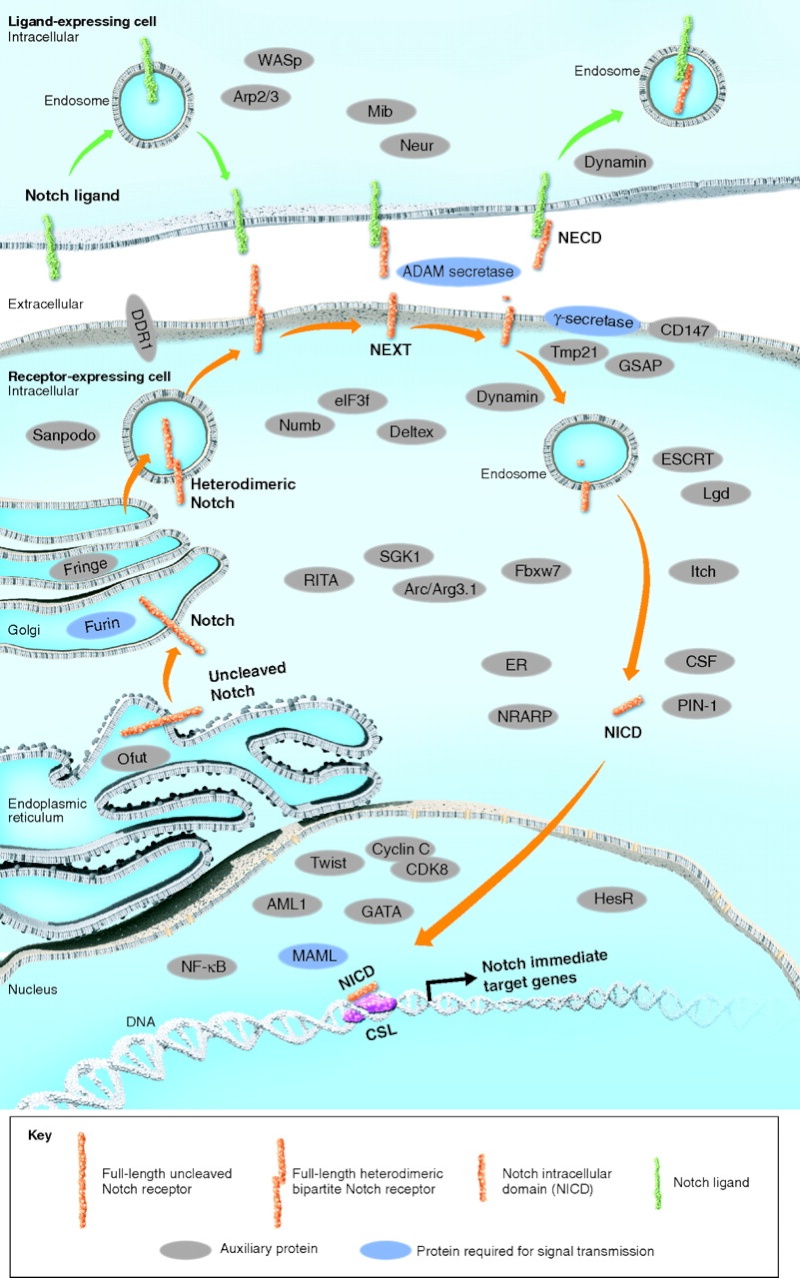
http://reasonandscience.heavenforum.org/t2181-cell-communication-and-signaling-evidence-of-design
Most signal-relay stations we know about were intelligently designed. Signal without recognition is meaningless. Communication implies a signalling convention (a “coming together” or agreement in advance) that a given signal means or represents something: e.g., that S-O-S means “Send Help!” The transmitter and receiver can be made of non-sentient materials, but the functional purpose of the system always comes from a mind. The mind uses the material substances to perform an algorithm that is not itself a product of the materials or the blind forces acting on them. Signal sequences may be composed of mindless matter, but they are marks of a mind behind the intelligent design.
How does interaction of the primary stimulus with the outside of the plasma membrane activate a process inside the cell without direct intracellular contact? If primary stimuli are able to trigger chemical changes within
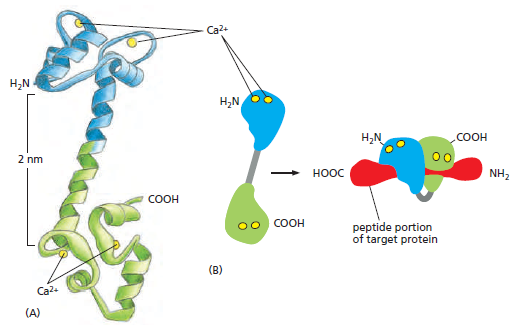
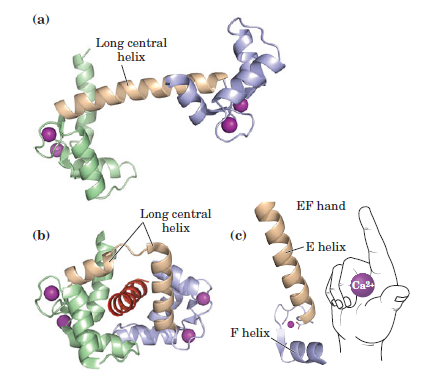
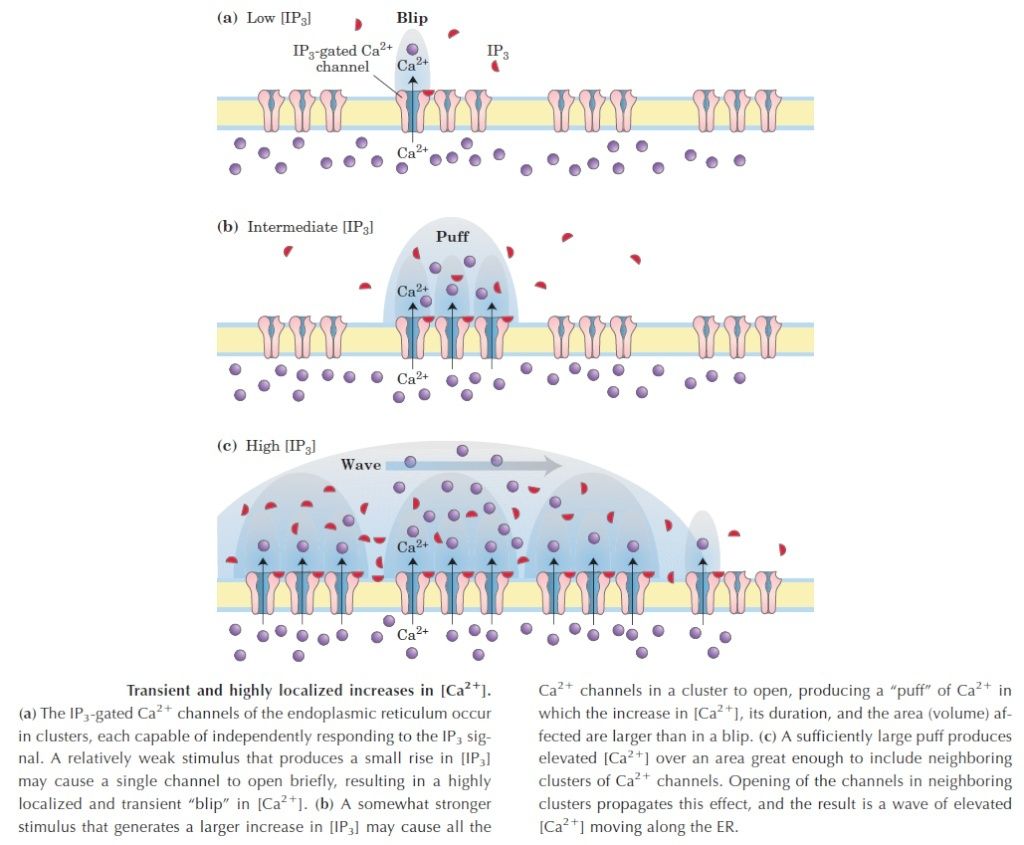
the cell, there must be a messenger within the cell, which transmits the information that the cell has been activated to the appropriate molecular machinery inside the cell that causes the cellular event. Ca2+ was the first such intracellular messenger to be identified.
