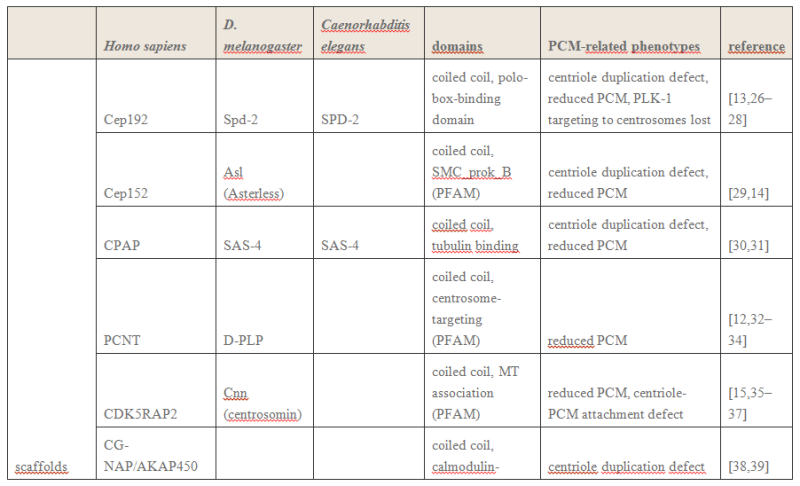
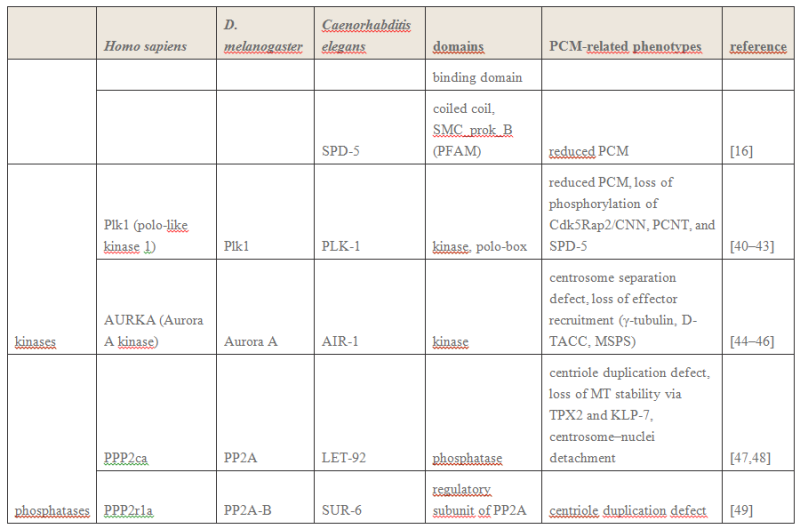
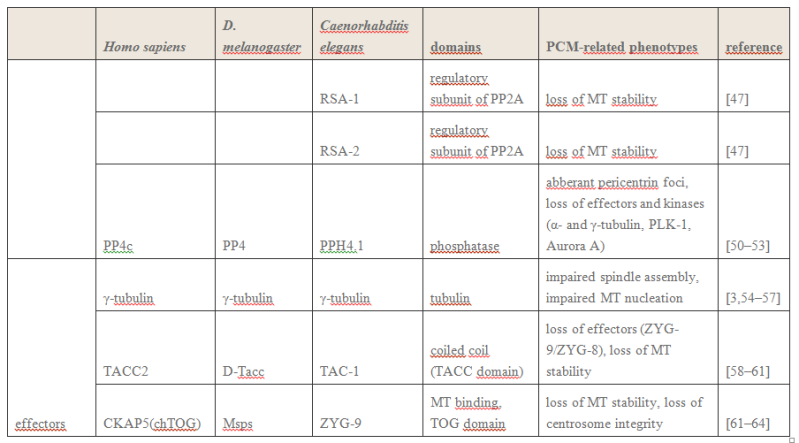
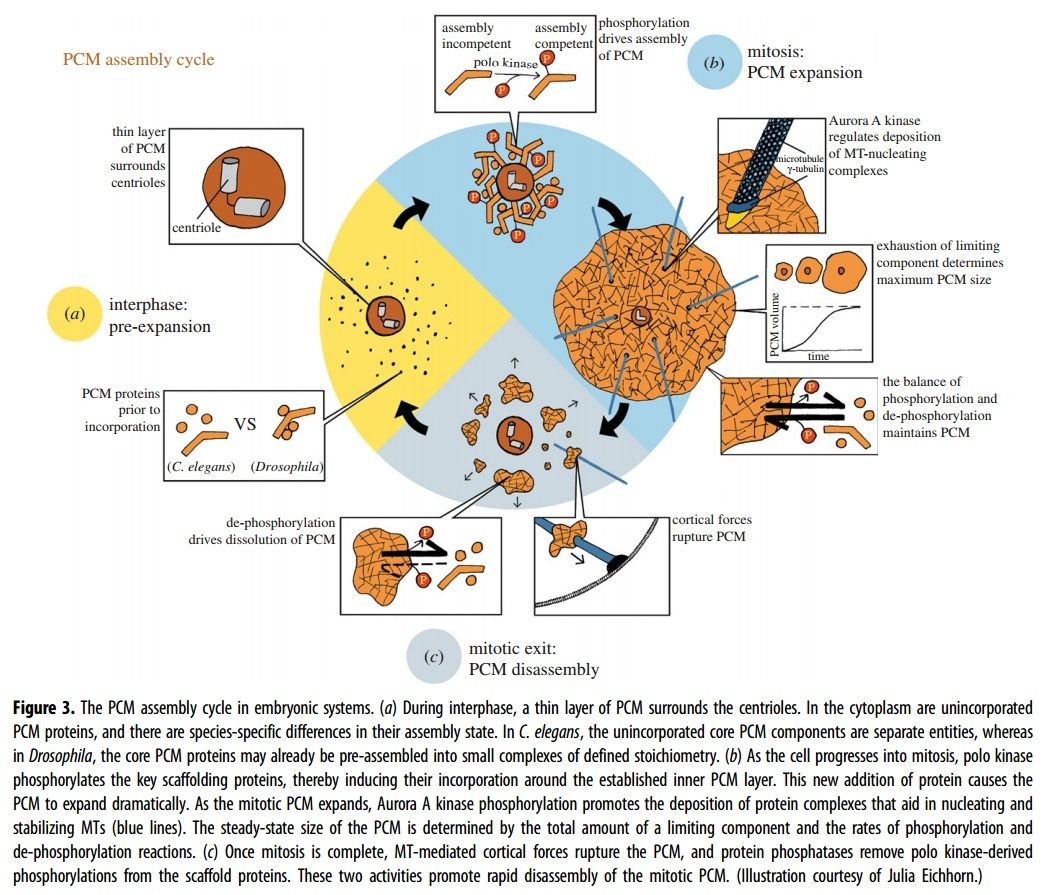
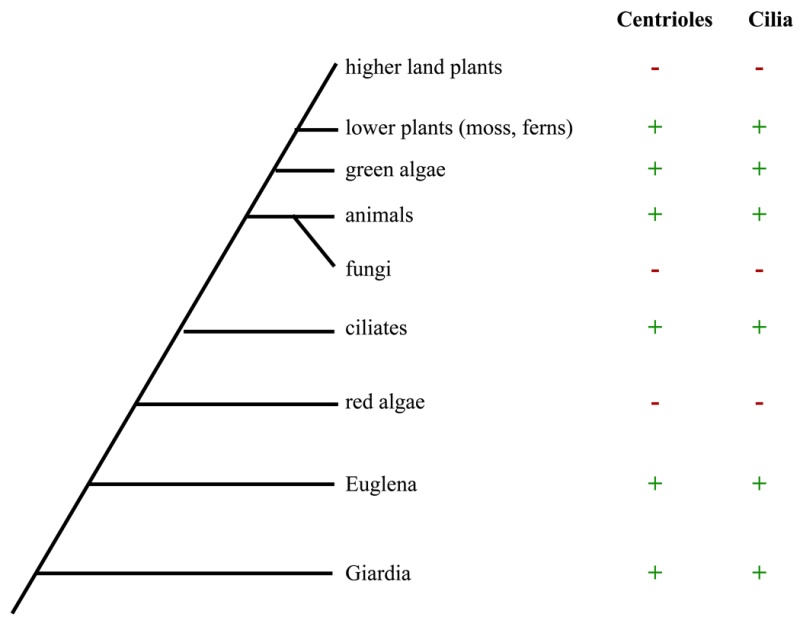
Electric fields generated by synchronized oscillations of microtubules, centrosomes and chromosomes regulate the dynamics of mitosis and meiosis 1
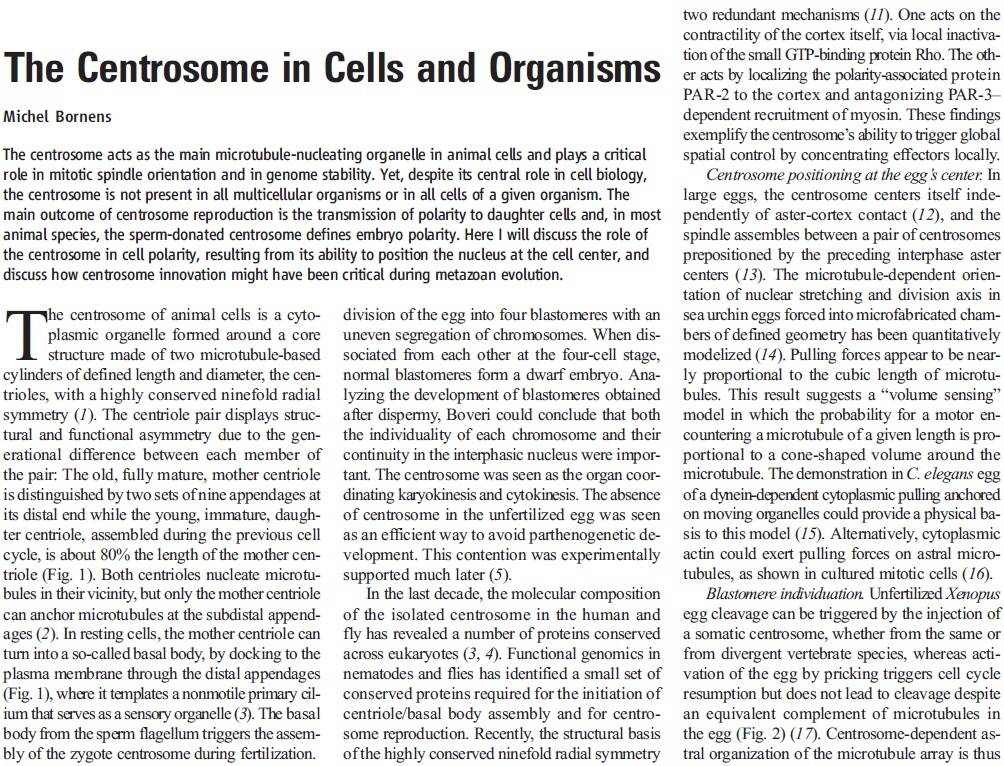
https://reasonandscience.catsboard.com/t2090-centriole-biogenesis-and-the-duplication-cycle-amazing-evidence-of-design#4888Background The choreography of microtubules, centrosomes and chromosomes during mitosis and meiosis is beautifully designed by nature. Finely regulated and synchronized movements of these super-macromolecular complexes against the entropic forces within a dividing cell ensure the fidelity of the genetic material in both daughter cells. Currently, several models exist for the mechanisms of chromosome congression and
spindle body assembly during M phase such as the search and capture model, kinetochoremediated k-fibre formation, kinetochore motors contributing to congression, and the polar wind model. The mechanisms evoked by these models probably overlap, so there is redundancy among them, since mutations in the genes involved have only mild effects on chromosome congression during mitosis [1]. Many open questions remain within these models. In the polar-wind model,
an unknown force (also known as the ejection force) generated by the spindle poles is considered to push the chromosomes to the spindle equator. Laser microsurgery experiments show that chromosome fragments without kinetochores are invariably expelled from the
spindle, and chromosomes without kinetochores can still move from the vicinity of the
spindle pole to the
spindle equator . The ejection force of the
spindle body is dependent on the polymerization of
spindle body microtubules, as depolymerization of astral microtubules by nocodazole or colcemid prevents the expulsion of severed chromosome arms from the
