https://reasonandscience.catsboard.com/t2036-the-complexity-of-transcription-through-rna-polymerase-enzymes-and-general-transcription-factors-in-eukaryotes
Without transcription from the genetic storage medium DNA into the current RNA world agents, no relevant genetic process can be initiated
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5398208/
Transcription of DNA requires 1. a promoter region, 2.a terminal signal, and 3. the transcription machinery that recognizes the promoter and terminator signal. Unless all three are present, no deal.
Transcribing the Codes In living cells, the first step of decoding the information in a genome is the transcription of its genes into RNA. In its simplest form, transcription requires the coordinated action of at least three essential components. A promoter region is necessary (although it may not be sufficient) for a segment of DNA to be recognized as a gene. A second segment of DNA is necessary (although it may not be sufficient) to serve as a transcription termination signal. Finally, transcription machinery made of multiple proteins must recognize the promoter and terminator and transcribe the gene.
The Central Dogma of Molecular Biology: "DNA makes RNA makes protein" Here the process begins. Transcription factors assemble at a specific promoter region along the DNA. The length of DNA following the promoter is a gene and it contains the recipe for a protein. A mediator protein complex arrives carrying the enzyme RNA polymerase. It manoeuvres the RNA polymerase into place... inserting it with the help of other factors between the strands of the DNA double helix. The assembled collection of all these factors is referred to as the transcription initiation complex... and now it is ready to be activated. The initiation complex requires contact with activator proteins, which bind to specific sequences of DNA known as enhancer regions. These regions may be thousands of base pairs distant from the start of the gene. Contact between the activator proteins and the initiation-complex releases the copying mechanism. The RNA polymerase unzips a small portion of the DNA helix exposing the bases on each strand. Only one of the strands is copied. It acts as a template for the synthesis of an RNA molecule which is assembled one sub-unit at a time by matching the DNA letter code on the template strand. The sub-units can be seen here entering the enzyme through its intake hole and they are joined together to form the long messenger RNA chain snaking out of the top.
The preinitiation complex (PIC) is a large complex of proteins that is necessary for the transcription of protein-coding genes in eukaryotes and archaea. 8


