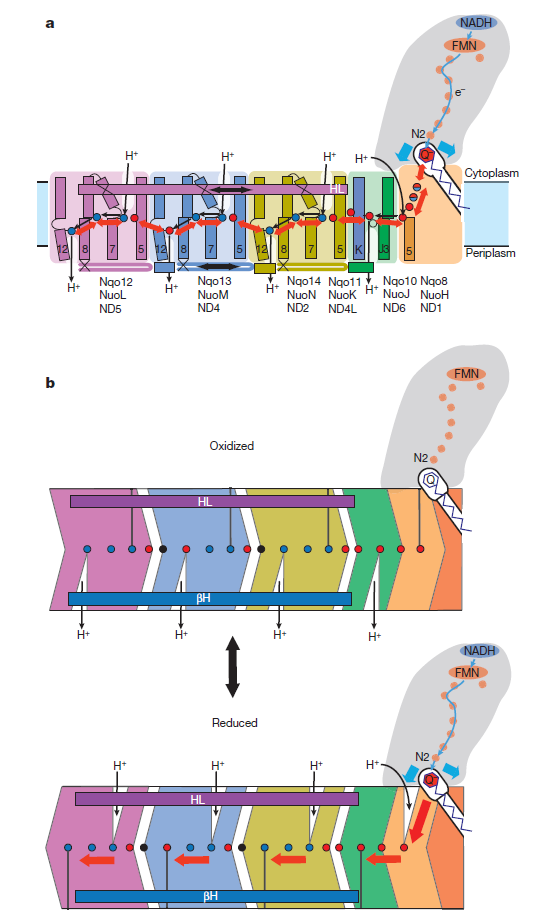
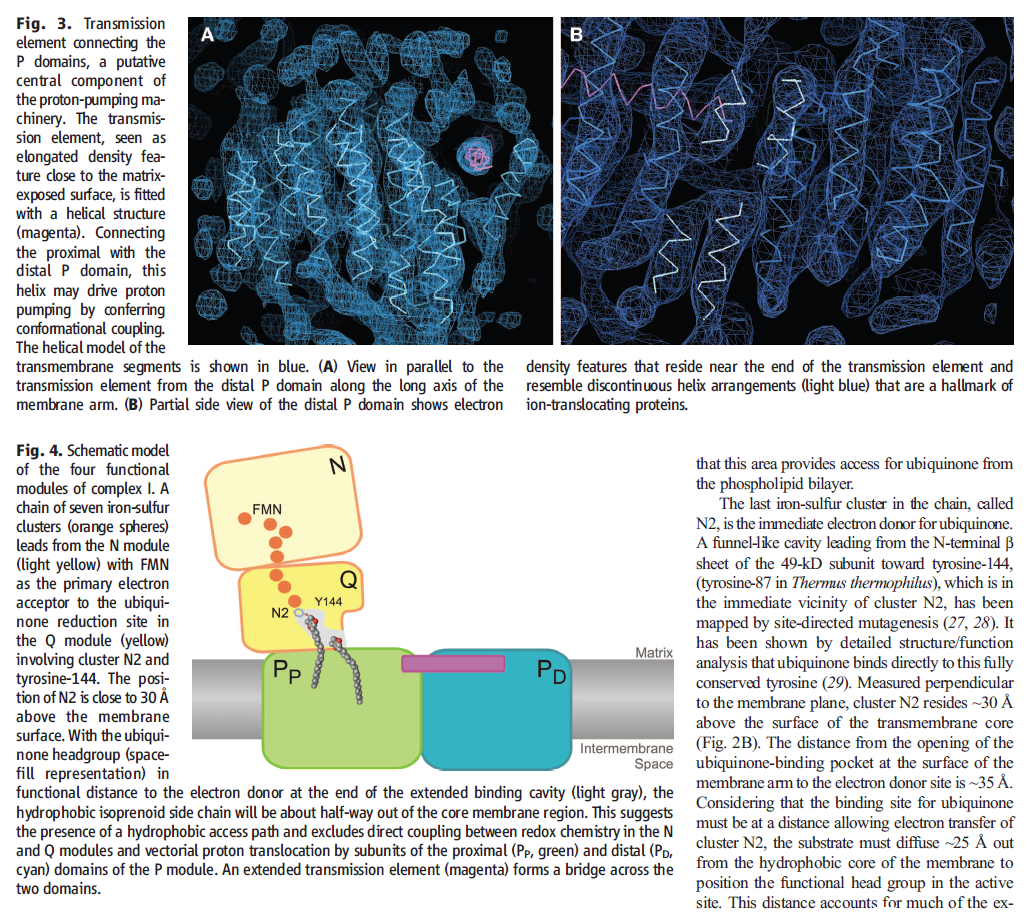
The NADH Dehydrogenase Complex Contains Separate Modules for Electron Transport and Proton Pumping
The NADH dehydrogenase complex is a massive assembly of membrane and nonmembrane proteins that receives electrons from NADH and passes them to ubiquinone. In animal mitochondria, it consists of more than 40 different protein subunits, with a molecular mass of nearly a million daltons. The x-ray structures of the NADH dehydrogenase complex from fungi and bacteria show that it is L-shaped, with both a hydrophobic membrane arm and a hydrophilic arm that projects into the mitochondrial matrix

The structure of NADH dehydrogenase. (A) The model of the mitochondrial complex shown here is based on
the x-ray structure of the smaller bacterial complex, which works in the same way. The matrix arm of NADH dehydrogenase
(also known as Complex I) contains eight iron–sulfur (FeS) clusters that appear to participate in electron transport. The
membrane contains more than 70 transmembrane helices, forming three distinct proton-pumping modules, while the matrix arm
contains the electron-transport cofactors. (B) NADH donates two electrons, via a bound flavin mononucleotide (FMN; yellow),
to a chain of seven iron–sulfur clusters (red and yellow spheres). From the terminal iron–sulfur cluster, the electrons pass to
ubiquinone (orange). Electron transfer results in conformational changes (black arrows) that are thought to be transmitted to a
long amphipathic α helix (purple) on the matrix side of the membrane arm, which pulls on discontinuous transmembrane helices
(red) in three membrane subunits, each of which resembles an antiporter (see Chapter 11). This movement is thought to change
the conformation of charged residues in the three proton channels, resulting in the translocation of three protons out of the
matrix. A fourth proton may be translocated at the interface of the two arms (dotted line). (C) This shows the symbol for NADH
dehydrogenase used throughout this chapter.
Electron transfer and proton pumping are physically separated in the NADH dehydrogenase complex, with electron transfer occurring in the matrix arm and proton pumping in the membrane arm. The NADH docks near the tip of the matrix arm, where it transfers its electrons via a bound flavin mononucleotide to a string of iron–sulfur clusters that runs down the arm, acting like a wire to carry electrons to a protein-bound molecule of ubiquinone. Electron transfer to the quinone is thought to trigger proton translocation in a set of proton pumps in the membrane arm, and for this to happen the two processes must be energetically and mechanically