spindle, whereas stabilization of microtubules by taxol drives chromosomes to the periphery of the astral array. In addition, the driving force responsible for the pole-ward flux of
spindle microtubules during metaphase remains uncharacterized. Cellular electric fields have been studied in various cell types, and several studies have reported the existence of dielectrophoretic forces around cells; electromagnetic interactions between cells have also been studied. Cifra et al. proposed that microtubules, which comprise heterodimers polymerized into a helical structure, can generate an electric field under intracellular energy excitation. Inhibition of microtubule polymerization by an external electromagnetic field has been reported by Kirson et al.. Pokorny´ et al. detected four peaks of electric field activity around yeast cells during M phase, which correlated with
spindle body assembly, kinetochore microtubule capture, and mitotic
spindle elongation during anaphase A and B, visualized by fluorescence microscopy. Comparing synchronized and unsynchronized tubulin mutants of yeast cells, Pokorny´ et al. verified that synchronized yeast cells show more electric activity during M phase than non-synchronized yeasts. Direct measurements of electric resonant oscillations in microtubules have been presented at conferences by A. Bandyopadhyay. The technical aspects of direct detection of electric fields within a living cell have been discussed in a recent review.
From our theoretical point of view,
many of the unidentified forces regulating major cellular dynamic events during mitosis are probably electric forces generated by the synchronized oscillation of the electric dipoles within these super-macro organelle structures. The electrical properties of microtubules and centrosomes The electric field of the microtubule is generated by the synchronized oscillation of α and β tubulins. These tubulins form electric dipoles during microtubule polymerization; under intracellular energy excitation, synchronized oscillation of α and β tubulin subunits generates a longitudinal electric field around the microtubule (Figure 1).