Furthermore, as Stephen Meyer explains in his book, Signature in the cell,
in both prokaryotes and eukaryotes, transcription constitutes a complex, functionally integrated process involving several specialized and individually necessary proteins. Yet production of each of these separate proteins is itself dependent on the very process of transcription that they make possible. To build RNA polymerase, for example, the cell must transcribe the genetic texts with the instructions for building RNA polymerase. Yet to transcribe this information requires RNA polymerase. The information necessary to manufacture RNA polymerase and all the other associated enzymes and protein cofactors of transcription is stored on the DNA template. But expressing that information on the DNA template for building the proteins of the transcription system requires most of the proteins of the transcription system.
What good is DNA for, if the transcription machinery is not in place in the cell to read the coded information stored in DNA? Must both not be in place and fully functional at the same time? and what good is the transcription machinery good for without DNA to read the message?
Discovery 7
In the 1950's, it was scientific belief that DNA was directly involved in protein synthesis, as the cellular function of RNA was not understood. However, based on the fact that reticulocytes and enucleated cells still produced proteins, some scientists deduced that DNA was not directly involved in protein synthesis. Polymerase phosphorylase was predicted to be responsible for cellular RNA synthesis (Losick et al 1976) until 1959 when Samuel B. Weiss and Leonard Gladstone first discovered DNA-dependent RNA polymerizing activity in the nuclear fraction of rat liver cells. Before then, active polymerase was always discovered in RNA extracts of bacterial RNA.
Weiss decided to run an experiment based on the assumption that RNA synthesis should be analogous to DNA synthesis (Losick et al 1976). He phosphorylated [32P]-labeled 5'CMP with ATP and crude brewer's yeast. The resultant [32P] was eluted on a Dowex-1-Cl column. A crude rat liver fraction isolated from the lower sedimenting fraction containing cell debris and nuclei showed incorporation of the [32P]-CTP into a TCA-precipitable fraction of RNA. Optimal incorporation of the labeled CTP occurred in the presence of other unlabeled ribonucleotide triphosphates as opposed to diphosphates. Addition of DNase slowed the incorporation, while addition of RNase significantly reduced the RNA chain length. However, Weiss and Gladstone did not indicate the presence of more than one class of RNA polymerase.
Later in 1969, Robert G. Roeder and William L Rutter separated and named the three mammalian RNA polymerases. They considered that there might be three different classes of RNA polymerases based on 3 assumptions:
1. Ribosomal RNA has a base composition different from other classes of RNA molecules.
2. Ribosomal genes are found in the nucleolus, while other classes of RNA molecules are dispersed in the nucleoplasm.
3. The regulation of ribosomal RNA synthesis and "DNA-like RNA," i.e. messenger RNA (mRNA) were independently regulated.
They isolated nuclei from rat liver cells. The RNA polymerase "activities" were dissolved and incompletely purified using sonication at a high ionic concentration, causing a high enzyme yield. They lowered the ionic concentration removed chromatin from the solution using centrifugation and ammonium sulfate fractionation. The enzymes were wholly purified using DEAE-Sephadex chromatography. Each polymerase was isolated and named according to its ammonium sulfate elution concentration. RNA polymerase I was isolated at the highest elution concentration. Polymerase II was isolated at medium salt concentration and polymerase III was isolated at the lowest concentration (Young 1991; Roeder and Rutter 1970).
The polymerases maintained functionally discrete properties after freezing, thawing, dilution, reconcentration, and second chromatographic purification. Polymerase II was characterized as easily soluble and found primarily in the nucleoplasm. Also, high ionic strength of Mn2+ increased mRNA synthesis and this activity was in the nucleoplasm. RNA polymerase I was localized in the nucleolus, while the functions of RNA polymerase III had not been characterized yet. In a subsequent experiment, the dynamic duo confirmed different activities for polymerases I and II at different stages of sea urchin embryo development. They found that during early cell division (blastula and postgastrula stages) synthesis of rRNA remains constant per cell attributed to the activity of RNA polymerase I, while mRNA synthesis increased in the cell. Thus, they confirmed that RNA polymerase II was responsible for the synthesis of "DNA-like RNA."
Transcription begins in the nucleus of the cell :