linked. A mechanical link is thought to be provided by a 6-nm long, amphipathic α helix that runs parallel to the membrane surface on the matrix side of the membrane arm. This helix may act like the connecting rod in a steam engine to generate a mechanical, energy-transducing power stroke that links the quinone-binding site to the proton-translocating modules in the membrane. The reduction of each quinone by the transfer of two electrons can cause four protons to be pumped out of the matrix into the crista space. In this way, NADH dehydrogenase generates roughly half of the total proton-motive force in mitochondria.
Nanomachines in the powerhouse of the cell 4
Scientists of the University of Freiburg and the University of Frankfurt have elucidated the architecture of the largest protein complex of the cellular respiratory chain.They discovered an unknown mechanism of energy conversion in this molecular complex. The mechanism is required to utilize the energy contained in food. After ten years of research work, the x-ray crystallographic analysis of the huge and most complicated protein complex of the mitochondrial respiratory chain was successful. The complex contains more than 40 different proteins, marks the entry to cellular respiration and is thus also called mitochondrial complex I. The results are published in the current online-edition of the journal “Science”.
A detailed understanding of the function of complex I is of special medical interest. Dysfunction of the complex is implicated in several neurodegenerative diseases such as Parkinson´s disease or Alzheimer´s disease, and also with the physiological processes of biological aging, in general.
The energy metabolism takes place in the so-called powerhouses of the cell, the mitochondria. They transduce the energy taken up as food into adenosine triphosphate, in short ATP, which is the universal energy currency of life. A chain of five complicated molecular machines in the mitochondrial membrane are responsible for the energy conversion. The production of ATP in mitochondria requires so many steps, as it is in principal a Knallgasreaction. In a laboratory experiment, hydrogen and oxygen gas would react in an explosion and the energy contained would be released as heat. In biological oxidation, the energy will be released by the membrane bound protein complexes of the respiratory chain in a controlled manner in small packages. Comparable to a fuel cell, this process generates an electrical membrane potential, which is the driving force of ATP synthesis. The total surface of all mitochondrial membranes in a human body covers about 14.000 square meter. This accounts for a daily production of about 65 kg of ATP.