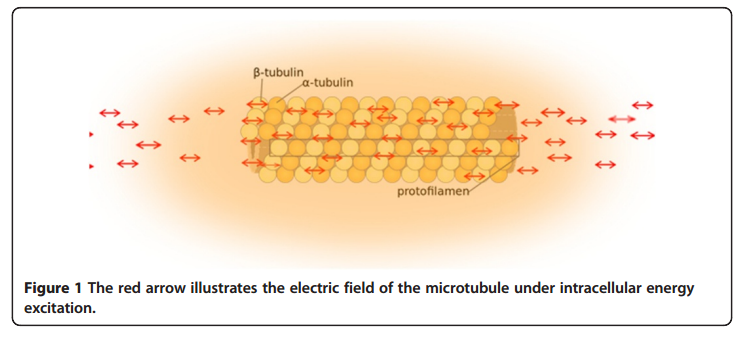
Cifra et al. suggested that the source of the energy excitation could be hydrolysis of guanosine triphosphate (GTP) during the process of dynamic instability of microtubules, and also energy transferred from the movement of motor proteins or released from mitochondria as “wasted” energy from the citric acid cycle. We propose that the overall entropic environment within a living cell could be the source of energy for electric oscillation of microtubules. Cancer cells have different entropic states from normal cells as a result of the Warburg effect, which can cause mitochondrial malfunctions and further lead to alteration of cytoskeleton-based cellular elastoelectrical oscillations [27]. The microtubule networks of cancer cells generate an electromagnetic field with different frequencies. Thus, specific electromagnetic frequencies have been used to diagnose specific cancers [21,28], and tumor-specific modulating electromagnetic fields have been used to treat patients with advanced cancer with positive results [22,29].
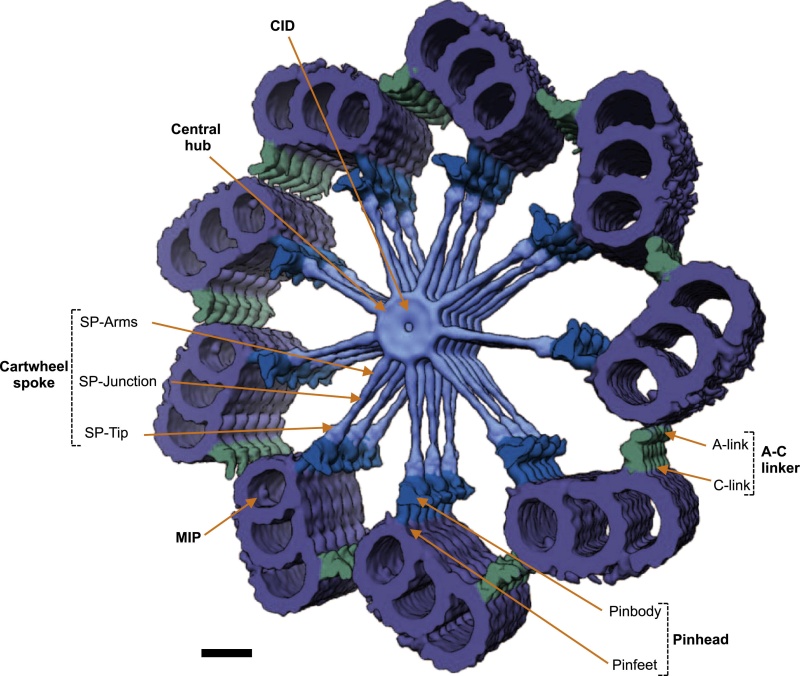
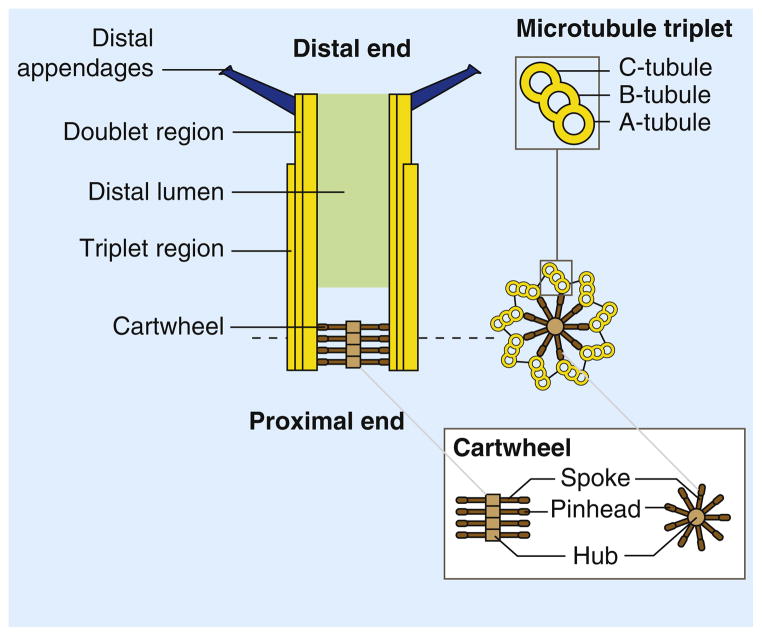
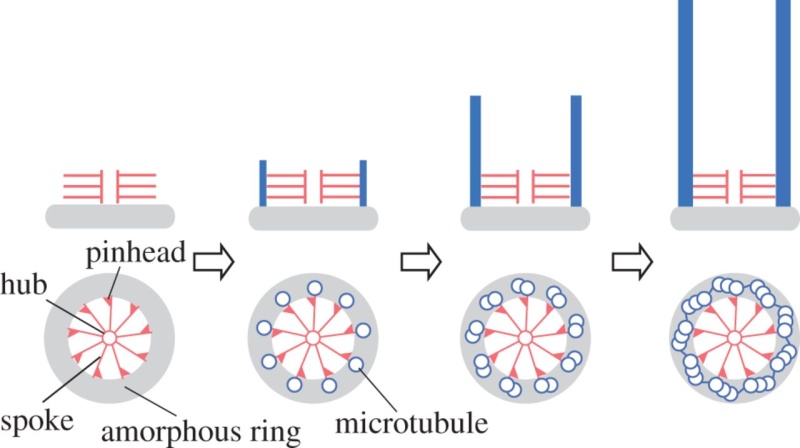
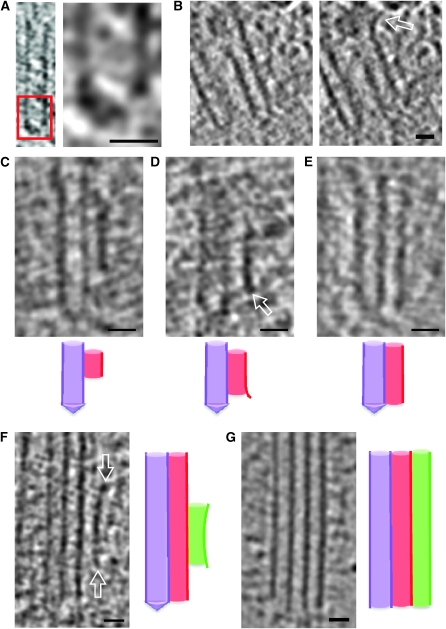
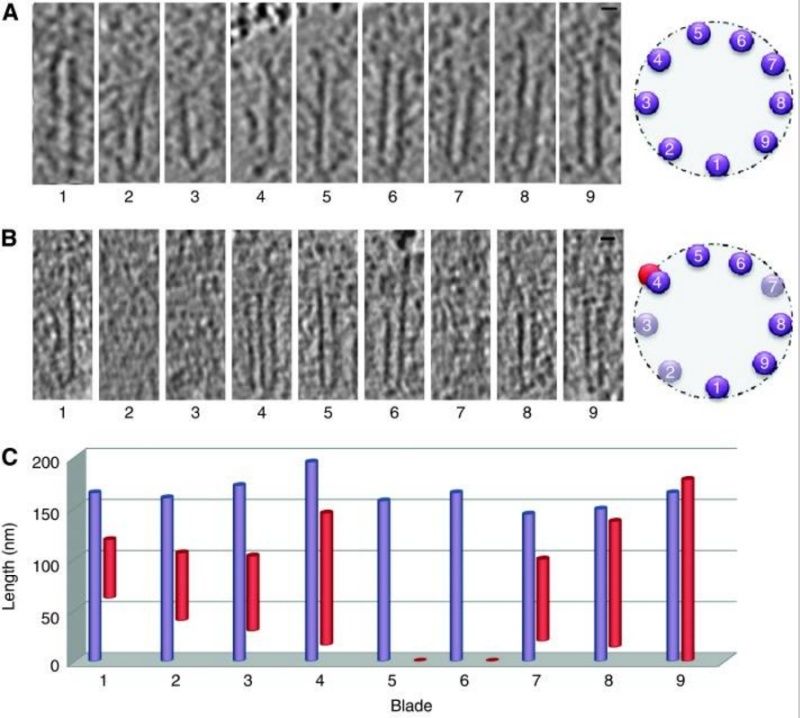
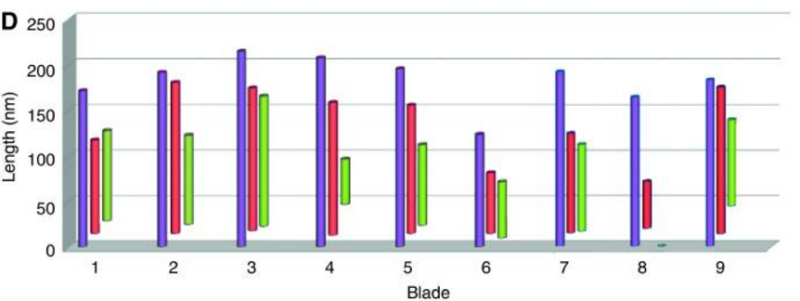
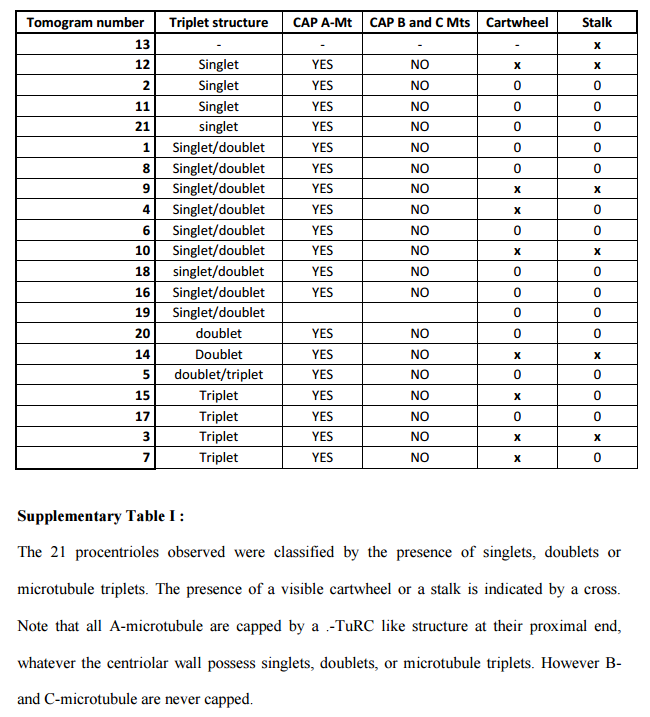
The centriole of the centrosome is composed of α, β and γ tubulins organized differently from the subunits of microtubules; each centrosome comprises two centrioles, which are composed of nine triplets of microtubules. The two centrioles are arranged perpendicularly and surrounded by an amorphous mass of dense material (the pericentriolar material) [30]. As in microtubules, an electric field would be generated by synchronized oscillation between the α and β tubulins within the microtubule triplet of the centrioles (Figure 2).
 Electric fields in centrosome separation and bipolar spindle body assembly
Electric fields in centrosome separation and bipolar spindle body assembly Mechanisms of centrosome separation and bipolar
spindle body assembly have been discussed in a recent review [31]. The process is still incompletely understood. Plus end-directed motor proteins such as kinesin 5 and minus end-directed motor proteins such as dynein are known to play dominant roles in centrosome separation and