Many different types of chemical reactions are required to produce a properly folded protein from the information contained in a gene, namely
INITIATION OF TRANSCRIPTION
CAPPING, ELONGATION, SPLICING
CLEAVAGE,POLYADENYLATION AND TERMINATION
EXPORT FROM THE NUCLEUS TO THE CYTOSOL
INITIATION OF PROTEIN SYNTHESIS (TRANSLATION)
COMPLETION OF PROTEIN SYNTHESIS AND PROTEIN FOLDING
PROTEIN DEGRADATION
Transcription :
1,2
Transcription is the process of making RNA from a DNA template. Several key factors are involved in this process. Including, DNA, transcription factors, RNA polymerase, and ATP.
This is an irreducibly complex system. DNA, transcription factors, RNA polymerase, and ATP must be present, otherwise, transcription cannot occur.
Transcription begins with a strand of DNA. It is divided into several important regions. The largest of these is the transcription unit. This portion of the DNA will be used to produce RNA. Upstream of the transcription unit is the TATA box. An enhancer region may also be involved.
Several complexes, known as transcription factors, are required for successful transcription. The first is TFIID, the largest of the general factors. A component of this factor, TBP, binds to the DNA using the TATA box to position TFIID near the transcription initiation site. Other transcription factors, including TFIIA and TFIIB, then attach.
These complexes prepare the DNA for the successful binding of RNA polymerase. One RNA polymerase is bound, other transcription factors complete the mature transcription complex.
Now, energy must be added to the system for transcription to begin. This energy is provided by the reduction of ATP into ADP and Pi.
RNA polymerase then synthesizes an RNA template from the strand of DNA. Most factors are released after transcription begins. When the end of the transcription unit is reached, the RNA polymerase dissociates, and the newly formed strand of RNA is released.
All the parts must come into existence at the same time, one has no function without the others. Also, the whole sequence of events must be coordinated, and all parts must fit precisely together. There is no feasible mechanism producing this complex system randomly and in a stepwise fashion. Evolution is not an option since transcription is required to make proteins, which are required to make replication work, which is essential upon which evolution works.
In contrast to bacteria, which contain a single type of RNA polymerase, eucaryotic nuclei have three: RNA polymerase I, RNA polymerase II, and RNA polymerase
III. RNA polymerase II transcribes most genes, including all those that encode proteins.
Lets concentrate just on RNA polymerase II
There are several important differences in the way in which the bacterial and eucaryotic enzymes function.
1.While bacterial RNA polymerase requires only a single additional protein (s factor) for transcription initiation to occur, eucaryotic RNA polymerases require many additional proteins.
2. Eucaryotic transcription initiation must deal with the packing of DNA into nucleosomes* and higher-order forms of chromatin structure, features absent from bacterial chromosomes. 3
Initiation of transcription of a eukaryotic gene by RNA polymerase II
To begin transcription, RNA polymerase requires several general transcription factors. The general transcription factors help to position eucaryotic RNA polymerase correctly at the promoter aid in pulling apart the two strands of DNA to allow transcription to begin, and release RNA polymerase from the promoter into the elongation mode once transcription has begun. The proteins are “general” because they are needed at nearly all promoters used by RNA polymerase II; consisting of a set of interacting proteins, they are designated as TFII (for transcription factor for polymerase II), and are denoted arbitrarily as TFIIB, TFIID, and so on.
Promoters contain specific DNA sequences such as response elements that provide a secure initial binding site for RNA polymerase and for proteins called transcription factors that recruit RNA polymerase. 4
In a broad sense, the eucaryotic general transcription factors carry out functions equivalent to those of the s factor in bacteria; indeed, portions of TFIIF
have the same three-dimensional structure as the equivalent portions of s.
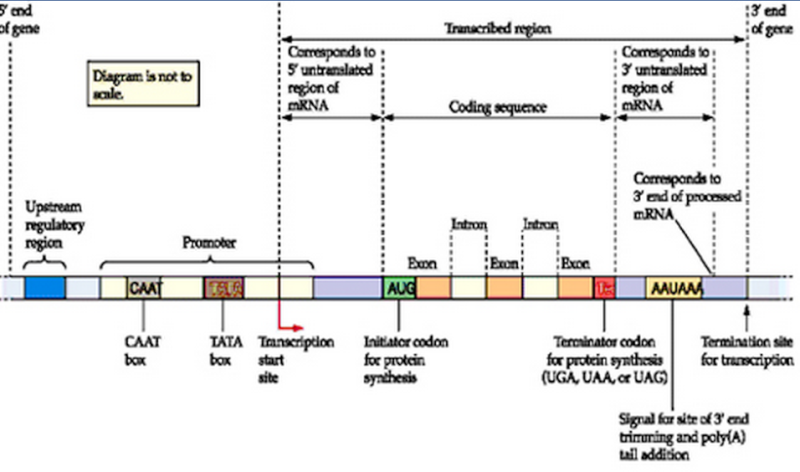
The above image illustrates the key elements of a typical eukaryotic gene. Transcription begins at the transcription start site and proceeds in the direction of the red arrow stopping at the termination site. This transcribed region will become pre-mRNA which includes introns, exons, a 5’ (read: "five prime") untranslated region (UTR) and a 3’ UTR. The initiator (start) codon and the terminator (stop) codon mark the limits of the protein coding sequence.
Following table summarizes their activities :

