https://reasonandscience.catsboard.com/t2096-the-astonishing-language-written-on-microtubules-amazing-evidence-of-design
The following information is truly mind-boggling. Take your time to read all through, and check the links. The creator of life has left a wealth of evidence of his existence in creation. A treasure grove to evidence intelligent design is every living cell. Its widely known that DNA is an advanced information storage device, encoding complex specified information to make proteins and directing many highly complex processes in the cell. What is less known, is that there are several other code systems as well, namely the histone binding code, transcription factor binding code, the splicing code, and the RNA secondary structure code. And there is another astonishing code system, called the tubulin code, which is being unraveled in recent scientific research. It is known so far that amongst other things, it directs and signals Kinesin and Myosin motor proteins precisely where and when to disengage from nanomolecular superhighways and deliver their cargo.
https://reasonandscience.catsboard.com/t1448-kinesin-motor-proteins-amazing-cargo-carriers-in-the-cell?highlight=kinesin
Recent research holds that this code in an amazing manner even stores our memories in the brain and makes them available in the long term.
https://reasonandscience.catsboard.com/t2182-heres-an-incredible-idea-for-how-memory-works#4032
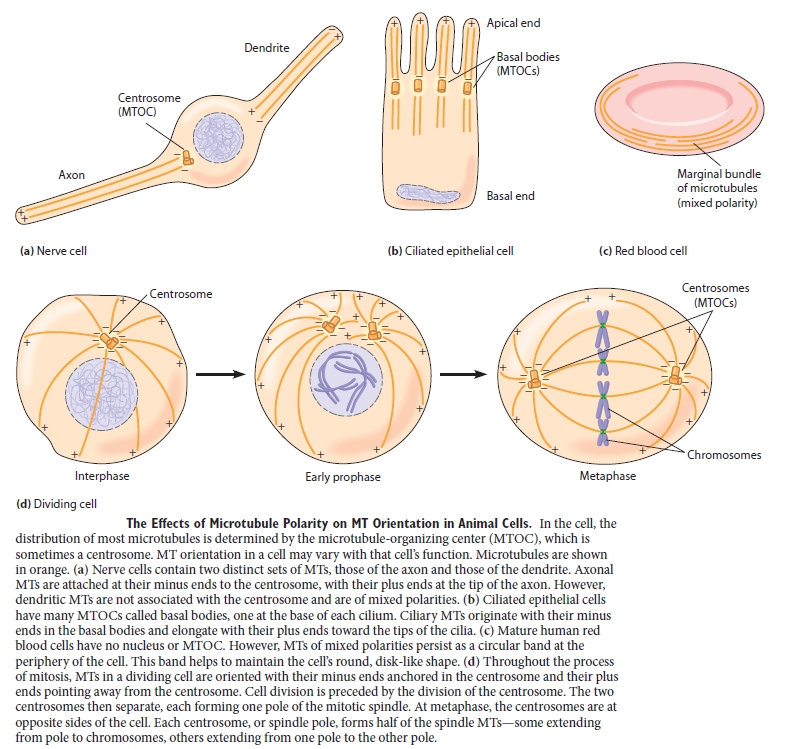
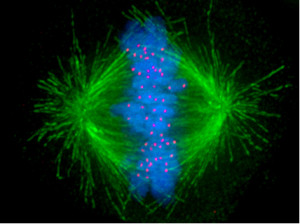
For cells to function properly, they must organize themselves and interact mechanically with each other and with their environment. They have to be correctly shaped, physically robust, and properly structured internally. Many have to change their shape and move from place to place. All cells have to be able to rearrange their internal components as they grow, divide, and adapt to changing circumstances. These spatial and mechanical functions depend on a remarkable system of filaments called the cytoskeleton. The cytoskeleton’s varied functions depend on the behavior of three families of protein filaments—actin filaments, microtubules, and intermediate filaments. Microtubules are very important in a number of cellular processes. They are involved in maintaining the structure of the cell and provide a platform for intracellular macromolecular assemblies through dynein and kinesin motors. They are also involved in chromosome separation (mitosis and meiosis) and are the major constituents of mitotic spindles, which are used to pull apart eukaryotic chromosomes. Mitotic cell division is the most fundamental task of all living cells. Cells have the intricate and tightly regulated machinery to ensure that mitosis occurs with appropriate frequency and high fidelity. If someone wants to explain the origin of eukaryotic cells, the arise of mitosis and its mechanism and involved cell organelles and proteins must be elucidated. The centrosome plays a crucial role: it functions as the major microtubule-organizing center and plays a vital role in guiding chromosome segregation during mitosis. In the centrosome, two centrioles reside at right angles to each other, connected at one end by fibers.