spindle assembly. However, centrosomal microtubules and microtubules of the nuclear envelope (NE) and cellular cortex need to move into close proximity for motor proteins to attach to both so they can generate the pulling forces. The current models assume a randomized mode of microtubule interaction, which is quite inefficient. For example, at a certain point a centrosome would have to stop moving until certain microtubules had grown sufficiently for appropriate bridging by motor proteins, particularly during prophase, when the centrosomes do not have many associated microtubules. When the electric fields of microtubules and centrosomes are considered, these structures are mutually attractive. Thus, centrosome movement along the microtubule networks of the cellular cortex and NE is more efficient. We can also envision a more autonomous mode of microtubule lattice formation within the cellular cortex and NE.
Centrosome Functions as a Molecular Dynamo in the Living Cell 2
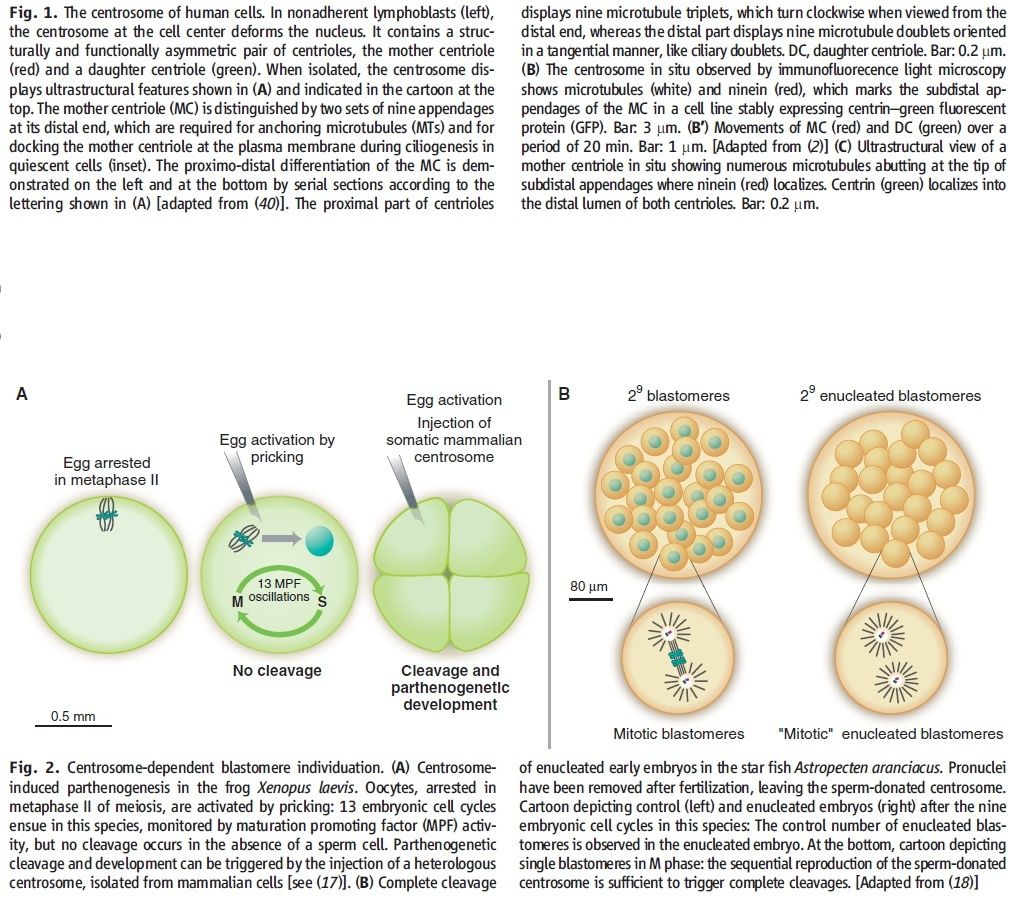
Recent development in the field of quantum biology highlights that the intracellular electromagnetic field (EMF) of microtubules plays an important role in many fundamental cellular processes such as mitosis. Here I propose an intriguing hypothesis that centrosome functions as molecular dynamo to generate electric flow over the microtubules, leading to the electric excitation of microtubule EMF that is required for spindle body microtubule self-assembly. With the help of motors proteins within the centrosome, centrosome transforms the energy from ATP into intracellular EMF in the living cell that shapes the functions of microtubules. There will be a general impact for the cell biology field to understand the mechanistic function of centrosome for the first time in correlation with its structural features. This hypothesis can be tested with technics such as super resolution live cell microscope.1. Introduction Centrosome was first discovered by Theodor Boveri in the 1880’s [1], it is the key organelle that is responsible for mitosis and meiosis in metazoan lineage of eukaryotic cells [2]. In animal cells, centrosome regulates the nucleation and spatial organization of microtubules, functioning as the primary microtubule-organizing center (MTOC) [3].