The assembly process begins when the general transcription factor TFIID binds to a short double-helical DNA sequence primarily composed of T and A nucleotides. For this reason, this sequence is known as the TATA sequence, or TATA box, and the subunit of TFIID that recognizes it is called TBP (for TATAbinding protein). The TATA box is typically located 25 nucleotides upstream from the transcription start site. It is not the only DNA sequence that signals the start of transcription , but for most polymerase II promoters it is the most important. The general transcription factors help to position eucaryotic RNA polymerase correctly at the promoter.
The binding of TFIID causes a large distortion in the DNA of the TATA box . This distortion is thought to serve as a physical landmark for the location of an active promoter in the midst of a very large genome, and it brings DNA sequences on both sides of the distortion together to allow for subsequent protein assembly steps. Other factors then assemble, along with RNA polymerase II, to form a complete transcription initiation complex TFIIH. Consisting of 9 subunits, it is nearly as large as RNA polymerase II itself
and, as we shall see shortly, performs several enzymatic steps needed for the initiation of transcription.
After forming a transcription initiation complex on the promoter DNA, RNA polymerase II must gain access to the template strand at the transcription start point. TFIIH, which contains a DNA helicase as one of its subunits, makes this step possible by hydrolyzing ATP and unwinding the DNA, thereby exposing the template strand. Next, RNA polymerase II, like the bacterial polymerase, remains at the promoter synthesizing short lengths of RNA until it undergoes a series of conformational changes that allow it to move away from the promoter and enter the elongation phase of transcription. A key step in this transition is the addition of phosphate groups to the “tail” of the RNA polymerase (known as the CTD or C-terminal domain). In humans, the CTD consists of 52 tandem repeats of a seven-amino-acid sequence, which extend from the RNA polymerase core structure. During transcription initiation, the serine located at the fifth position in the repeat sequence (Ser5) is phosphorylated by TFIIH, which contains a protein kinase in another of its subunits.
The polymerase can then disengage from the cluster of general transcription factors. During this process, it undergoes a series of conformational changes that tighten its interaction with DNA, and it acquires new proteins that allow it to transcribe for long distances, and in some cases for many hours, without dissociating from DNA. Once the polymerase II has begun elongating the RNA transcript, most of the general transcription factors are released from the DNA so that they are available to initiate another round of transcription with a new RNA polymerase molecule. As we see shortly, the phosphorylation of the tail of RNA polymerase II also causes components of the RNA-processing machinery to load onto the polymerase and thus be positioned to modify the newly transcribed RNA as it emerges from the polymerase.
Polymerase II Also Requires Activator, Mediator, and Chromatin-Modifying Proteins
DNA in eucaryotic cells is packaged into nucleosomes, which are further arranged in higher-order chromatin structures. As a result, transcription initiation in a eucaryotic cell is
more complex and requires even more proteins than it does on purified DNA. First, gene regulatory proteins known as transcriptional activators must bind to specific sequences in DNA and help to attract RNA polymerase II to the start point of transcription.

For RNA polymerase to successfully bind to a eukaryotic promoter and initiate transcription, a set of proteins called transcription factors must first assemble on the promoter.
The assembly process begins upstream from the transcription start site, where proteins called basal factors bind to a short TATA sequence in the promoter.
Other basal factor proteins then bind, eventually forming a full transcription factor complex able to capture the RNA polymerase.
Basal factors are essential for transcription but cannot by themselves increase or decrease its rate. A second set of transcription factors called coactivators link the basal factors with a third set of transcription factor proteins called activators.
Activators are regulatory proteins that bind to sequences on DNA called enhancers. Enhancers are located at sites that are distant from the promoter.
The interaction of activator proteins with transcription factor subunits increases the rate of transcription.
Many enhancers, scattered around the chromosome, can bind different activators, which provide a variety of responses to various signals.
When a second kind of regulatory protein called a repressor binds to a "silencer" sequence located adjacent to or overlapping an enhancer sequence, the corresponding activator is no longer able to bind to the DNA.