The now presented structural model provides important and unexpected insights for the function of complex I. A special type of „transmission element“, which is not known from any other protein, appears to be responsible for the energy transduction within the complex by mechanical nanoscale coupling. Transferred to the technical world, this could be described as a power transmission by a coupling rod, which connects for instance the wheels of a steam train. This new nano-mechanical principle will now be analysed by additional functional studies and a refined structural analysis.
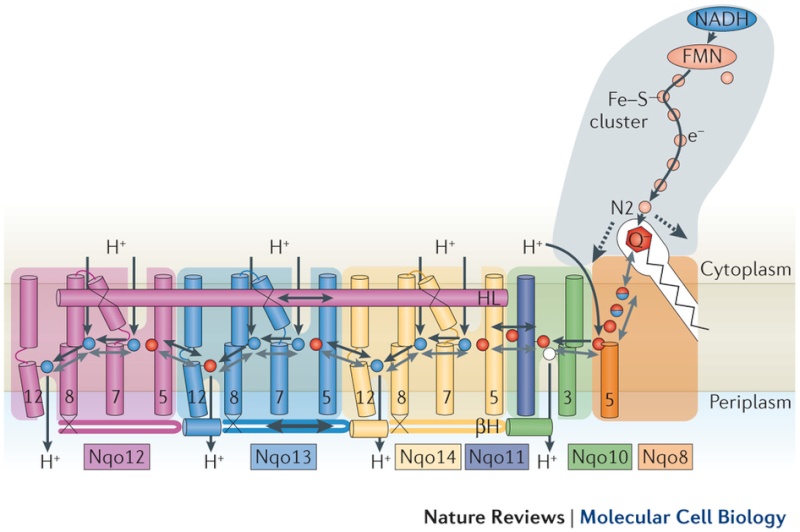
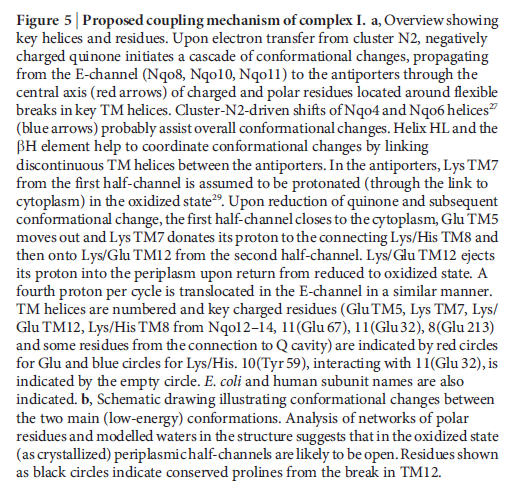
Key helices and residues of complex I are depicted schematically. Upon electron transfer from the Fe–S cluster N2, negatively charged quinone (or charged residues nearby) initiates a cascade of conformational changes, propagating from the E-channel (at Nqo8, Nqo10 and Nqo11) to the antiporters via the central axis (indicated by grey arrows) comprising charged and polar residues that are located around flexible breaks in key transmembrane helices (TMHs). Cluster N2-driven shifts (dashed arrows) of Nqo4 and Nqo6 helices33 (not shown) are likely to assist overall conformational changes. Helix HL and the βH element help to coordinate conformational changes by linking discontinuous TMHs between the antiporters. Key charged residues can be protonated from the cytoplasm through several possible pathways, including inter-subunit transfer (indicated by black arrows) (Fig. 3). Following the reduction of quinone and completion of conformational changes, Lys or GluTM12 in the antiporters and Glu32 from Nqo11 in the E-channel each eject a proton into the periplasm. TMHs are numbered and key charged residues (that is, GluTM5, LysTM7, Lys or HisTM8 and Lys or GluTM12 from Nqo12–Nqo14, as well as Glu67 and Glu32 from Nqo11, which interacts with Tyr59 from Nqo10, Glu213 from Nqo8 and some residues from the connection to the quinone cavity) are indicated by red circles for Glu, blue circles for Lys or His, and white circle for Tyr. FMN, flavin mononucleotide. Figure from Ref. 9, Nature Publishing Group.