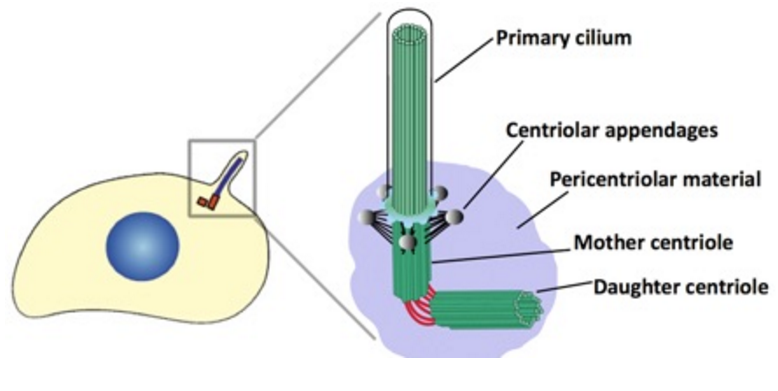
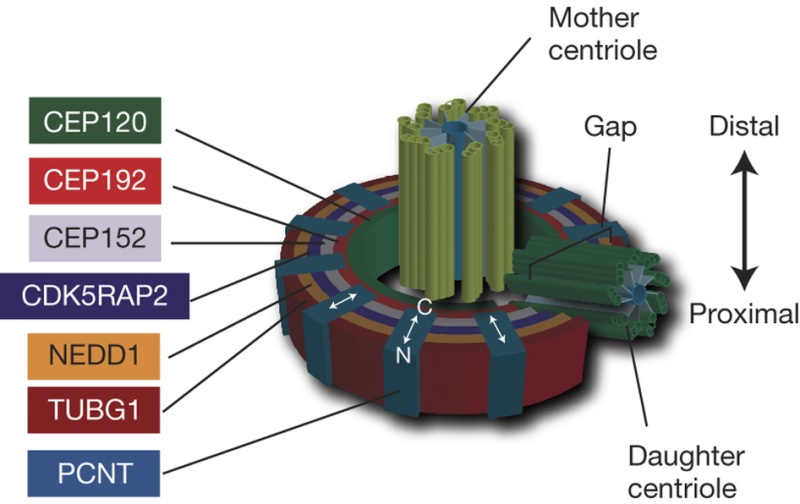
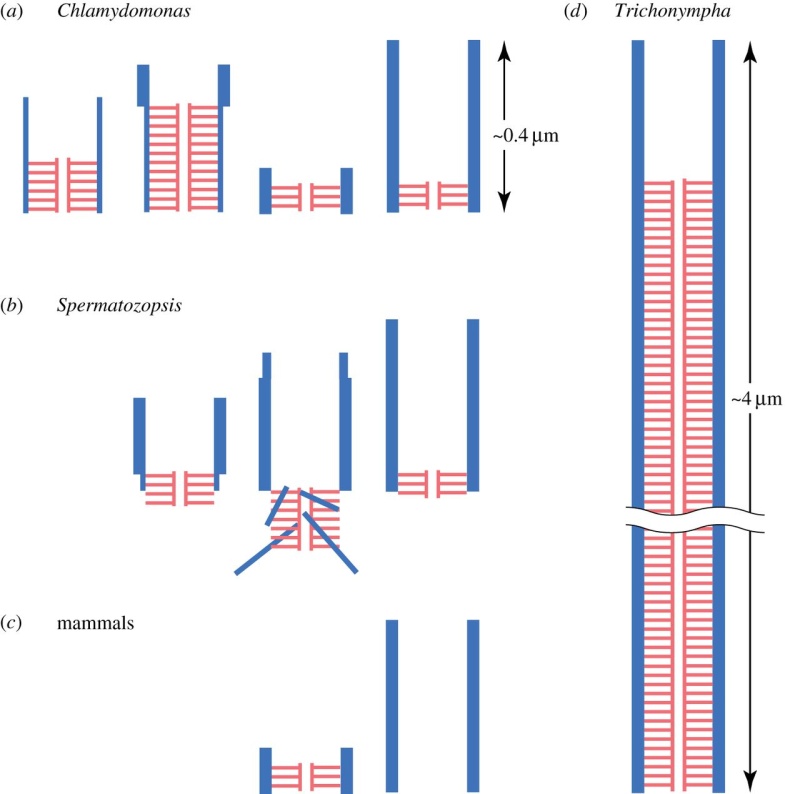
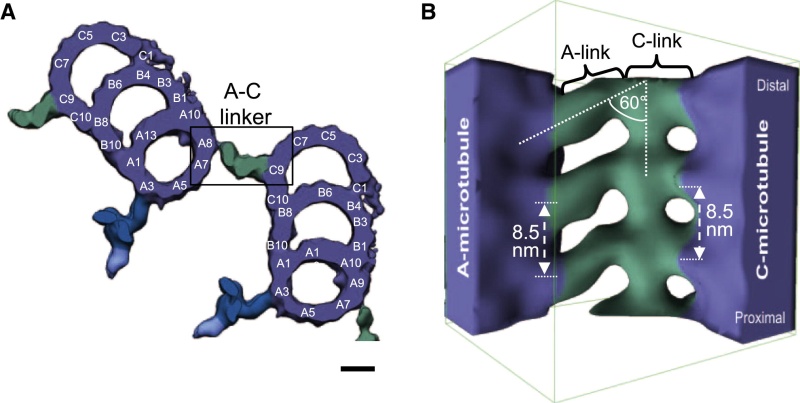
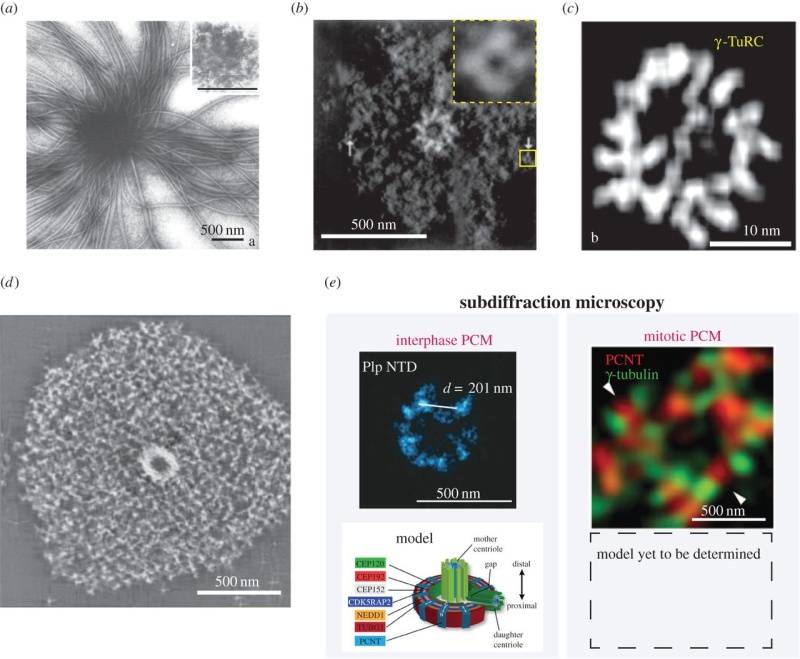
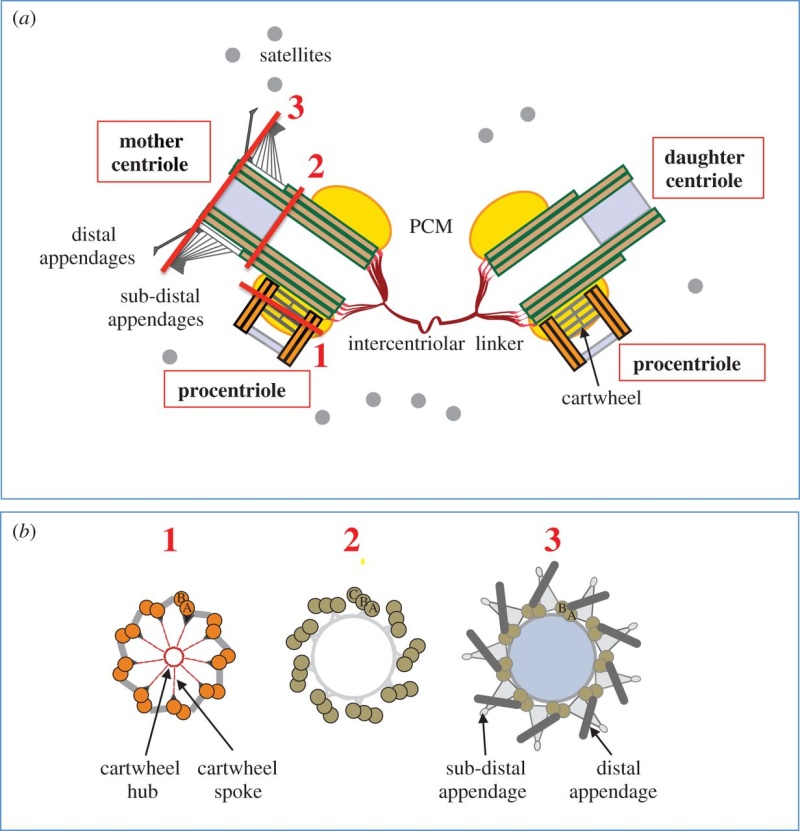
The centrosome is comprised of two centrioles that are surrounded by pericentriolar material (PCM). The two centrioles are perpendicularly arranged, one centriole has additional appendages at the end farthest from the other centriole (distal) and is called the mother or maternal centriole, the subdistal appendages of the maternal centriole also act like microtubule-anchoring sites [4] [5]. Pelletier et al. reported that subdiffraction imaging of centrosomes revealed pericentriolar material which had higher-order organizational features. Centrosome components adopt a toroidal pattern with progressively larger, overlapping diameters around the proximal end of the mother centriole in interphase cells. On one side, the toroid is slightly opened (gap) in the area where the daughter centriole is positioned [6], this higher order structural feature of centrosome may help the mother and daughter centriole to form an orthogonal configuration. In most of cases, each centriole is composed with 9 MT triplets and is ~0.5 μm in length and 0.2 μm in diameter [7] [8]. Recent studies in the field of quantum biology point to the possibility that electric magnetic interactions may involve in many fundamental cellular processes [9] [10]. In particular,
the electromagnetic property of microtubule has been reported with both computation modelling and experimental evidences, Cifra et al. used computation model to simulate the electric pulse moving along microtubules, Bandyopadhyay et al. reported that nano sized electric pump was required for the self-assembly of microtubules in live cell simulator[11]-[13]. Medical treatment of cancer with cancer cell specific interfering EMF has been developed to disrupt the mitotic