These architecturally perfect structures are essential in many animal cells and plants (though not in flowering plants or fungi, or in prokaryotes). They help organize the centrosomes, whose spindles of microtubules during cell division reach out to the lined-up chromosomes and pull them into the daughter cells.
https://reasonandscience.catsboard.com/t2090-centriole-centrosome-the-centriole-spindle-the-most-complex-machine-known-in-nature?highlight=spindle
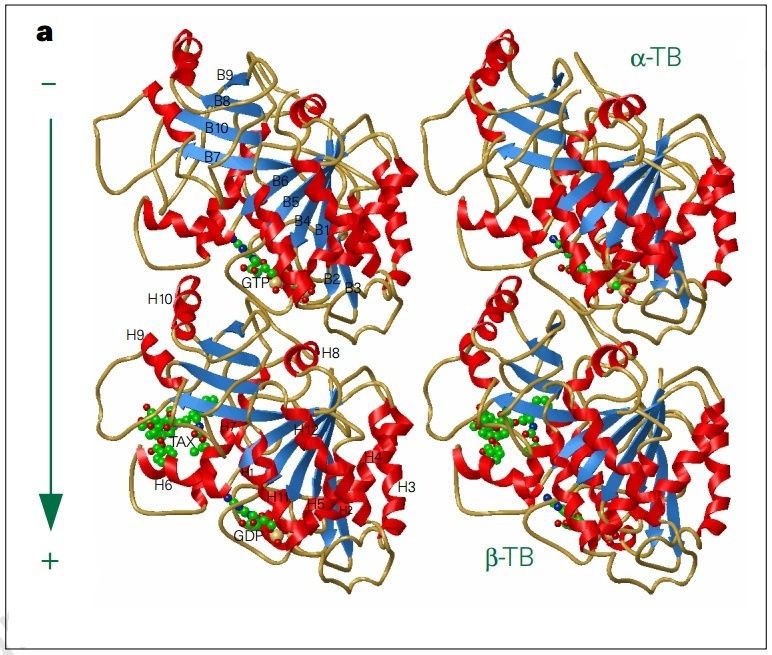
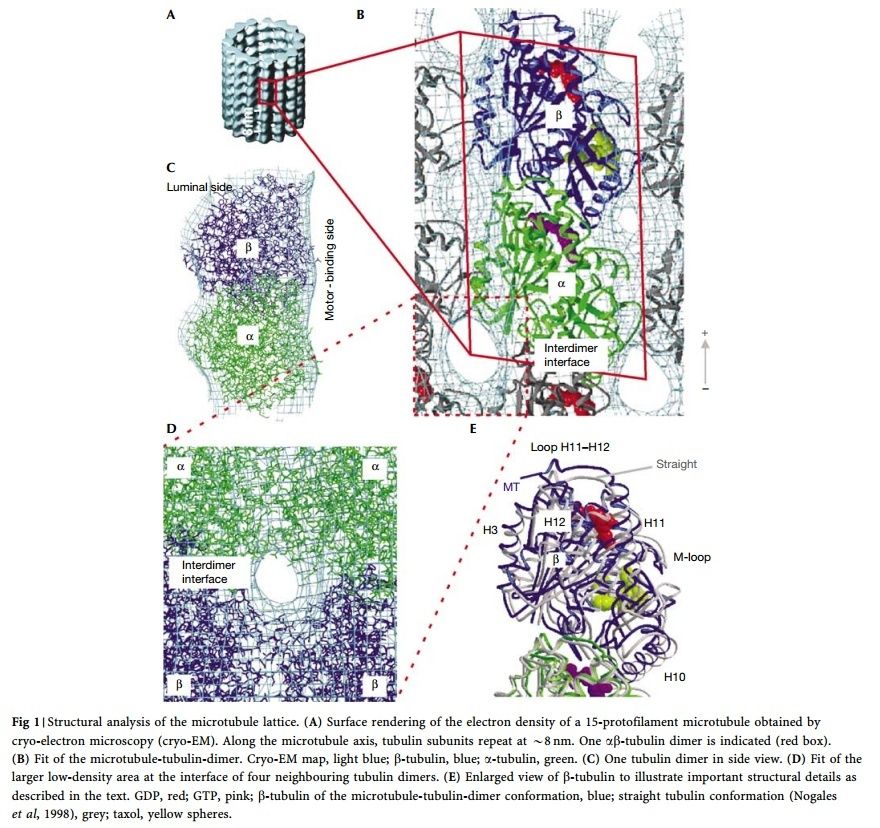
α- and β-tubulin heterodimers are the structural subunits of microtubules. The structure is divided in the amino-terminal domain containing the nucleotide-binding region, an intermediate domain containing the Taxol-binding site, and the carboxy-terminal domain, which probably constitutes the binding surface for motor proteins. Unless all 3 functional domains were fully functional right from the beginning, tubulins would have no useful function. There would be no reason for the Taxol-binding site to be without motor proteins existing. Dynamic instability, the stochastic switching between growth and shrinkage, is essential for microtubule function.