The presence of activators on DNA is required for transcription initiation in a eukaryotic cell. Second, eucaryotic transcription initiation in vivo requires the presence of a protein complex known as Mediator, which allows the activator proteins to communicate properly with the polymerase II and with the general transcription factors. Finally, transcription initiation in a eucaryotic cell typically requires the local recruitment of chromatin modifying enzymes, including chromatin remodelling complexes and histone-modifying enzymes.
Many proteins (well over 100 individual subunits) must assemble at the start point of transcription to initiate transcription in a eucaryotic cell. The order of assembly of these proteins does not seem to follow a prescribed pathway; rather, the order differs from gene to gene. Indeed, some of these different protein complexes may interact with each other away from the DNA and be brought to DNA as preformed subassemblies.

1.Transcription begins with a strand of DNA. This simplified DNA model has been color-coded to show regions with important roles in transcription.

2.The largest key region is the transcription unit.

3.Upstream of the transcription unit is the TATA-box, a smaller section that helps to position the complexes involved in transcription.The assembly process begins when the general transcription factor TFIID binds to a short double-helical DNA sequence primarily composed of T and A nucleotides. For this reason, this sequence is known as the TATA sequence, or TATA box

4.The final high-lighted region is the enhancer.
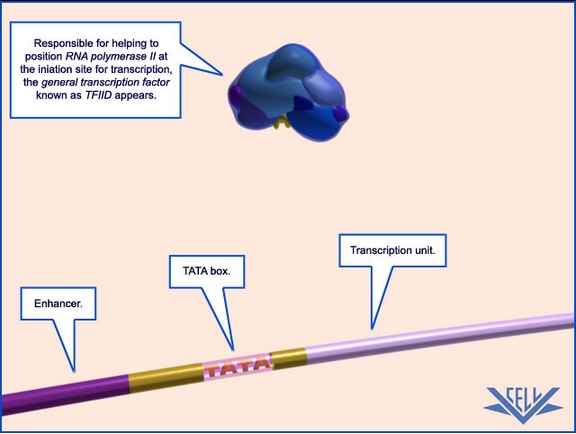
5. TFIID, a general transcription factor, is shown approaching the strand of DNA.

6. TFIID is the largest of the general transcription factors involved in eukaryotic transcription. The yellow part of the complex is called TBP. (for TATAbinding protein)

7. TBP (yellow) binds to the DNA, using the TATA-box to position itself near the iniation site of transcription.Through its subunit
TBP, TFIID recognizes and binds the TATA box, which then enables the adjacent binding of TFIIB


8. When the TBP portion of the TFIID molecule attaches to the TATA-box, its shape causes the DNA to bend.

9.Two smaller general transcription factors are shown coming into view: TFIIA (orange) and TFIIB (red).

10. TFIIA binds to the TATA-box near TFIID.
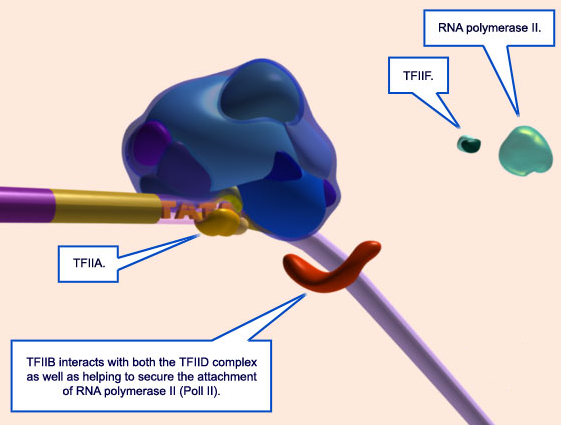
11. TFIIB approaches the TATA-box. The Pol II complex is being assembled in the distance.
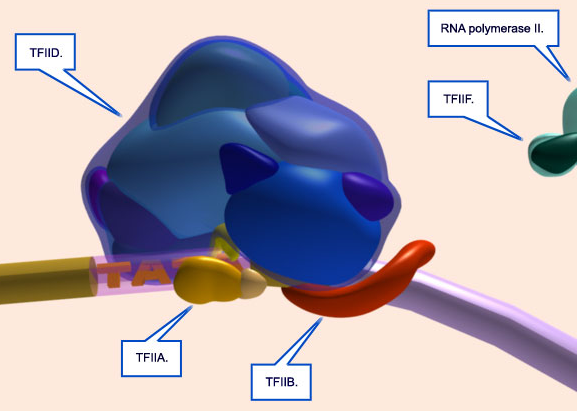
12.TFIIB binds to the TATA-box and TFIID. It is thought to help the Pol II complex bind correctly.