spindle microtubules of cancer cells [14]. Centriole produces an electromagnetic field apparently due to the longitudinal oscillation of its microtubules (MTs). Centrosome clustering is a hallmark of cancer cells. A cluster of centrioles is therefore presumed to produce an enhanced electromagnetic field. It is possible to target cancer cells using nano particles based on the enhanced electromagnetic property of cancer cells [15]. However, some important questions remain unanswered, what is the energy source of the intracellular electric field and what is the molecular mechanism that leads to the excitation of intracellular electric field? ATP is the most common cellular energy source.
To transform the chemical energy in ATP into electric magnetic field within the living cell, cell needs to have a molecular dynamo to transform the mechanistic movement of protein complexes to directional movements of intracellular electrons, leading to the electric excitation of the spindle body microtubules as well as the M phase chromosomes, which is essential for mitosis [9].
2. Hypothesis Here I present a novel hypothesis that centrosome functions as a molecular dynamo in the living cell to generate electric current from the cytosol electrolyte to the spindle body and M phase chromosome, leads to the electric excitation of the spindle body and chromosome during mitosis. Based upon the structure of the centrosome, there is one microtubule in the center, and 9 microtubule triplicate outside, connected by motor proteins such as dynein and kinesin [3]
https://vimeo.com/58347006
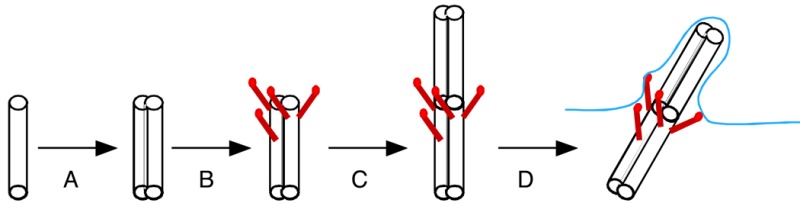
The mechanistic movement of these motor proteins will trigger the rotation of the microtubules triplets forming the barrel structure of the centriole to rotate around the center microtubule. The rotation and electric oscillation of each centriole will generate a dynamic electromagnetic field that mimic the physical structure of the centriole, and the orthogonal arrangement of centrioles of each centrosome will result in the microtubules of the barrel structure of each centriole to cut the electromagnetic field generated by the other centriole when rotating (Figure 1).