https://reasonandscience.catsboard.com/t2096-the-cytoskeleton-microtubules-and-post-translational-modification#4033
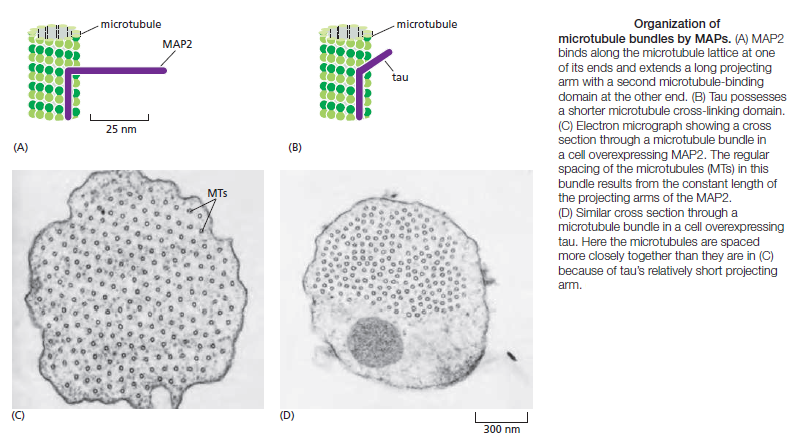
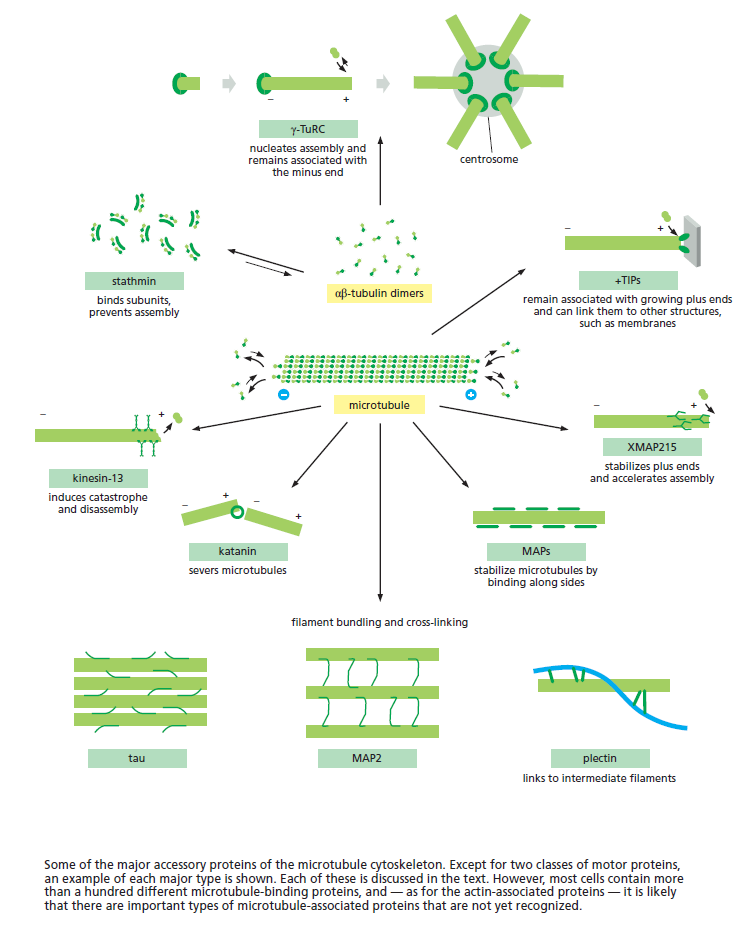
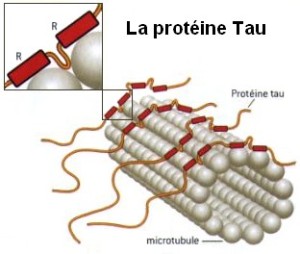
Microtubule dynamics inside the cell are governed by a variety of proteins that bind tubulin dimers or microtubules. Proteins that bind to microtubules are collectively called microtubule-associated proteins, or MAPs.The MAP family includes large proteins like MAP-1A, MAP-1B, MAP-1C, MAP-2, and MAP-4 and smaller components like tau and MAP-2C.
This is highly relevant. Microtubules depend on microtubule-associated proteins for proper function. Interdependence is a hallmark of intelligent design and strong evidence that both, microtubules, and MAP's had to emerge together, at the same time, since one depends on the other for proper function. But more than that. Microtubules are essential to form the cytoskeleton, which is essential for cell shape and structure. In a few words, No MAP's, no proper function of microtubules. No microtubules, no proper function of the cytoskeleton. No cytoskeleton, no proper functioning cell. The evidence is very strong, that all these elements had to arise together at once. Kinesin and Dynein belong to MAP proteins. Kinesin-13 proteins contribute to microtubule depolymerizing activity to the centrosome and centromere during mitosis. These activities have been shown to be essential for spindle morphogenesis and chromosome segregation. A step-wise evolutionary emergence of eukaryotic cells is not feasible since several parts of the call can only work if interacting together in an interlocked fully developed system.