The term ‘second messenger’ was introduced by Earl Sutherland (1915–1974) and colleagues to describe cyclic AMP, responsible for initiating the pathway leading to release of glucose from glycogen in the liver, which is activated by hormones such as adrenaline (epinephrine) and glucagon, binding receptors on the hepatocyte cell surface. Cyclic AMP also is the second messenger initiating triglyceride breakdown in adipose tissue. Adrenaline also causes a rise in cyclic AMP in skeletal muscle, when we run. This also promotes the breakdown of glycogen to glucose, to help make more ATP. But cyclic AMP is not the second messenger responsible for triggering contraction. This is Ca2+ through binding to its intracellular target – troponin C. Yet, in many non-excitable cells Ca2+ is really a ‘third’ messenger.
When things change, cells respond. Every cell, from the humble bacterium to the most sophisticated eukaryotic cell, monitors its intracellular and extracellular environment, processes the information it gathers, and responds accordingly. Unicellular organisms, for example, modify their behavior in response to changes in environmental nutrients or toxins. The cells of multicellular organisms detect and respond to countless internal and extracellular signals that control their growth, division, and differentiation during development, as well as their behavior in adult tissues. At the heart of all these communication systems are regulatory proteins that produce chemical signals, which are sent from one place to another in the body or within a cell, usually being processed along the way and integrated with other signals to provide clear and effective communication. The study of cell signaling has traditionally focused on the mechanisms by which eukaryotic cells communicate with each other using extracellular signal molecules such as hormones and growth factors. In this chapter, we describe the features of some of these cell–cell communication systems, and we use them to illustrate the general principles by which any regulatory system, inside or outside the cell, is able to generate, process, and respond to signals. Our main focus is on animal cells, but we end by considering the special features of cell signaling in plants.
PRINCIPLES OF CELL SIGNALING
Long before multicellular creatures roamed the Earth, unicellular organisms had developed mechanisms for responding to physical and chemical changes in their environment. These almost certainly included mechanisms for responding to the presence of other cells. Evidence comes from studies of present-day unicellular organisms such as bacteria and yeasts. Although these cells lead mostly independent lives, they can communicate and influence one another’s behavior. Many bacteria, for example, respond to chemical signals that are secreted by their neighbors and accumulate at higher population density. This process, called quorum sensing, allows bacteria to coordinate their behavior, including their motility, antibiotic production, spore formation, and sexual conjugation. Similarly, yeast cells communicate with one another in preparation for mating. The budding yeast Saccharomyces cerevisiae provides a well-studied example: when a haploid individual is ready to mate, it secretes a peptide mating factor that signals cells of the opposite mating type to stop proliferating and prepare to mate. The subsequent fusion of two haploid cells of opposite mating type produces a diploid zygote. Intercellular communication achieved an astonishing level of complexity during the arise of multicellular organisms. These organisms are tight-knit societies of cells, in which the well-being of the individual cell is often set aside for the benefit of the organism as a whole. Complex systems of intercellular communication have emerged to allow the collaboration and coordination of different tissues
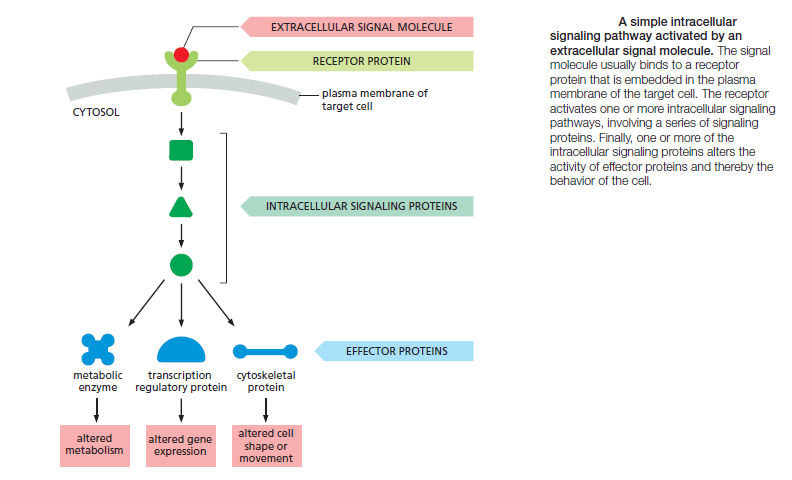
and cell types. Bewildering arrays of signaling systems govern every conceivable feature of cell and tissue function during development and in the adult. Communication between cells in multicellular organisms is mediated mainly by extracellular signal molecules. Some of these operate over long distances,signaling to cells far away; others signal only to immediate neighbors. Most cells in multicellular organisms both emit and receive signals. Reception of the signals depends on receptor proteins, usually (but not always) at the cell surface, which bind the signal molecule. The binding activates the receptor, which in turn activates one or more intracellular signaling pathways or systems. These systems depend on intracellular signaling proteins, which process the signal inside the receiving cell and distribute it to the appropriate intracellular targets. The targets that lie at the end of signaling pathways are generally called effector proteins, which are altered in some way by the incoming signal and implement the appropriate change in cell behavior. Depending on the signal and the type and state of the receiving cell, these effectors can be transcription regulators, ion channels, components of a metabolic pathway, or parts of the cytoskeleton
The fundamental features of cell signaling have been conserved throughout the evolution of the eukaryotes. In budding yeast, for example, the response to mating factor depends on cell-surface receptor proteins, intracellular GTP-binding proteins, and protein kinases that are clearly related to functionally similar proteins in animal cells. Through gene duplication and divergence, however, the signaling systems in animals have become much more elaborate than those in yeasts; the human genome, for example, contains more than 1500 genes that encode receptor proteins, and the number of different receptor proteins is further increased by alternative RNA splicing and post-translational modifications.