13. The Pol II complex has been assembled and is approaching the start site for transcription.
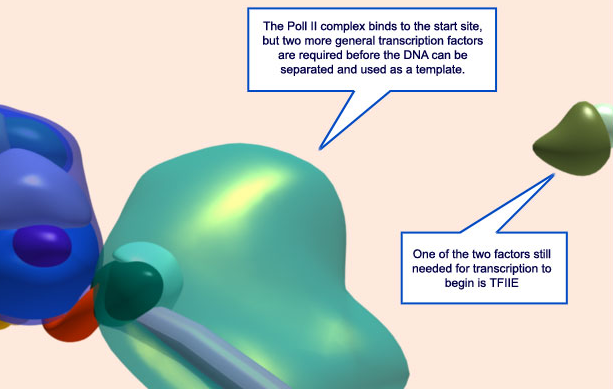
14. Aided by the general transcription factors already in place, Pol II binds to the DNA strand at the start site for transcription. TFIIE is show approaching from the right.
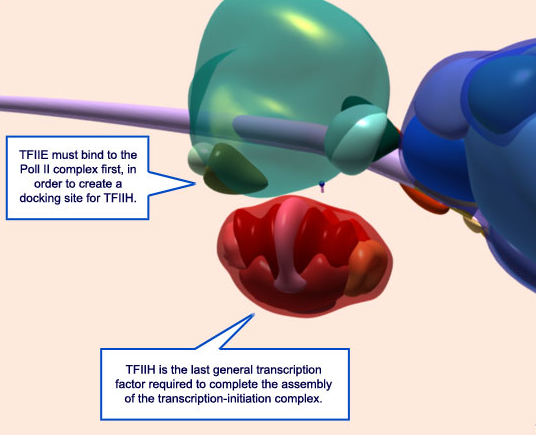
15. Additional factors must still bind to the complex in order to start transcription. Here, TFIIE has already bound (olive green) and TFIIH (red) is preparing to do the same.

16. Once all of the general transcription factors are bound, energy, in the form of ATP (blue/pink), is needed to activate the Pol II complex.

17. Once the ATP have been added, an 'eye' opens in the DNA giving access to the DNA template, and the creation of mRNA can begin.

18. Once mRNA elongation begins, TFIIE (olive green) and TFIIH (red) are released. Also released at this time are TFIIA and TFIIB (not shown).

19. TFIIE is shown moving into the distance, and the mRNA strand is elongating rapidly.

20. Elongation of the mRNA stops when the end of the transcription unit is reached.
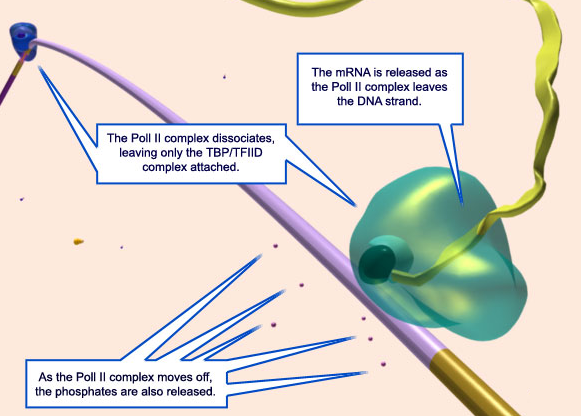
21. As elongation stops, the DNA 'eye' closes.