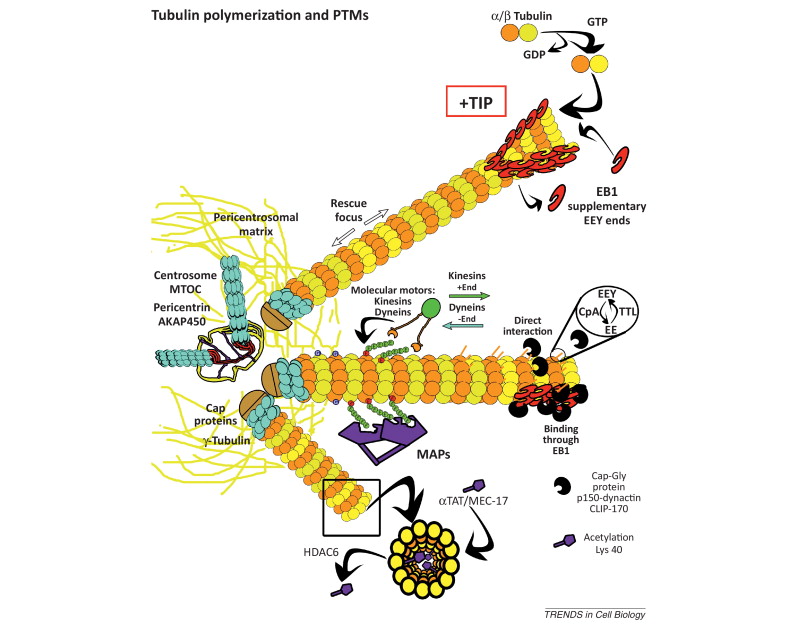
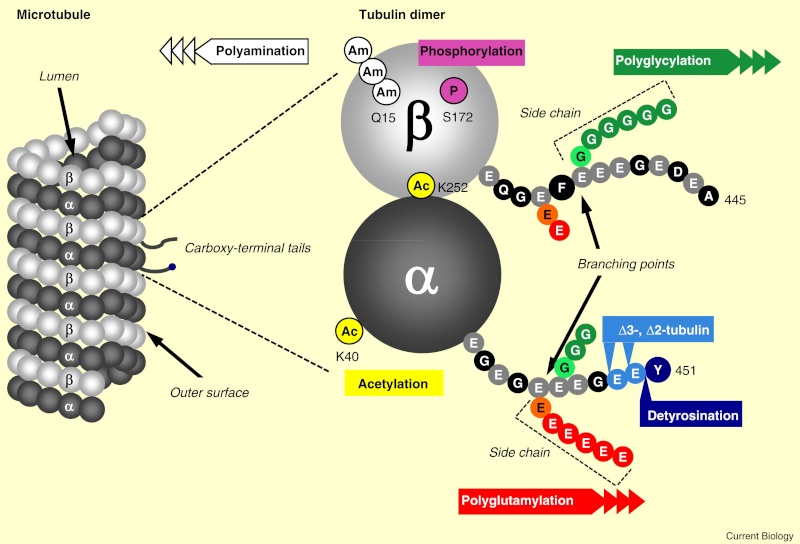
When incorporated into microtubules, tubulin accumulates a number of post-translational modifications, many of which are unique to these proteins. These modifications include detyrosination, acetylation, polyglutamylation, polyglycylation,phosphorylation, ubiquitination, sumoylation, and palmitoylation. The α- and β-tubulin heterodimer undergoes multiple post-translational modifications (PTMs). The modified tubulin subunits are non-uniformly distributed along microtubules. Analogous to the model of the ‘histone code’ on chromatin, diverse PTMs are proposed to form a biochemical ‘tubulin code’ that can be ‘read’ by factors that interact with microtubules.
This is a relevant and amazing fact and raises the question of how the " tubulin code " beside the several other codes in the cell emerged. In my view, once more this shows that intelligence was required to create these amazing biomolecular structures; the formation of coded information has always shown to be able only to be produced by intelligent minds. What good would the tubulin code be for, if no specific goal was foreseen, that is, it acts as an emitter of information, and if there is no destination and receiver of the information, there is no reason for the code to arise in the first place? So both, sender and receiver, must exist first as hardware, that is the microtubules with the post-transcriptional modified tubulin units in a specified coded conformation, and then the receiver, which can be MAP's in general, or Kinesin or Myosin motor proteins, which are directed to the right destination to fulfill specific tasks, or other proteins directed for specific jobs.