Extracellular Signals Can Act Over Short or Long Distances
Many extracellular signal molecules remain bound to the surface of the signaling cell and influence only cells that contact it
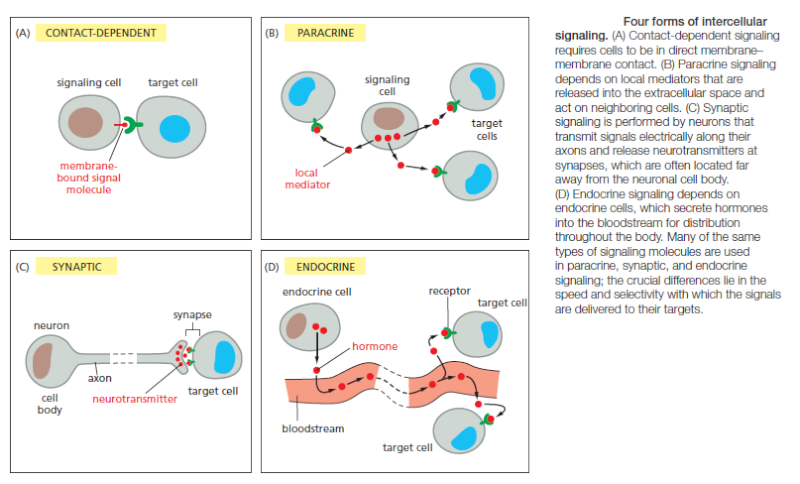
Such contactdependent signaling is especially important during development and in immune responses. Contact-dependent signaling during development can sometimes operate over relatively large distances if the communicating cells extend long thin processes to make contact with one another. In most cases, however, signaling cells secrete signal molecules into the extracellular fluid. Often, the secreted molecules are local mediators, which act only on cells in the local environment of the signaling cell. This is called paracrine signaling (Figure B). Usually, the signaling and target cells in paracrine signaling are of different cell types, but cells may also produce signals that they themselves respond to: this is referred to as autocrine signaling. Cancer cells, for example, often produce extracellular signals that stimulate their own survival and proliferation.
Large multicellular organisms like us need long-range signaling mechanisms to coordinate the behavior of cells in remote parts of the body. Thus, they have cell types specialized for intercellular communication over large distances. The most sophisticated of these are nerve cells, or neurons, which typically extend long, branching processes (axons) that enable them to contact target cells far away, where the processes terminate at the specialized sites of signal transmission known as chemical synapses. When a neuron is activated by stimuli from other nerve cells, it sends electrical impulses (action potentials) rapidly along its axon; when the impulse reaches the synapse at the end of the axon, it triggers secretion of a chemical signal that acts as a neurotransmitter. The tightly organized structure of the synapse ensures that the neurotransmitter is delivered specifically to receptors on the postsynaptic target cell (Figure C). A quite different strategy for signaling over long distances makes use of endocrine cells, which secrete their signal molecules, called hormones, into the bloodstream. The blood carries the molecules far and wide, allowing them to act on target cells that may lie anywhere in the body (Figure D).
Extracellular Signal Molecules Bind to Specific Receptors
Cells in multicellular animals communicate by means of hundreds of kinds of extracellular signal molecules. These include proteins, small peptides, amino acids, nucleotides, steroids, retinoids, fatty acid derivatives, and even dissolved gases such as nitric oxide and carbon monoxide. Most of these signal molecules are released into the extracellular space by exocytosis from the signaling cell. Some, however, are emitted by diffusion through the signaling cell’s plasma membrane, whereas others are displayed on the external surface of the cell and remain attached to it, providing a signal to other cells only when they make contact. Transmembrane signal proteins may operate in this way, or their extracellular domains may be released from the signaling cell’s surface by proteolytic cleavage and then act at a distance. Regardless of the nature of the signal, the target cell responds by means of a receptor, which binds the signal molecule and then initiates a response in the target cell. The binding site of the receptor has a complex structure that is shaped to recognize the signal molecule with high specificity, helping to ensure that the receptor responds only to the appropriate signal and not to the many other signaling molecules surrounding the cell.
In most cases, receptors are transmembrane proteins on the target-cell surface. When these proteins bind an extracellular signal molecule (a ligand), they become activated and generate various intracellular signals that alter the behavior of the cell. In other cases, the receptor proteins are inside the target cell, and the signal molecule has to enter the cell to bind to them: this requires that the signal molecule be sufficiently small and hydrophobic to diffuse across the target cell’s plasma membrane
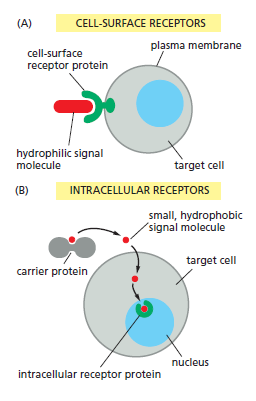
The binding of extracellular signal molecules to either cell-surface or intracellular receptors.
(A) Most signal molecules are hydrophilic and are therefore unable to cross the target cell’s plasma membrane directly; instead, they bind to cell-surface receptors, which in turn generate signals inside the target cell.
(B) Some small signal molecules, by contrast, diffuse across the plasma membrane and bind to receptor proteins inside the target cell—either in the cytosol or in the nucleus (as shown here). Many of these small signal molecules are hydrophobic and poorly soluble in aqueous solutions; they are therefore transported in the bloodstream and other extracellular fluids bound to carrier proteins, from which they dissociate before entering the target cell.
Each Cell Is Programmed to Respond to Specific Combinations of Extracellular Signals
A typical cell in a multicellular organism is exposed to hundreds of different signal molecules in its environment. The molecules can be soluble, bound to the extracellular matrix, or bound to the surface of a neighboring cell; they can be stimulatory or inhibitory; they can act in innumerable different combinations; and they can influence almost any aspect of cell behavior. The cell responds to this blizzard of signals selectively, in large part by expressing only those receptors and intracellular signaling systems that respond to the signals that are required for the regulation of that cell. Most cells respond to many different signals in the environment, and some of these signals may influence the response to other signals. One of the key challenges in cell biology is to determine how a cell integrates all of this signaling information in order to make decisions—to divide, to move, to differentiate, and so on. Many cells, for example, require a specific combination of extracellular survival factors to allow the cell to continue living; when deprived of these signals,
the cell activates a suicide program and kills itself—usually by apoptosis, a form of programmed cell death. Cell proliferation often depends on a combination of signals that promote both cell division and survival, as well as signals that stimulate cell growth
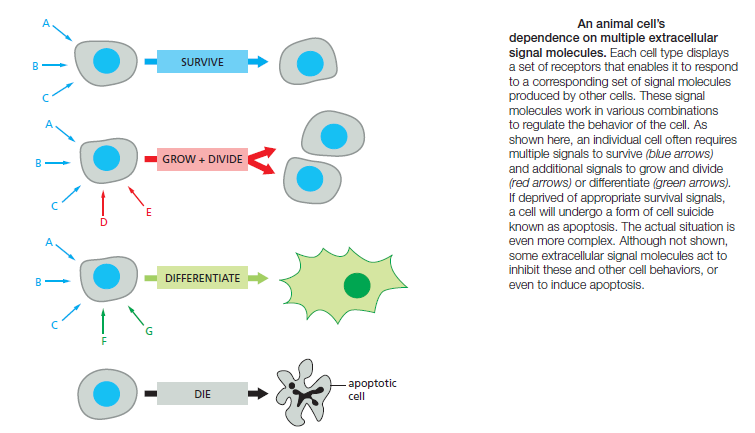
On the other hand, differentiation into a nondividing state (called terminal differentiation) frequently requires a different combination of survival and differentiation signals that must override any signal to divide. In principle, the hundreds of signal molecules that an animal makes can be used in an almost unlimited number of combinations to control the diverse behaviors of its cells in highly specific ways. Relatively small numbers of types of signal molecules and receptors are sufficient. The complexity lies in the ways in
which cells respond to the combinations of signals that they receive. A signal molecule often has different effects on different types of target cells.The neurotransmitter acetylcholine
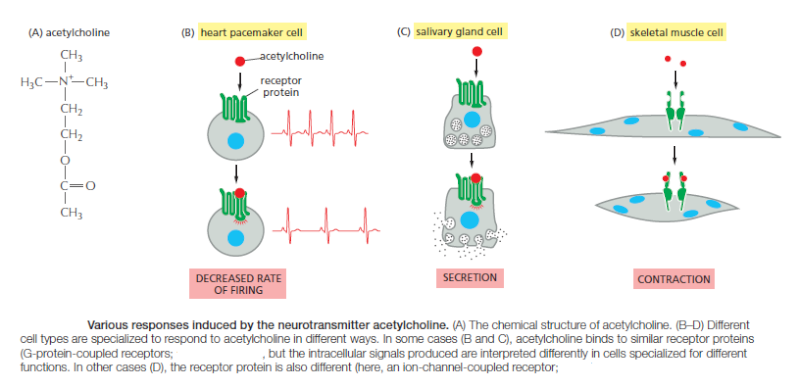
(Figure A) above, for example, decreases the rate of action potential firing in heart pacemaker cells (Figure B) and stimulates the production of saliva by salivary gland cells (Figure C), even though the receptors are the same on both cell types. In skeletal muscle, acetylcholine causes the cells to contract by binding to a different receptor protein (Figure D). The different effects of acetylcholine in these cell types result from differences in the intracellular signaling proteins, effector proteins, and genes that are activated. Thus, an extracellular signal itself has little information content; it simply induces the cell to respond according to its predetermined state, which depends on the cell’s developmental history and the specific genes it expresses.