 Such a natural design makes centrosome to function as a molecular dynamo, generating directional electron flow through the dipolar structure of each individual microtubule in the centrosome, transforming the energy from ATP to electric current.
Such a natural design makes centrosome to function as a molecular dynamo, generating directional electron flow through the dipolar structure of each individual microtubule in the centrosome, transforming the energy from ATP to electric current. During mitosis, centrosome is known to locate at the microtubule organizing center (MTOC), and only the mother centriole contains the sub-distal appendages that connect with
spindle body microtubules, which allow the electrons to move from centrosome to the
spindle body microtubules. During mitosis chromosomes are connected with
spindle body microtubules with K-fibres, which are microtubule bundles that join kinetochores to the
spindle poles.
Pericentriolar material (PCM) is composed primarily of hyaluronic acid (HA) and has a similar negative charge density as DNA [16].
The electrons flow over the spindle body leads to the electric excitation of the spindle body and chromosome, generating an enhanced intracellular electromagnetic field during mitosis, which is consistent with the observation of yeast cell at M phase [17] [18].
Bandyopadhyay et al. reported nano-sized electromagnetic pumping is required for microtubule self-assembly [13], the electric excitation of
spindle body microtubules is required for the self-assembly and growth of the
spindle body microtubules during M phase. The basal body of cilium is a centrosome like cellular organelle [19], similar molecular mechanism is applied for the basal body centriole to generate electric flow over the cilium microtubules, forming the nano electromagnetic field that is required for the self-assembly of cilium microtubule. Collectively,
my hypothesis proposes centrosome is at the center of the electric network that continuously drawn the cellular chemical energy from ATP to feed the intracellular electromagnetic field. On Centrioles, Microtubules, and Cellular Electromagnetism 3
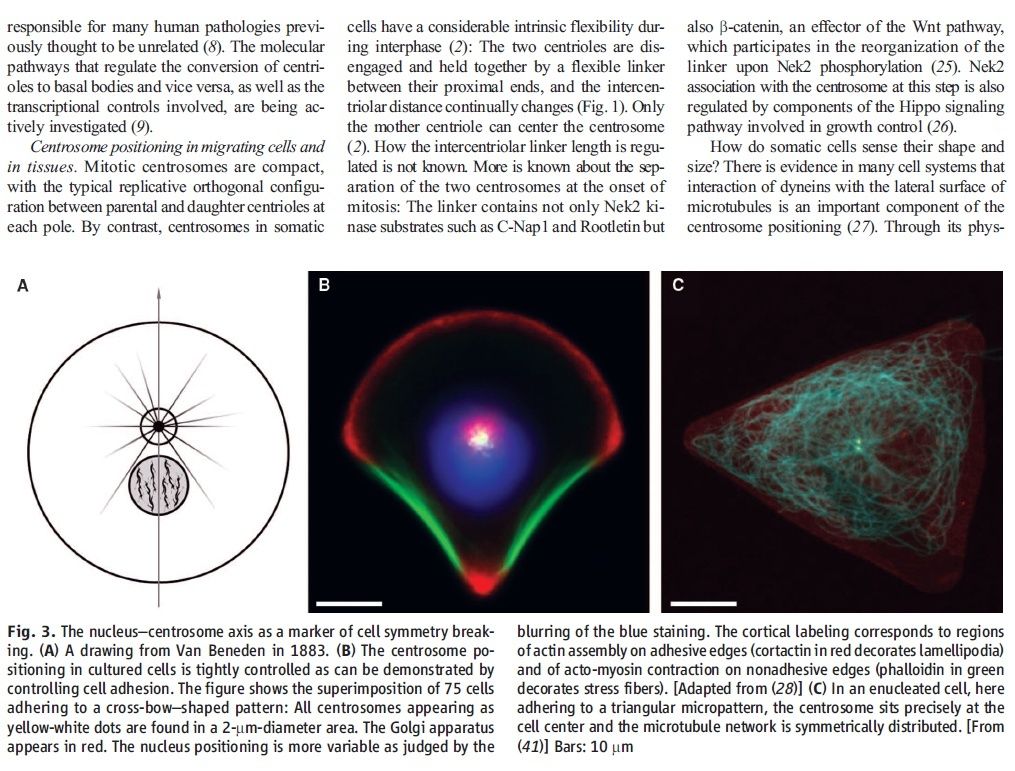
1 Introduction Centrioles are tiny organelles lying adjacent to the nucleus of human and animal (eukaryotic) cells (see Fig. 1). Although their existence has been known for now over 100 years [1–7], centrioles have only recently been studied in any significant detail. This recent interest has been stimulated by:
(1) dramatic advances in imaging technologies resulting in a better understanding of centriole geometry;
(2) improved understanding of the important role of centrioles in cell mechanics—particularly in mitosis; and
(3) the increasing belief that centriole malfunction occurs in virtually all solid tumor cancers. It is now known that centrioles have the following properties:
(1) They have precise geometry, occurring in pairs as small annular cylinders (approximately 400 nm of long and 200 nm in diameter), with cylinder axes perpendicular to one another.
(2) As with deoxyribonucleic acid (DNA) centrioles are selfduplicating and they duplicate at approximately the same time as DNA duplicates.
(3) Centrioles are the only organelles without a membrane cover.
(4) Centrioles are active participants in the cell division (mitosis) process.
(5) During the S-phase of normal cell mitosis, each centriole duplicates once and only once. In abnormal mitosis, centrioles may duplicate multiple times and with disrupted geometry.
 Figure 2
Figure 2 provides a drawing of a centriole pair. Interestingly, approximately 45 years ago, Dr. Paul Schafer, while working at the Veterans Administration Hospital in Washington, D.C., discovered and reported that esophageal cancer cells have distorted centriole geometries. He suggested that cancers develop due to abnormalities in the electromagnetic fields surrounding the centrioles. Unfortunately, as with Boveri, Schafer’s work has received relatively little attention until recently. The following paragraphs summarize recent and state-of-the-art findings about centrioles, their inner workings, and their overall effect upon cell electropolarity. How these findings could be an aid in cell research is also discussed—particularly in solving the problem of in vivo insertion of therapeutic nanoparticles .
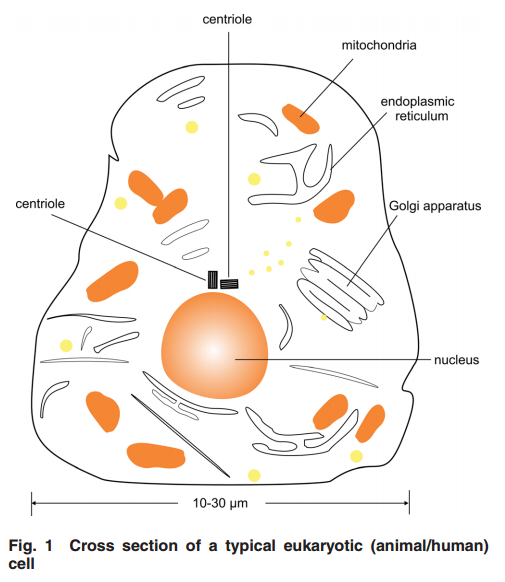 2 Centriole GeometryFigure 1
2 Centriole GeometryFigure 1 provides a typical drawing of a cell interior. The centrioles are seen as the small perpendicular organelles adjacent to the nucleus. In Fig. 2, we have a closer view of the centriole pair. long as the mother. The daughter centriole is attached to the base, or proximal, end of the mother centriole. In this configuration, the axis of the daughter intersects the axis of the mother, with the axes of the two centrioles being perpendicular. Surrounding the centriole pair, at the adjoining base, is an amorphous cloud of numerous proteins known as the MT organizing center (MTOC) . The centrioles together with the MTOC are known as the centrosome. The centrosome, being roughly spherical, has a diameter of approximately 4 lm. The MTOC is believed to be “electron dense” . In this regard, with the MTOC at the intersecting bases of the centriole
pair, the proximal or base end of the centriole is given negative polarity or potential. The distal ends are thus positive.