Taken together, multiple and complex tubulin PTMs provide a myriad of combinatorial possibilities to specifically ‘tag’ microtubule subpopulations in cells, thus destining them for precise functions. How this tubulin or microtubule code allows cells to divide, migrate, communicate, and differentiate in an ordered manner is an exciting question that needs to be answered in the near future. Initial insights have already revealed the potential roles of tubulin PTMs in a number of human pathologies, like cancer, neurodegeneration, and ciliopathies. This raises the question: If PTM's are not precise and fully functioning, they cause diseases. What about if the MAP's are not fully specified and evolved? There is a threshold, a dividing line between a non-functional protein - amino acid sequence that is non-functional, and when it has enough residues to fold properly and become functional. How proteins arose in the first place is a mystery for the proponents of natural mechanisms..... Not only does it have to be elucidated how this tubulin or microtubule code allows cells to do all these tasks, but also what explains best it's arising and encoding. Most of these enzymes are specific to tubulin and microtubule post-translational modifications. They have only use if microtubules exist. Microtubules, however, require these enzymes to modify their structures. It can, therefore, be concluded that they are interdependent and could not arise independently by natural evolutionary mechanisms.
An emerging hypothesis is that tubulin modifications specify a code that dictates biological outcomes through changes in higher-order microtubule structure and/or by recruiting and interacting with effector proteins. This hypothesis is analogous to the histone code hypothesis ‑ that modifications on core histones, acting in a combinatorial or sequential fashion, specify multiple functions of chromatin such as changes in higher-order chromatin structure or selective activation of transcription. The apparent parallels between these two types of structural frameworks, chromatin in the nucleus and microtubules in the cytoplasm, are intriguing
Isn't that striking evidence of a common designer that invented both codes?
https://reasonandscience.catsboard.com/t2096-the-cytoskeleton-microtubules-and-post-translational-modification#4035
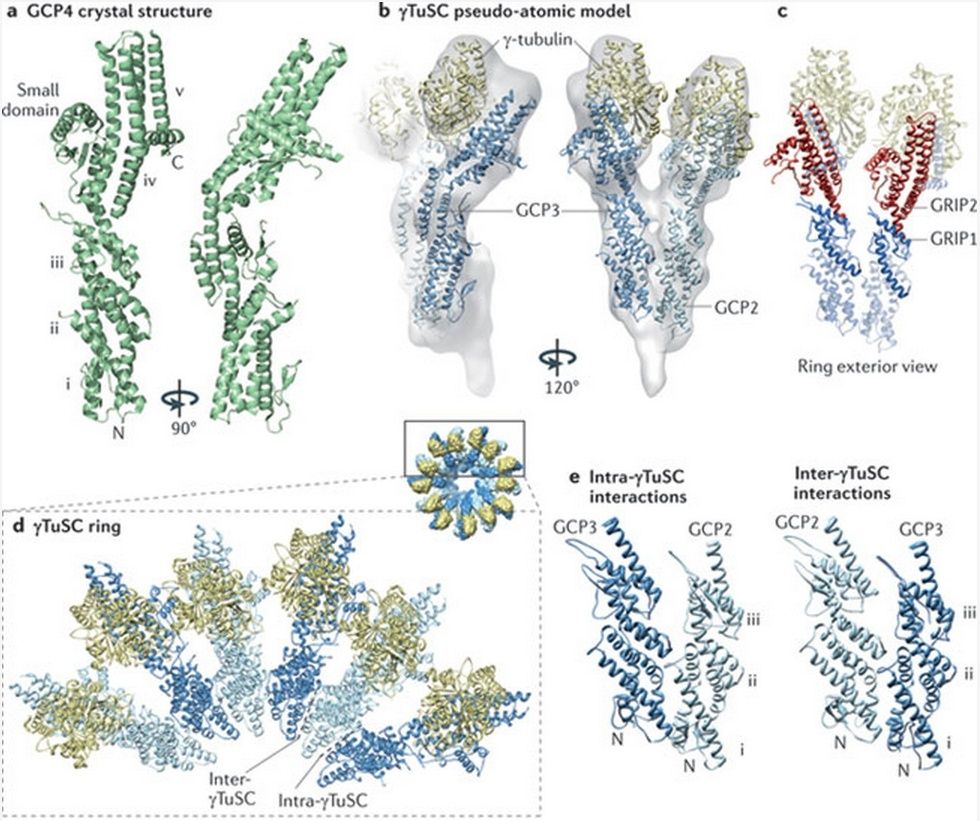
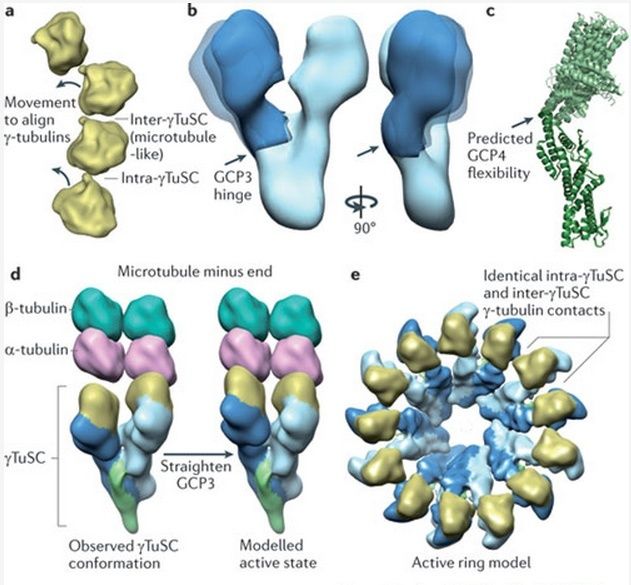
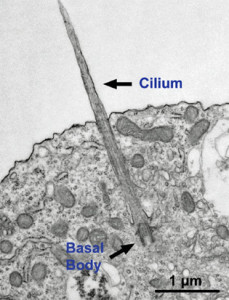
Microtubules are typically nucleated and organized by dedicated organelles called microtubule-organizing centres (MTOCs). Contained within the MTOC is another type of tubulin, γ-tubulin, which is distinct from the α- and β-subunits of the microtubules themselves. The γ-tubulin combines with several other associated proteins to form a lock washer-like structure known as the γ-tubulin ring complex" (γ-TuRC). This complex acts as a template for α/β-tubulin dimers to begin polymerization; it acts as a cap of the (−) end while microtubule growth continues away from the MTOC in the (+) direction. The γ-tubulin small complex (γTuSC) is the conserved, essential core of the microtubule nucleating machinery, and it is found in nearly all eukaryotes.