There Are Three Major Classes of Cell-Surface Receptor Proteins
Most extracellular signal molecules bind to specific receptor proteins on the surface of the target cells they influence and do not enter the cytosol or nucleus. These cell-surface receptors act as signal transducers by converting an extracellular ligand-binding event into intracellular signals that alter the behavior of the target cell. Most cell-surface receptor proteins belong to one of three classes, defined bytheir transduction mechanism. Ion-channel-coupled receptors, also known as transmitter-gated ion channels or ionotropic receptors, are involved in rapid synaptic signaling between nerve cells and other electrically excitable target cells such as nerve and muscle cells
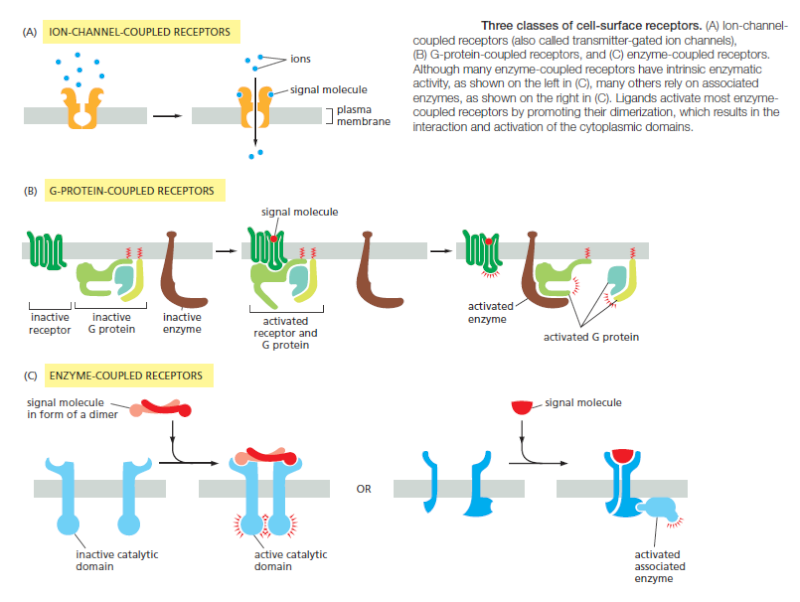
This type of signaling is mediated by a small number of neurotransmitters that transiently open or close an ion channel formed by the protein to which they bind, briefly changing the ion permeability of the plasma membrane and thereby changing the excitability of the postsynaptic target cell. Most ion-channel-coupled receptors belong to a large family ofhomologous, multipass transmembrane proteins. G-protein-coupled receptors act by indirectly regulating the activity of a separate plasma-membrane-bound target protein, which is generally either an enzyme or an ion channel. A trimeric GTP-binding protein (G protein) mediates the interaction between the activated receptor and this target protein (Figure B above). The activation of the target protein can change the concentration of one or more small intracellular signaling molecules (if the target protein is an enzyme), or it can change the ion permeability of the plasma membrane (if the target protein is an ion channel). The small intracellular signaling molecules act in turn to alter the behavior of yet other signaling proteins in the cell. Enzyme-coupled receptors either function as enzymes or associate directly with enzymes that they activate (Figure C). They are usually single-pass transmembrane proteins that have their ligand-binding site outside the cell and their catalytic or enzyme-binding site inside. Enzyme-coupled receptors are heterogeneous in structure compared with the other two classes; the great majority, however, are either protein kinases or associate with protein kinases, which phosphorylate specific sets of proteins in the target cell when activated. There are also some types of cell-surface receptors that do not fit easily into any of these classes but have important functions in controlling the specialization of different cell types during development and in tissue renewal and repair in adults. We discuss these in a later section, after we explain how G-protein-coupled receptors and enzyme-coupled receptors operate. First, we continue our general discussion of the principles of signaling via cell-surface receptors.
Cell-Surface Receptors Relay Signals Via Intracellular Signaling Molecules
Numerous intracellular signaling molecules relay signals received by cell-surface receptors into the cell interior. The resulting chain of intracellular signaling events ultimately alters effector proteins that are responsible for modifying the behavior of the cell.Some intracellular signaling molecules are small chemicals, which are often called second messengers (the “first messengers” being the extracellular signals). They are generated in large amounts in response to receptor activation and diffuse away from their source, spreading the signal to other parts of the cell. Some, such as cyclic AMP and Ca2+, are water-soluble and diffuse in the cytosol, while others, such as diacylglycerol, are lipid-soluble and diffuse in the plane of the plasma membrane. In either case, they pass the signal on by binding to and altering the behavior of selected signaling or effector proteins.
Most intracellular signaling molecules are proteins, which help relay the signal into the cell by either generating second messengers or activating the next signaling or effector protein in the pathway. Many of these proteins behave like molecular switches. When they receive a signal, they switch from an inactive to an active state, until another process switches them off, returning them to their inactive state. The switching off is just as important as the switching on. If a signaling pathway is to recover after transmitting a signal so that it can be ready to transmit another, every activated molecule in the pathway must return to its original, unactivated state. The largest class of molecular switches consists of proteins that are activated or inactivated by phosphorylation . For these proteins, the switch is thrown in one direction by a protein kinase, which covalently adds one or more phosphate groups to specific amino acids on the signaling protein, and in the other direction by a protein phosphatase, which removes the phosphate groups (Figure A below).
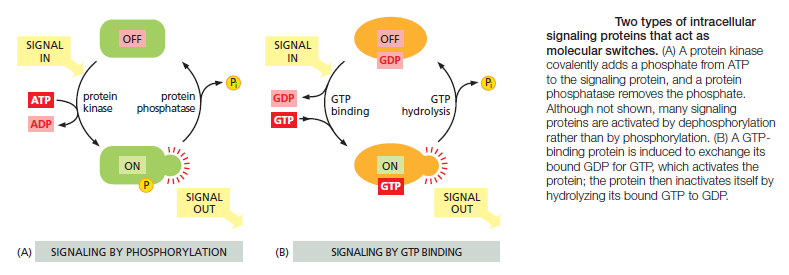
The activity of any protein regulated by phosphorylation depends on the balance between the activities of the kinases that phosphorylate it and of the phosphatases that dephosphorylate it. About 30–50% of human proteins contain covalently attached phosphate, and the human genome encodes about 520 protein kinases and about 150 protein phosphatases. A typical mammalian cell makes use of hundreds of distinct types of protein kinases at any moment.