The authors of the paper did not mention evolution. The only oblique reference is that the working parts are “highly conserved” (unevolved) throughout the entire realm of life:
Fourteen central subunits are highly conserved among eukaryotes and prokaryotes. They form the structural core of the two arms of the complex and are essential for its bioenergetic functions. 26 accessory subunits that are not found in prokaryotes endosymbiosis hello ??!! are arranged around this core and presumably function in assembly, stabilization, regulation and additional metabolic pathways not directly linked to energy conservation.
Four other times the paper mentioned that key elements of the enzyme are conserved or highly conserved. There was no attempt to explain how this “nano-mechanical principle” emerged or evolved. They did mention, though, that mutations and dysfunctions in Complex I that allow the formation of ROS are implicated in debilitating conditions like Parkinson’s and Alzheimer’s disease – and perhaps in the aging process itself.
Isn’t this wonderful information? Now we see that the respiratory transport chain in mitochondria includes coupling rods that act like little locomotives. Those rods must be moving incredibly fast. They are pumping protons like gangbusters, 24x7, all the years of your life. This mechanical wonder is only one amazing device in the first stage of a respiratory chain that includes some 40 enzymes. The machinery dazzles and boggles the mind as it continues on its way to the climax of ATP synthase, one of the most elegant and perfect molecular machines.
Over and over again, we find researchers ignoring Darwinism as they uncover the workings of molecular machines in the cell. Darwin himself could never have imagined that life at its foundations would be this complex, this mechanical. It has all the appearance of Paley’s pocket watch – only more elegant, more efficient, and more beautiful at an unimaginably small scale. And this is just one of thousands of such machines. Remember the other locomotives, the machines that transport cargo down your molecular railroad?
Notice how scientists in a recent paper in PNAS employed “engineering models to understand the control principles” of a biological phenomenon.) Fire the storytellers! Train engineers! (Catch the pun?) When science discovers powered locomotives at work in the simplest organisms, it no longer needs storytellers with loco motives.
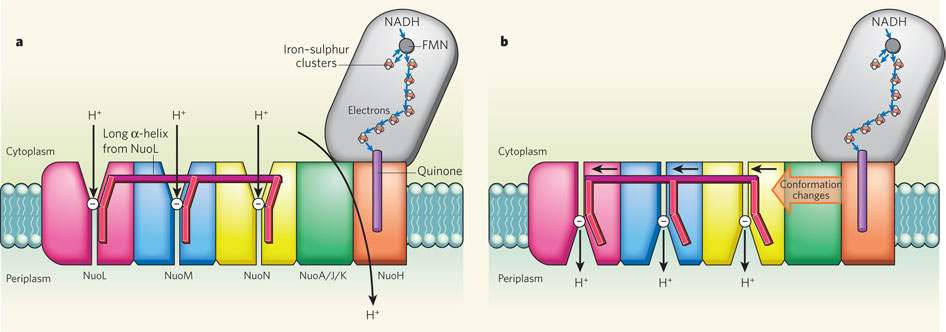
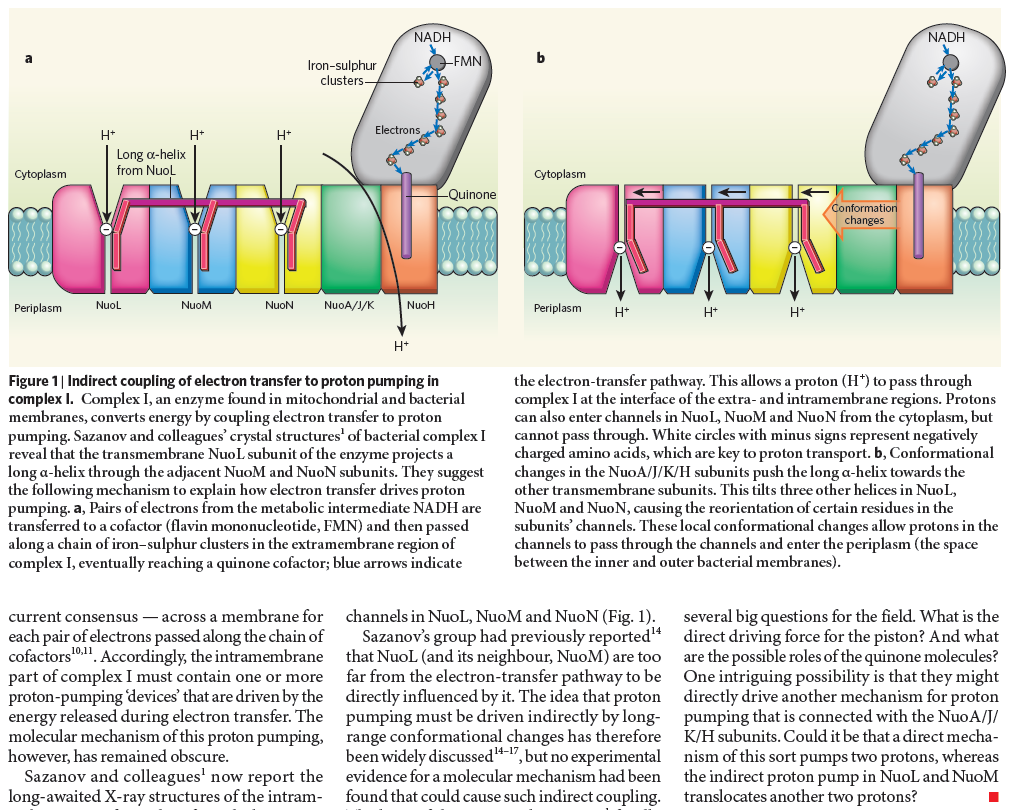
Complex I, an enzyme found in mitochondrial and bacterial membranes, converts energy by coupling electron transfer to proton pumping. Sazanov and colleagues' crystal structures1 of bacterial complex I reveal that the transmembrane NuoL subunit of the enzyme projects a long α-helix through the adjacent NuoM and NuoN subunits. They suggest the following mechanism to explain how electron transfer drives proton pumping. a, Pairs of electrons from the metabolic