This γ-tubulin ring complex is a striking example of a purposeful design that is required to nucleate the microtubules into the right shape. There would be no function for the γ-tubulin ring complex to emerge without microtubules since it would have no function on its own. Furthermore, it is made of several subunits that are indispensable for proper use, that is, for example, the attachment factors, accessory proteins, and γ-tubulins, which constitute an irreducible γ-tubulins ring complex, made of several interlocked parts, which could not emerge by natural selection. The complex has only a purposeful function when microtubules have to be assembled. So the γ-tubulins ring complex and microtubules are interdependent.
See its striking structure here :
https://reasonandscience.catsboard.com/t2096-the-cytoskeleton-microtubules-and-post-translational-modification#4040
Here’s an Incredible Idea For How Memory Works
Cytoskeletal Signaling: Is Memory Encoded in Microtubule Lattices by CaMKII Phosphorylation?
how the brain could store information long-term has been something of a mystery. But now researchers have developed a very interesting idea of how the brain’s neurons could store information using, believe it or not, a binary encoding scheme based on phosphorylation:
Memory is attributed to strengthened synaptic connections among particular brain neurons, yet synaptic membrane components are transient, whereas memories can endure. This suggests synaptic information is encoded and ‘hard-wired’ elsewhere, e.g. at molecular levels within the post-synaptic neuron. In long-term potentiation (LTP), a cellular and molecular model for memory, post-synaptic calcium ion (Ca2+) flux activates the hexagonal Ca2+-calmodulin dependent kinase II (CaMKII), a dodacameric holoenzyme containing 2 hexagonal sets of 6 kinase domains.
This enzyme has a astonishing and remarkable configuration and functionality :
Each kinase domain can either phosphorylate substrate proteins, or not (i.e. encoding one bit). Thus each set of extended CaMKII kinases can potentially encode synaptic Ca2+ information via phosphorylation as ordered arrays of binary ‘bits’. Candidate sites for CaMKII phosphorylation-encoded molecular memory include microtubules (MTs), cylindrical organelles whose surfaces represent a regular lattice with a pattern of hexagonal polymers of the protein tubulin. Using molecular mechanics modeling and electrostatic profiling, we find that spatial dimensions and geometry of the extended CaMKII kinase domains precisely match those of MT hexagonal lattices. This suggests sets of six CaMKII kinase domains phosphorylate hexagonal MT lattice neighborhoods collectively, e.g. conveying synaptic information as ordered arrays of six “bits”, and thus “bytes”, with 64 to 5,281 possible bit states per CaMKII-MT byte. Signaling and encoding in MTs and other cytoskeletal structures offer rapid, robust solid-state information processing which may reflect a general code for MT-based memory and information processing within neurons and other eukaryotic cells.
Size and geometry of the activated hexagonal CaMKII holoenzyme and the two types of hexagonal lattices (A and B) in MTs are identical. 6 extended kinases can interface collectively with 6 tubulins
Is the precise interface matching striking coincidence, or purposeful design ? Either a intelligent , goal oriented creator made the correct size, where CaMKII would fit and match the hexagonal lattices, or that is the result of unguided, random, evolutionary processes. What explanation makes more sense ?