Protein kinases attach phosphate to the hydroxyl group of specific amino acids on the target protein. There are two main types of protein kinase. The great majority are serine/threonine kinases, which phosphorylate the hydroxyl groups of serines and threonines in their targets. Others are tyrosine kinases, which phosphorylate proteins on tyrosines. The two types of protein kinase are closely related members of a large family, differing primarily in the structure of their protein substrate binding sites. Many intracellular signaling proteins controlled by phosphorylation are themselves protein kinases, and these are often organized into kinase cascades. In such a cascade, one protein kinase, activated by phosphorylation, phosphorylates the next protein kinase in the sequence, and so on, relaying the signal onward and, in some cases, amplifying it or spreading it to other signaling pathways. The other important class of molecular switches consists of GTP-binding proteins. These proteins switch between an “on” (actively signaling) state when GTP is bound and an “off” state when GDP is bound. In the “on” state, they usually have intrinsic GTPase activity and shut themselves off by hydrolyzing their bound GTP to GDP (Figure 15–7B). There are two major types of GTP-binding proteins. Large, trimeric GTP-binding proteins (also called G proteins) help relay signals from G-protein-coupled receptors that activate them (see Figure B). Small monomeric GTPases (also called monomeric GTP-binding proteins) help relay signals from many classes of cell-surface receptors. Specific regulatory proteins control both types of GTP-binding proteins. GTPase-activating proteins (GAPs) drive the proteins into an “off” state by increasing the rate of hydrolysis of bound GTP. Conversely, guanine nucleotide exchange factors (GEFs) activate GTP-binding proteins by promoting the release of bound GDP, which allows a new GTP to bind. In the case of trimeric G proteins, the activated receptor serves as the GEF
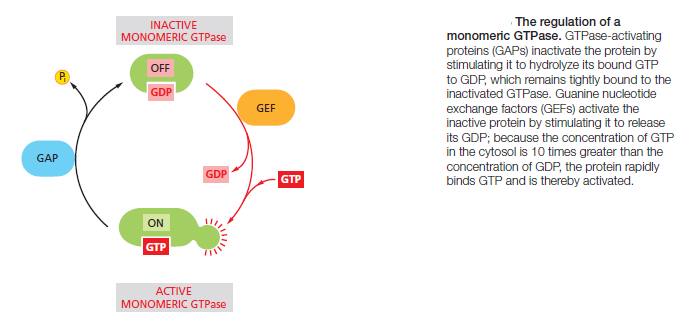
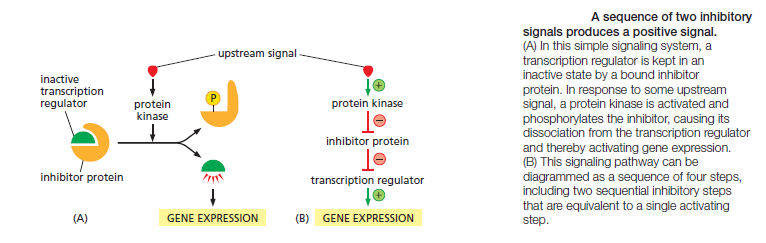
Not all molecular switches in signaling systems depend on phosphorylation or GTP binding. Some signaling proteins are switched on or off by the binding of another signaling protein or a second messenger such as cyclic AMP or Ca2+, or by covalent modifications other than phosphorylation or dephosphorylation, such as ubiquitylation. For simplicity, we often portray a signaling pathway as a series of activation steps . It is important to note, however, that most signaling pathways contain inhibitory steps, and a sequence of two inhibitory steps can have the same effect as one activating step. This double-negative activation is very common in signaling systems
Intracellular Signals Must Be Specific and Precise in a Noisy Cytoplasm
Ideally, an activated intracellular signaling molecule should interact only with the appropriate downstream targets, and, likewise, the targets should only be activated by the appropriate upstream signal. In reality, however, intracellular signaling molecules share the cytoplasm with a crowd of closely related signaling molecules that control a diverse array of cellular processes. It is inevitable that an occasional signaling molecule will bind or modify the wrong partner, potentially creating unwanted cross-talk and interference between signaling systems. How does a signal remain strong, precise, and specific under these noisy conditions? The first line of defense comes from the high affinity and specificity of the interactions between signaling molecules and their correct partners compared to the relatively low affinity of the interactions between inappropriate partners.
Question: How could evolution have provided the programming and fine tuning of the correct partners, and provided the right specificity and high affinity ? trial and error ? What would have happened while the communication system was not fully developed and set up ? Would the cell not have died, if not able to react appropriately to the environment changes ?
The binding of a signaling molecule to the correct target is determined by precise and complex interactions between complementary surfaces on the two molecules. Protein kinases, for example, contain active sites that recognize a specific amino acid sequence around the phosphorylation site on the correct target protein, and they often contain additional docking sites that promote a specific, high-affinity interaction with the target. These and related mechanisms help provide a strong and persistent interaction between the correct partners, reducing the likelihood of inappropriate interactions with other proteins. Another important way that cells avoid responses to unwanted background signals depends on the ability of many downstream target proteins to simply ignore such signals. These proteins respond only when the upstream signal reaches a high concentration or activity level. Consider a signaling pathway in which a protein kinase activates some downstream target protein by phosphorylation. If a response is triggered only when more than half of the target proteins are phosphorylated, then there will be little harm done if a small number of them are occasionally phosphorylated by some inappropriate protein kinase. Furthermore, constitutively active protein phosphatases will further reduce the impact of background phosphorylation by rapidly removing much of it. In these and other ways,
intracellular signaling systems filter out noise, generating little or no response to low levels of stimuli. Cells in a population often exhibit random variation in the concentration or
activity of their intracellular signaling molecules. Similarly, individual molecules in a large population of molecules vary in their activity or interactions with other molecules. This signal variability introduces another form of noise that can interfere with the precision and efficiency of signaling. Most signaling systems, however, are built to generate remarkably robust and precise responses even when upstream signals are variable or even when some components of the system are disabled.In many cases, this robustness depends on the presence of backup mechanisms: for example, a signal might employ two parallel pathways to activate a single common downstream target protein, allowing the response to occur even if one pathway is crippled.