22. The Pol II complex is now released from the DNA, along with the phosphates added to it by the ATP.

23.As the Pol II complex dissociates from the DNA, the mRNA strand is released. At this time, TFIID also unbinds from the DNA, allowing the DNA to return to its normal shape.
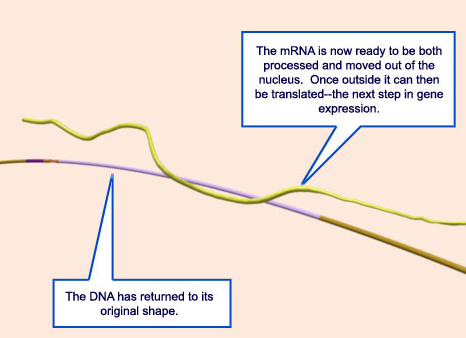
24.Transcription is now complete. An mRNA copy has been produced and is now ready to be moved outside of the nucleus and be used in the translation process.
Transcription preinitiation complex
The initial event of transcription of any gene requires the formation of a pre-initiation complex which associates with the gene’s promoter region. Once activation of this complex has occurred, the RNA polymerase leaves the promoter region and begins the process of elongation of nascent RNA chains. 4
RNA polymerase II (pol II) is capable of RNA synthesis but is unable to recognize a promoter or to initiate transcription. For these essential functions, a set of general transcription factors (GTFs)—termed TFIIB, -D, -E, -F, and -H—is required. 6
The minimal preinitiation complex (abbreviated PIC) includes six general transcription factors: TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH. Additional regulatory complexes (co-activators and chromatin-remodeling complexes) could also be components of the PIC.
Assembly
A classical view of PIC formation at the promoter involves the following steps:
TATA-binding protein (TBP, a subunit of TFIID) binds the promoter, creating a sharp bend in the promoter DNA.
TBP-TFIIA interactions recruit TFIIA to the promoter.
TBP-TFIIB interactions recruit TFIIB to the promoter.
TFIIB-RNA polymerase II and TFIIB-TFIIF interactions recruit RNA polymerase II and TFIIF to the promoter.
TFIIE joins the growing complex and recruits TFIIH.
Subunits within TFIIH that have ATPase and helicase activity create negative superhelical tension in the DNA.
Negative superhelical tension causes approximately one turn of DNA to unwind and form the transcription bubble.
The template strand of the transcription bubble engages with the RNA polymerase II active site.
RNA synthesis begins.
After synthesis of ~10 nucleotides of RNA, and an obligatory phase of several abortive transcription cycles, RNA polymerase II escapes the promoter region to transcribe the remainder of the gene.
An alternative hypothesis of PIC assembly postulates the recruitment of a pre-assembled "RNA Polymerase II holoenzyme" directly to the promoter (composed of all, or nearly all GTFs and RNA polymerase II and regulatory complexes), in a manner similar to the bacterial RNA polymerase (RNAP).
Question: How could the proteins above involved in transcription preinitiation arise by natural processes? They are only functional if working as a whole in a highly coordinated manner ?! The above process could not arise in a stepwise manner but works only if fully functioning right from the start. The process could not be the result of evolution since, in order for evolution to work, transcription must already be in place?
http://mol-biol4masters.masters.grkraj.org/html/Gene_Expression_II4-RNAP_III_Promoter.htm
* Nucleosomes: http://reasonandscience.heavenforum.org/t2017-the-nucleosomes-amazing-design#3392
https://www.youtube.com/watch?v=XzVXhemtwmA
1) https://www.youtube.com/watch?v=WsofH466lqk
2) http://vcell.ndsu.nodak.edu/animations/transcription/movie-flash.htm
3) http://reasonandscience.heavenforum.org/t2017-the-nucleosomes-amazing-design#3392
4) http://en.wikipedia.org/wiki/Promoter_%28genetics%29
5) http://www.biology.emory.edu/research/Lucchesi/html/research.html
6) http://www.sciencemag.org/content/342/6159/1238724
7) http://www.biochem.umd.edu/biochem/kahn/molmachines/newpolII/discovery.html
8 ) https://en.wikipedia.org/wiki/RNA_polymerase_II_holoenzyme
Last edited by Otangelo on Sat May 28, 2022 12:32 pm; edited 38 times in total









 5
5