3 Centriole Duplication and Mitosis Centrioles are the principal drivers of cell division and duplication (mitosis). Mitosis is typically described as occurring in four phases: In the first phase (the “prophase”), each centriole becomes a new “mother” centriole supporting the creation of a new daughter centriole on its side and at its proximal end. As these daughter centrioles develop and grow, the original centriole pair becomes two pair, with the original mother centriole having a new daughter, and the original daughter having a new daughter of its own .
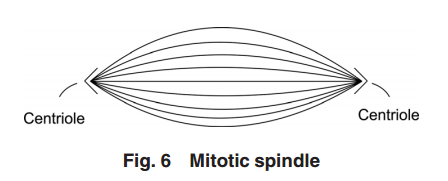
 Figure 5
Figure 5 illustrates the two-pair configuration.
During the time that the centrioles are duplicating, the nucleus membrane begins to soften, break down, and the DNA condenses in preparation for division. Finally, the connection between the centriole pairs stretches and the pairs move apart. Interestingly, the original mother centriole with its new daughter remains somewhat stationary while the less mature daughter-new daughter centriole pair moves away, around the nucleus to the opposite side. During the movement, this immature centriole pair becomes mature. Even though the centriole pairs are separating, they remain connected by MTs about the collapsing nucleus—almost in the shape of a football. Taken as a group, these MTs are known as the “mitotic
spindle” (see Fig. 6).
The remaining three phases (metaphase, anaphase, and teleophase) complete the cell separation process with the metaphase being the creation of a symmetric structure between the two pairs of centrioles; the anaphase being the separation in the midplane; and finally, the teleophase being the formation of new nuclear membranes about the separated parts. The two centriole pairs each align themselves adjacent to the separated nuclei respectively. The separated nuclii then move further apart, each taking with it approximately half of the original cell organelles and half of the remaining cell material (the cytoplasm). Thus the original cell becomes two cells.
4 Cell Electropolarity via Longitudinal MT Vibration Numerous studies have shown that the MTs within a centriole generate an electromagnetic field about the centriole. This field is believed to occur due to longitudinal (axial) vibration of the MT filaments. Consider again Fig. 3 showing again a sketch of an MT, (1 of 27), parallel to the axis of the centriole barrel. The proximal (or base) end of the MT is immersed in the electron dense material of the MTOC. With this proximal end being negative, and thus the distal end being positive, this positive distal end is immersed in the cloud of material of the centrosome, or pericentriolar material. As noted earlier, the MT filaments are composed of alternate a-tubulin and b-tubulin protein pairs (“dimers”).
A single dimer has a high electric charge difference (polarity) along its axis. Then with the filament dimers being arranged in a series of dimers along the filament length, there is a voltage change from the proximal (negative) to the distal (positive) end of the filament. It is the tubulin that is the source of this potential difference. An MT filament surface is relatively smooth, allowing for relatively free longitudinal, oscillatory movement (vibration) of the filament. Then
taken together the longitudinal, or axial, vibration of the 13 filaments of an MT and then the 27 MTs making up the centriole barrel produce the electromagnetic field surrounding the centriole . Interestingly, this field is also found to be ferromagnetic. Also of interest, the fundamental vibration frequency of an MT filament is approximately 465 MHz, although this frequency is continually changing due to the ongoing length changing of the filaments.
The electropolarity of the centrioles enables them to exert forces at a distance—that is, forces without physical contact. Also, it is this high electropolarity of the centriole, lying next to the nucleus, which produces the overall electropolarity of the cell. The transepithelial potentials, that is, the potential difference across a cell membrane cover may range from a few millivolts to tens of millivolts. Finally, as one would expect, the peak of cell electropolarity occurs during mitosis, when there are four, instead of two, centrioles. Correspondingly, the MTs then orient the mitotic
spindle along the axis of cell polarity.
Findings (1)
Electromagnetics play an important role in cell functioning and especially in cell duplication and division (mitosis).
(2) Cell electropolarity arises from the centrioles via the MTs of the centriole blades.
(3) The MTs obtain their polarity via a, b dimers arranged in series along the filaments of the MT wall.
(4) Overall cell electropolarity is increased during mitosis due to the presence of two pairs of centrioles.
(5) Overall cell electropolarity is even greater in cancer cells due to the presence of supernumerary centrioles and centriole clustering. Indeed, supernumerary centrioles tend to cluster together—apparently due to electromagnetic attraction [70].
(6) Clinically, it is found that breast cancer tissue has sufficiently large electrical polarity that it can be detected relative to surrounding tissue at the breast surface, and the aggressiveness of breast cancer is proportional to the degree of centrosome amplification.
(7) In general, there is a small but steady extracellular voltage gradient between cancer tissue and neighboring normal tissue .
(8 Centrosomal amplification is regarded as a “hallmark” of cancer cells —that is, all major cancer cells have centrosomal amplification .
(9)
In an early stage of mitosis, electromagnetic forces send the less mature pair of centrioles around the nucleus to the other side. (10) The electropolarity associated with centriole clustering may be useful for tumor identification and treatment using charged nanoparticles
Open Questions and Needed Data (1)
Why is centriole geometry so precise and why does it have the particular form that it has? (2) During centriole duplication, why does the daughter centriole arise and develop on the side and at the base of the mother centriole?
(3) What is the magnitude of the electropolarity change along a centriole length, across a centriole pair, and across a duplicating pair of centrioles?
(4) What is the magnitude of the electropolarity change across a centriole cluster of a cancer cell?
(5) Is the electropolarity of a centriole cluster sufficiently strong to attract charged nanoparticles?
(6) What is the range of frequencies of the longitudinal vibrations of MTs with variable lengths?
(7) What is the three-dimensional map of potential of a normal cell? Additional experimentation will be required to answer these questions. To this end, it now appears that the best approach is to use current nanotechnology including the use of atomic force microscopy with magnetic sensitive probing tips. This is a subject of ongoing local research.
1)http://download.springer.com/static/pdf/238/art%253A10.1186%252F1742-4682-9-26.pdf?originUrl=http%3A%2F%2Flink.springer.com%2Farticle%2F10.1186%2F1742-4682-9-26&token2=exp=1464118570~acl=%2Fstatic%2Fpdf%2F238%2Fart%25253A10.1186%25252F1742-4682-9-26.pdf%3ForiginUrl%3Dhttp%253A%252F%252Flink.springer.com%252Farticle%252F10.1186%252F1742-4682-9-26*~hmac=84a38aee1ee3fcaf89567ff0515b79c6436aad91a7cf73d89a1a084def147165
2) http://file.scirp.org/pdf/ABB_2015072015020110.pdf
3) file:///D:/Downloads/nano_005_03_031003.pdf