Intracellular Signaling Complexes Form at Activated Receptors
One simple and effective strategy for enhancing the specificity of interactions between signaling molecules is to localize them in the same part of the cell or even within large protein complexes, thereby ensuring that they interact only with each other and not with inappropriate partners. Such mechanisms often involve scaffold proteins, which bring together groups of interacting signaling proteins into signaling complexes, often before a signal has been received (Figure A below)
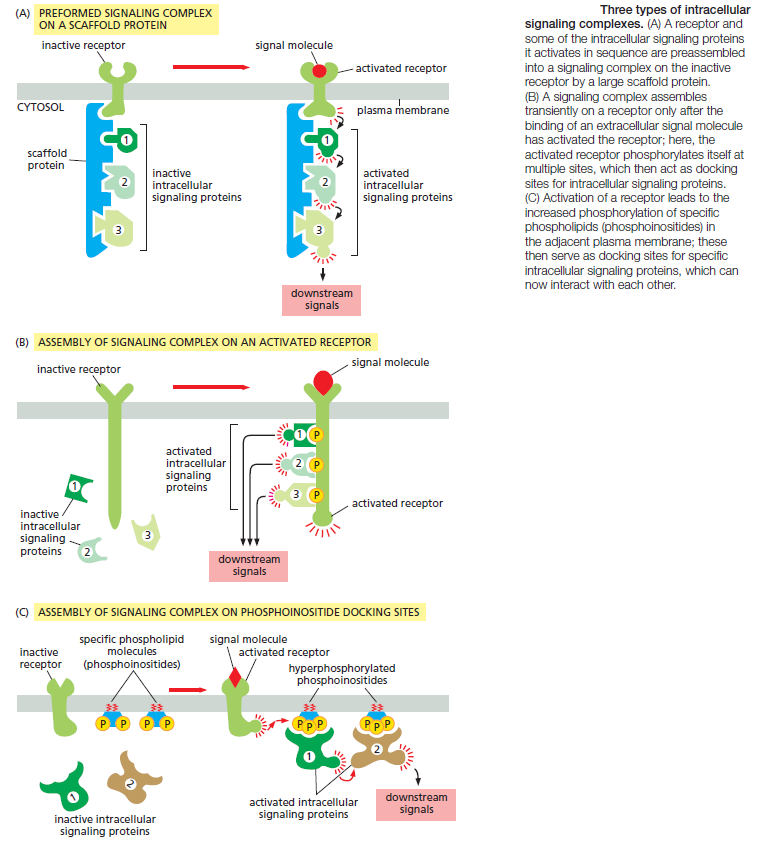
Because the scaffold holds the proteins in close proximity, they can interact at high local concentrations and be sequentially activated rapidly, efficiently, and selectively in response to an appropriate extracellular signal, avoiding unwanted cross-talk with other signaling pathways. In other cases, such signaling complexes form only transiently in response to an extracellular signal and rapidly disassemble when the signal is gone. They often assemble around a receptor after an extracellular signal molecule has activated
it. In many of these cases, the cytoplasmic tail of the activated receptor is phosphorylated during the activation process, and the phosphorylated amino acids then serve as docking sites for the assembly of other signaling proteins (Figure B). In yet other cases, receptor activation leads to the production of modified phospholipid molecules (called phosphoinositides) in the adjacent plasma membrane, which then recruit specific intracellular signaling proteins to this region of membrane, where they are activated (Figure C).
Modular Interaction Domains Mediate Interactions Between Intracellular Signaling Proteins
Simply bringing intracellular signaling proteins together into close proximity is sometimes sufficient to activate them. Thus, induced proximity, where a signal triggers assembly of a signaling complex, is commonly used to relay signals from protein to protein along a signaling pathway. The assembly of such signaling complexes depends on various highly conserved, small interaction domains, which are found in many intracellular signaling proteins. Each of these compact protein modules binds to a particular structural motif in another protein or lipid. The recognized motif in the interacting protein can be a short peptide sequence, a covalent modification (such as a phosphorylated amino acid), or another protein domain. The use of modular interaction domains facilitates signaling pathways; it can be inserted at many locations in a protein without disturbing the protein’s folding or function, a new interaction domain added to an existing signaling protein could connect the protein to additional signaling pathways. There are many types of interaction domains in signaling proteins. Src homology 2 (SH2) domains and phosphotyrosine-binding (PTB) domains, for example, bind to phosphorylated tyrosines in a particular peptide sequence on activated receptors or intracellular signaling proteins. Src homology 3 (SH3) domains bind to short, proline-rich amino acid sequences. Some pleckstrin homology (PH) domains bind to the charged head groups of specific phosphoinositides that are produced in the plasma membrane in response to an extracellular signal; they enable the protein they are part of to dock on the membrane and interact with other similarly recruited signaling proteins (see Figure C). Some signaling proteins consist solely of two or more interaction domains and function only as adaptors to link two other proteins together in a signaling pathway. Interaction domains enable signaling proteins to bind to one another in multiple specific combinations. Like Lego
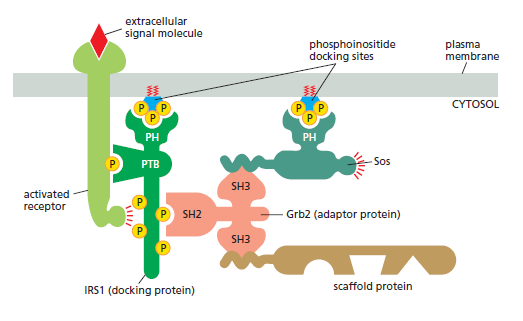
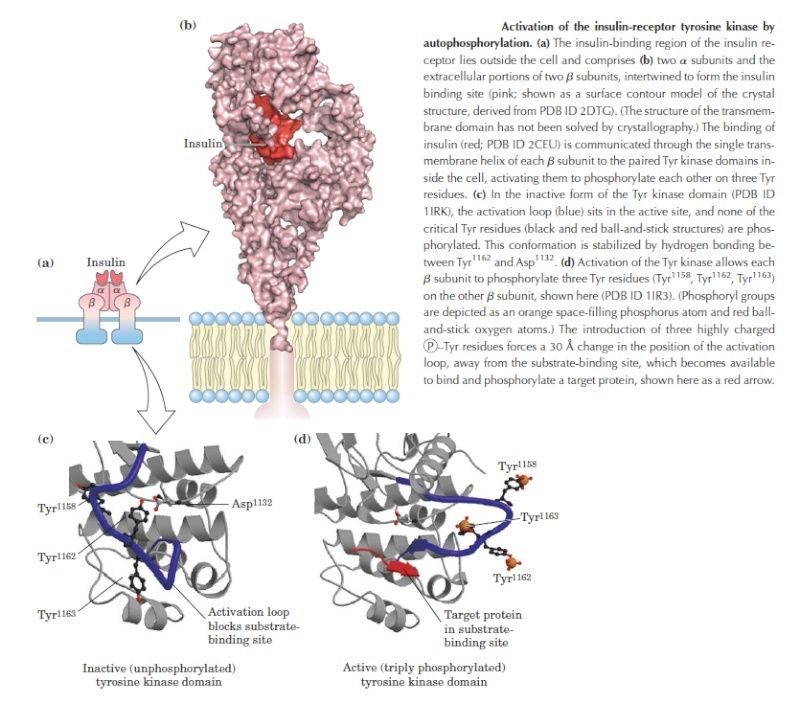
A specific signaling complex formed using modular interaction domains. This example is based on the insulin receptor, which is an enzyme-coupled receptor (a receptor tyrosine kinase, discussed later). First, the activated receptor phosphorylates itself on tyrosines, and one of the phosphotyrosines then recruits a docking protein called insulin receptor substrate-1 (IRS1) via a PTB domain of IRS1; the PH domain of IRS1 also binds to specific phosphoinositides on the inner surface of the plasma membrane. Then, the activated receptor phosphorylates IRS1 on tyrosines, and one of these phosphotyrosines binds the SH2 domain of the adaptor protein Grb2. Next, Grb2 uses one of its two SH3domains to bind to a proline-rich region of a protein called Sos, which relays the signal downstream by acting as a GEF to activate a monomeric GTPase called Ras (not shown). Sos also binds to phosphoinositides in the plasma membrane via its PH domain. Grb2 uses its other SH3 domain to bind to a proline-rich sequence in a scaffold protein. The scaffold protein binds several other signaling proteins, and the other phosphorylated tyrosines on IRS1 recruit additional signaling proteins that have SH2 domains (not shown).