intermediate NADH are transferred to a cofactor (flavin mononucleotide, FMN) and then passed along a chain of iron–sulphur clusters in the extramembrane region of complex I, eventually reaching a quinone cofactor; blue arrows indicate the electron-transfer pathway. This allows a proton (H+) to pass through complex I at the interface of the extra- and intramembrane regions. Protons can also enter channels in NuoL, NuoM and NuoN from the cytoplasm, but cannot pass through. White circles with minus signs represent negatively charged amino acids, which are key to proton transport. b, Conformational changes in the NuoA/J/K/H subunits push the long α-helix towards the other transmembrane subunits. This tilts three other helices in NuoL, NuoM and NuoN, causing the reorientation of certain residues in the subunits' channels. These local conformational changes allow protons in the channels to pass through the channels and enter the periplasm (the space between the inner and outer bacterial membranes).
Structure of the membrane domain of respiratory complex I 5
The structure of the membrane domain of respiratory complex I from E. coli is shown in colour and the aligned hydrophilic domain of complex I from Thermus thermophilus in grey. In the three antiporter-like subunits the two symmetry-related inverted domains, involved in proton translocation, are shown in shades of green. The two connecting elements are shown in yellow (helix HL) and blue (b-hairpin-helix element). Complex I is a large molecular proton-pumping machine. Its structure is strongly suggestive of a mechanism that involves conformational coupling via connecting elements acting like a coupling rod in a steam engine, driving symmetry-related domains instead of wheels. To highlight a remarkable analogy between the independent creations of man and nature, the background shows a drawing by James Watt of his steam engine developed between 1787-1800 (The image of steam engine is from the book: "James Watt, Volume 3: Triumph through Adversity, 1785-1819" by Rev. Dr. Richard L. Hills (Landmark Publishing Ltd., 2006)). Credits: Rouslan Efremov and Leonid Sazanov, MRC.
Visualize an old locomotive train roaring down the tracks. One of the characteristic images that surely comes to mind is the oscillating motion of the coupling rods on the wheels. The long rods that connected the wheels provided a way to convert heat energy from the steam into mechanical energy
1) http://creationsafaris.com/crev201009.htm
2) https://web.archive.org/web/20170708104125/http://creationsafaris.com/crev201007.htm
3) https://www.sfb746.uni-freiburg.de/Publications/Hunte/hunte-2010.pdf
4) http://www.uni-freiburg.de/news/news_050710_2_en
5) http://www.esrf.eu/news/general/respiratory-complex1/index_html
6) http://www.nature.com/scitable/topicpage/why-are-cells-powered-by-proton-gradients-14373960
7) http://www.ebi.ac.uk/pdbe/quips?story=CXI
Last edited by Admin on Mon Feb 11, 2019 8:47 pm; edited 10 times in total