The electrostatic pattern formed by a neighborhood of tubulin dimers on a microtubule ( MT ) surface shows highly negative charged regions surrounded by a less pronounced positive background, dependent on the MT lattice type . These electrostatic fingerprints are complementary to those formed by the 6 CaMKII holoenzyme kinase domains making the two natural substrates for interaction. Alignment of the CaMKII holoenzyme with tubulin dimers in the A-lattice MT arrangement yields converging electric field lines indicating a mutually attractive interaction.
So additionally to the precise interface matching significant association of the CaMKII holoenzyme with the MT through electrostatic forces indicates cumulative evidence of design.
there are 26 possible encoding states for a single CaMKII-MT interaction resulting in the storage of 64 bits of information. This case, however, only accounts for either α- or β-tubulin phosphorylation, not both. In the second scenario each tubulin dimer is considered to have three possible states – no phosphorylation (0), β-tubulin phosphorylation (1), or α-tubulin phosphorylation (2) (see Figure 5 B). These are ternary states, or ‘trits’ (rather than bits). Six possible sites on the A-lattice yield 36 = 729 possible states. The third scenario considers the 9-tubulin B-lattice neighborhood with ternary states. As in the previous scenarios the central dimer is not considered available for phosphorylation. In this case, 6 tubulin dimers out of 8 may be phosphorylated in three possible ways. The total number of possible states for the B lattice neighborhood is thus 36–28−8(27) = 5281 unique states.
So thirdly we have here a advanced encoding mechanism of information, which adds to the precise interface and electrostatic force interactions, which adds further cumulative evidence of design.
https://reasonandscience.catsboard.com/t2181-cell-communication-and-signalling-evidence-of-design#4019
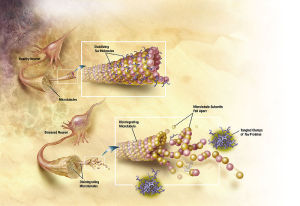
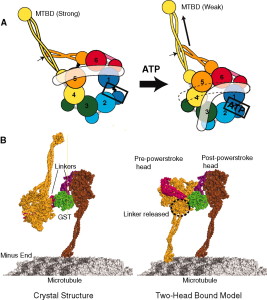
Motor proteins dynein and kinesin move along microtubules (using ATP as fuel) to transport and deliver components and precursors to specific synaptic locations. While microtubules are assumed to function as passive guides, like railroad tracks for motor proteins, the guidance mechanism seems to be through CaMKII kinase enzymes which "write" on microtubules through phosphorylation and encode the way to regulate motor protein transport along microtubules directly, and signal motor proteins precisely where and when to disengage from microtubules and deliver their cargo. There needs to be programming all the way along. Programming to make the specific enzymes, and how they have to operate. That constitutes in my view another amazing argument for intelligent design.
The Cytoskeleton, microtubules, and post-translational modification
The Cytoplasm Is Organized by the Cytoskeleton and Is Highly Dynamic
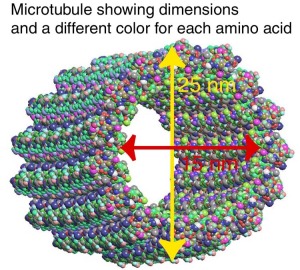
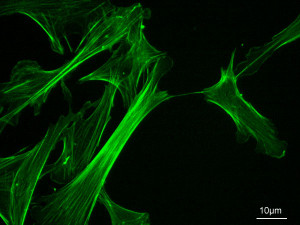
Fluorescence microscopy reveals several types of protein filaments crisscrossing the eukaryotic cell, forming an interlocking three-dimensional meshwork, the cytoskeleton. There are three general types of cytoplasmic filaments—actin filaments, microtubules, and intermediate filaments.