Another way of bringing receptors and intracellular signaling proteins together is to concentrate them in a specific region of the cell. An important example is the primary cilium that projects like an antenna from the surface of most vertebrate cells . It is usually short and nonmotile and has microtubules in its core, and a number of surface receptors and signaling proteins are concentrated there. Light and smell receptors are also highly concentrated in specialized cilia.
The Relationship Between Signal and Response Varies in Different Signaling Pathways
The function of an intracellular signaling system is to detect and measure a specific stimulus in one location of a cell and then generate an appropriately timed and measured response at another location. The system accomplishes this task by sending information in the form of molecular “signals” from the sensor to the target, often through a series of intermediaries that do not simply pass the signal along but process it in various ways. All signaling systems do not work in precisely the same way: each has specialized behaviors that produce a response that is appropriate for the cell function that system controls. In the following paragraphs, we list some of these behaviors and describe how they vary in different systems, as a foundation for more detailed discussions later.
1. Response timing varies dramatically in different signaling systems, according to the speed required for the response. In some cases, such as synaptic signaling, the response can occur within milliseconds.In other cases, as in the control of cell fate by morphogens during development, a full response can require hours or days.
2. Sensitivity to extracellular signals can vary greatly. Hormones tend to act at very low concentrations on their distant target cells, which are therefore highly sensitive to low concentrations of signal. Neurotransmitters, on the other hand, operate at much higher concentrations at a synapse, reducing the need for high sensitivity in postsynaptic receptors. Sensitivity is often controlled by changes in the number or affinity of the receptors on the target cell. A particularly important mechanism for increasing the sensitivity
of a signaling system is signal amplification, whereby a small number of activated cell-surface receptors evoke a large intracellular response either by producing large amounts of a second messenger or by activating many copies of a downstream signaling protein.
3. Dynamic range of a signaling system is related to its sensitivity. Some systems, like those involved in simple developmental decisions, are responsive over a narrow range of extracellular signal concentrations. Other systems, like those controlling vision or the metabolic response to some hormones, are highly responsive over a much broader range of signal strengths. We will see that broad dynamic range is often achieved by adaptation mechanisms that adjust the responsiveness of the system according to the prevailing
amount of signal.
4. Persistence of a response can vary greatly. A transient response of less than a second is appropriate in some synaptic responses, for example, while a prolonged or even permanent response is required in cell fate decisions during development. Numerous mechanisms, including positive feedback, can be used to alter the duration and reversibility of a response.
5. Signal processing can convert a simple signal into a complex response. In many systems, for example, a gradual increase in an extracellular signal is converted into an abrupt, switchlike response. In other cases, a simple input signal is converted into an oscillatory response, produced by a repeating series of transient intracellular signals. Feedback usually lies at the heart of biochemical switches and oscillators.
6. Integration allows a response to be governed by multiple inputs. As discussed earlier, for example, specific combinations of extracellular signals are generally required to stimulate complex cell behaviors such as cell survival and proliferation. The cell therefore has to integrate information coming from multiple signals, which often depends on intracellular coincidence detectors; these proteins are equivalent to AND gates in the microprocessor of a computer, in that they are only activated if they receive multiple converging signals.
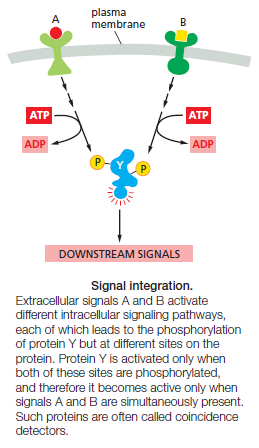
7. Coordination of multiple responses in one cell can be achieved by a single extracellular signal. Some extracellular signal molecules, for example, stimulate a cell to both grow and divide. This coordination generally depends on mechanisms for distributing a signal to multiple effectors, by creating branches in the signaling pathway. In some cases, the branching of signaling pathways can allow one signal to modulate the strength of a response to other signals. Given the complexity that arises from behaviors like signal integration, distribution, and feedback, it is clear that signaling systems rarely depend on a simple linear sequence of steps but are often more like a network, in which informationflows not just forward but in multiple directions—and sometimes even backward. A major research challenge is to understand the nature of these networks and the response behaviors they can achieve.
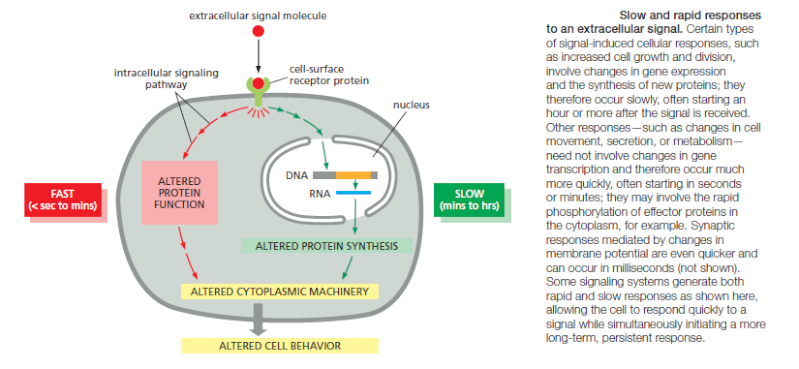
The Speed of a Response Depends on the Turnover of Signaling Molecules
The speed of any signaling response depends on the nature of the intracellular signaling molecules that carry out the target cell’s response. When the response requires only changes in proteins already present in the cell, it can occur very rapidly: an allosteric change in a neurotransmitter-gated ion channel, for example, can alter the plasma membrane electrical potential in milliseconds, and responses that depend solely on protein phosphorylation can occur within seconds. When the response involves changes in gene expression and the synthesis of new proteins, however, it usually requires many minutes or hours, regardless of the mode of signal delivery
It is natural to think of intracellular signaling systems in terms of the changes produced when an extracellular signal is delivered. But it is just as important to consider what happens when the signal is withdrawn. During development, transient extracellular signals often produce lasting effects: they can trigger a change in the cell’s development that persists indefinitely through cell memory mechanisms. In most cases in adult tissues,however, the response fades when a signal ceases. Often the effect is transient because the signal exerts its effects by altering the concentrations of intracellular molecules that are short-lived (unstable), undergoing continual turnover. Thus, once the extracellular signal is gone, the degradation of the old molecules quickly wipes out all traces of the signal’s action. It follows that the speed with which a cell responds to signal removal depends on the rate of destruction, or turnover, of the intracellular molecules that the signal affects. It is also true, although much less obvious, that this turnover rate can determine the promptness of the response when an extracellular signal arrives. Consider, for example, two intracellular signaling molecules, X and Y, both of which are normally maintained at a steady-state concentration of 1000 molecules per cell. The cell synthesizes and degrades molecule Y at a rate of 100 molecules per second, with each molecule having an average lifetime of 10 seconds. Molecule X has a turnover rate that is 10 times slower than that of Y: it is both synthesized and degraded at a rate of 10 molecules per second, so that each molecule has an average lifetime in the cell of 100 seconds. If a signal acting on the cell causes a tenfold increase in the synthesis rates of both X and Y with no change in the molecular lifetimes, at the end of 1 second the concentration of Y will have increased by nearly 900 molecules per cell (10 × 100 – 100), while the concentration of X will have increased by only 90 molecules per cell. In fact, after a molecule’s synthesis rate has been either increased or decreased abruptly, the time required for the molecule to shift halfway from its old to its new equilibrium concentration is equal to its half-life—that is, equal to the time that would be required for its concentration to fall by half if all synthesis were stopped. The same principles apply to proteins and small molecules, whether the molecules are in the extracellular space or inside cells. Many intracellular proteins have short half-lives, some surviving for less than 10 minutes. In most cases, these are key regulatory proteins whose concentrations are rapidly controlled in the cell by changes in their rates of synthesis. As we have seen, many cell responses to extracellular signals depend on the conversion of intracellular signaling proteins from an inactive to an active form, rather than on their synthesis or degradation. Phosphorylation or the binding of GTP, for example, commonly activates signaling proteins. Even in these cases, however, the activation must be rapidly and continuously reversed (by dephosphorylation or GTP hydrolysis to GDP, respectively, in these examples) to make rapid signaling possible. These inactivation processes play a crucial part in determining the magnitude, rapidity, and duration of the response.