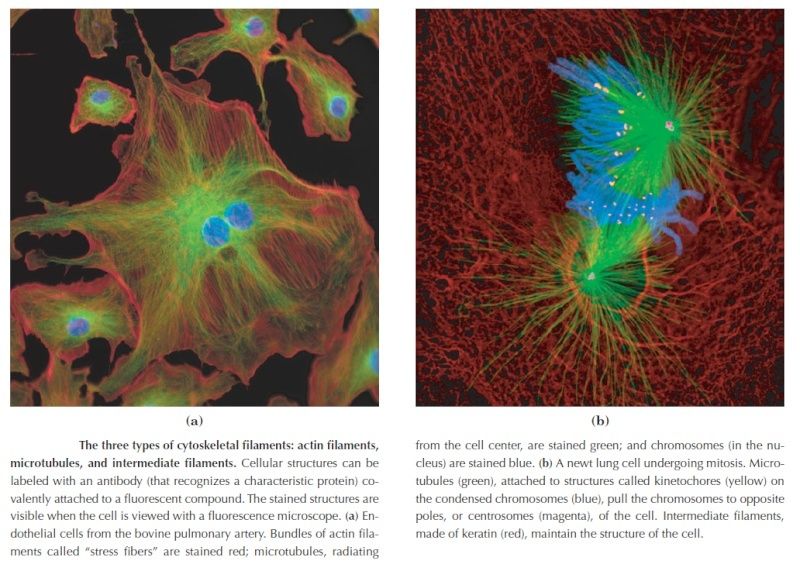
differing in width (from about 6 to 22 nm), composition, and specific function. All types provide structure and organization to the cytoplasm and shape to the cell. Actin filaments and microtubules also help to produce the motion of organelles or of the whole cell. Each type of cytoskeletal component is composed of simple protein subunits that associate noncovalently to form filaments of uniform thickness. These filaments are not permanent structures; they undergo constant disassembly into their protein subunits and reassembly into filaments. Their locations in cells are not rigidly fixed but may change dramatically with mitosis, cytokinesis, amoeboid motion, or changes in cell shape. The assembly, disassembly, and location of all types of filaments are regulated by other proteins, which serve to link or bundle the filaments or to move cytoplasmic organelles along the filaments. The picture that emerges from this brief survey of eukaryotic cell structure is of a cell with a meshwork of structural fibers and a complex system of membraneenclosed compartments. The filaments disassemble and then reassemble elsewhere. Membranous vesicles bud from one organelle and fuse with another. Organelles move through the cytoplasm along protein filaments, their motion powered by energy-dependent motor proteins. The endomembrane system segregates specific metabolic processes and provides surfaces on which certain enzyme-catalyzed reactions occur. Exocytosis and endocytosis, mechanisms of transport (out of and into cells, respectively) that involve
membrane fusion and fission, provide paths between the cytoplasm and surrounding medium, allowing for secretion of substances produced in the cell and uptake of extracellular materials.
Although complex, this organization of the cytoplasm is far from random. The motion and positioning of organelles and cytoskeletal elements are under tight regulation, and at certain stages in its life, a eukaryotic cell undergoes dramatic, finely orchestrated reorganizations, such as the events of mitosis. The interactions between the cytoskeleton and organelles are noncovalent, reversible, and subject to regulation in response to various intracellular and extracellular signals.
The Cytoskeleton
For cells to function properly, they must organize themselves in space and interact mechanically with each other and with their environment. They have to be correctly shaped, physically robust, and properly structured internally. Many have to change their shape and move from place to place. All cells have to be able to rearrange their internal components as they grow, divide, and adapt to changing circumstances. These spatial and mechanical functions depend on a remarkable system of filaments called the cytoskeleton. The cytoskeleton’s varied functions depend on the behavior of three families of protein filaments—actin filaments, microtubules, and intermediate filaments. Each type of filament has distinct mechanical properties, dynamics, and biological roles, but all share certain fundamental features. Just as we require our ligaments, bones, and muscles to work together, so all three cytoskeletal filament systems must normally function collectively to give a cell its strength, its shape, and its ability to move. In this chapter, we describe the function and conservation of the three main filament systems. We explain the basic principles underlying filament assembly and disassembly, and how other proteins interact with the filaments to alter their dynamics, enabling the cell to establish and maintain internal order, to shape and remodel its surface, and to move organelles in a directed manner from one place to another. Finally, we discuss how the integration and regulation of the cytoskeleton allows a cell to move to new locations.
FUNCTION AND ORIGIN OF THE CYTOSKELETON
The three major cytoskeletal filaments are responsible for different aspects of the cell’s spatial organization and mechanical properties. Actin filaments determine the shape of the cell’s surface and are necessary for whole-cell locomotion; they also drive the pinching of one cell into two. Microtubules determine the positions of membrane-enclosed organelles, direct intracellular transport, and form the mitotic spindle that segregates chromosomes during cell division. Intermediate filaments provide mechanical strength. All of these cytoskeletal filaments interact with hundreds of accessory proteins that regulate and link the filaments to other cell components, as well as to each other. The accessory proteins are essential for the controlled assembly of the cytoskeletal filaments in particular locations, and they include the motor proteins, remarkable molecular machines that convert the energy of ATP hydrolysis into mechanical force that can either move organelles along the filaments or move the filaments themselves. In this section, we discuss the general features of the proteins that make up the filaments of the cytoskeleton. We focus on their ability to form intrinsically polarized and self-organized structures that are highly dynamic, allowing the cell to rapidly modify cytoskeletal structure and function under different conditions.
https://sci-hub.tw/https://www.annualreviews.org/doi/full/10.1146/annurev-cellbio-100818-125149
Last edited by Admin on Mon Jul 27, 2020 6:44 am; edited 29 times in total