SIGNALING THROUGH G-PROTEIN-COUPLED RECEPTORS
G-protein-coupled receptors (GPCRs) form the largest family of cell-surface receptors, and they mediate most responses to signals from the external world, as well as signals from other cells, including hormones, neurotransmitters, and local mediators. Our senses of sight, smell, and taste depend on them. There are more than 800 GPCRs in humans, and in mice there are about 1000 concerned with the sense of smell alone. The signal molecules that act on GPCRs are as varied in structure as they are in function and include proteins and small peptides, as well as derivatives of amino acids and fatty acids, not to mention photons of light and all the molecules that we can smell or taste. The same signal molecule can activate many different GPCR family members; for example, adrenaline activates at least 9 distinct GPCRs, acetylcholine another 5, and the neurotransmitter serotonin at least 14. The different receptors for the same signal are usually expressed in different cell types and elicit different responses. Despite the chemical and functional diversity of the signal molecules that activate them, all GPCRs have a similar structure. They consist of a single polypeptide chain that threads back and forth across the lipid bilayer seven times, forming a cylindrical structure, often with a deep ligand-binding site at its center
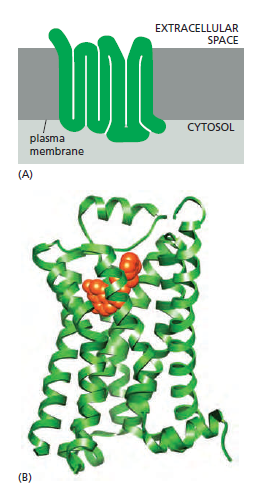
A G-protein-coupled receptor (GPCR).
(A) GPCRs that bind small ligands such as adrenaline have small extracellular domains, and the ligand usually binds deep within the plane of the membrane to a site that is formed by amino acids from several transmembrane segments. GPCRs that bind protein ligands have a large extracellular domain (not shown here) that contributes to ligand binding.
(B) The structure of the β2-adrenergic receptor, a receptor for the neurotransmitter adrenaline, illustrates the typical cylindrical arrangement of the seven transmembrane helices in a GPCR. The ligand (orange) binds in a pocket between the helices, resulting in conformational changes on the cytoplasmic surface of the receptor that promote G-protein activation (not shown). (PDB code: 3P0G.)
In addition to their characteristic orientation in the plasma membrane, they all use G proteins to relay the signal into the cell interior. The GPCR superfamily includes rhodopsin, the light-activated protein in the vertebrate eye, as well as the large number of olfactory receptors in the vertebrate nose. Other family members are found in unicellular organisms: the receptors in yeasts that recognize secreted mating factors are an example. It is likely that the GPCRs that mediate cell–cell signaling in multicellular organisms evolved from the sensory receptors in their unicellular eukaryotic ancestors. It is remarkable that almost half of all known drugs work through GPCRs or the signaling pathways GPCRs activate. Of the many hundreds of genes in the human genome that encode GPCRs, about 150 encode orphan receptors, for which the ligand is unknown. Many of them are likely targets for new drugs that remain to be discovered.
Trimeric G Proteins Relay Signals From GPCRs
When an extracellular signal molecule binds to a GPCR, the receptor undergoes a conformational change that enables it to activate a trimeric GTP-binding protein (G protein), which couples the receptor to enzymes or ion channels in the membrane. In some cases, the G protein is physically associated with the receptor before the receptor is activated, whereas in others it binds only after receptor activation. There are various types of G proteins, each specific for a particular set of GPCRs and for a particular set of target proteins in the plasma membrane. They all have a similar structure, however, and operate similarly. G proteins are composed of three protein subunits—α, β, and γ. In the unstimulated state, the α subunit has GDP bound and the G protein is inactive
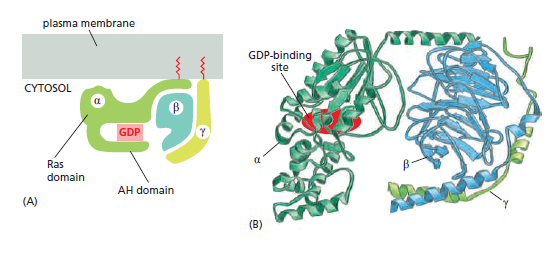
Many G-protein-coupled receptors have a large extracellular ligand-binding domain. 1When an appropriate protein ligand binds to this domain, the receptor undergoes a conformational change that is transmitted to its cytosolic regions, which now activate a trimeric GTP-binding protein (or G protein for short). As the name implies, a trimeric G protein is composed of three protein subunits called alpha, beta, and gamma. Both the alpha and gamma subunits have covalently attached lipid tails that help anchor the G protein in the plasma membrane.
In the absence of a signal, the alpha subunit has a GDP bound, and the G protein is inactive. In some cases, the inactive G protein is associated with the inactive receptor, while, in other cases, as shown here, it only binds after the receptor is activated. In either case, an activated receptor induces a conformational change in the alpha subunit, causing the GDP to dissociate.
GTP, which is abundant in the cytosol, can now readily bind in place of the GDP. GTP binding causes a further conformational change in the G protein, activating both the alpha subunit and beta-gamma complex. The activated alpha subunit dissociates from the activated beta–gamma complex, whereas in other cases the two activated components stay together.
In either case, both of the activated components can now regulate the activity of target proteins in the plasma membrane, as shown here for a GTP-bound alpha subunit. The activated target proteins then relay the signal to other components in the signaling cascade. Eventually, the alpha subunit hydrolyses its bound GTP to GDP, which inactivates the subunit. This step is often accelerated by the binding of another protein, called a regulator of G-protein signaling (or RGS). The inactivated, GDP- bound alpha subunit now reforms an inactive G protein with a beta–gamma complex, turning off other downstream events.
As long as the signaling receptor remains stimulated, it can continue to activate G proteins. Upon prolonged stimulation, however, the receptors eventually inactivate, even if their activating ligands remain bound. In this case, a receptor kinase phosphorylates the cytosolic portions of the activated receptor. Once a receptor has been phosphorylated in this way, it binds with high affinity to an arrestin protein, which inactivates the receptor by preventing its interaction with G proteins.
When a GPCR is activated, it acts like a guanine nucleotide exchange factor (GEF) and induces the α subunit to release its bound GDP, allowing GTP to bind in its place. GTP binding then causes an activating conformational change in the Gα subunit, releasing the G protein from the receptor and triggering dissociation of the GTP-bound Gα subunit from the Gβγ pair—both of which then interact with various targets, such as enzymes and ion channels in the plasma membrane, which relay the signal onward
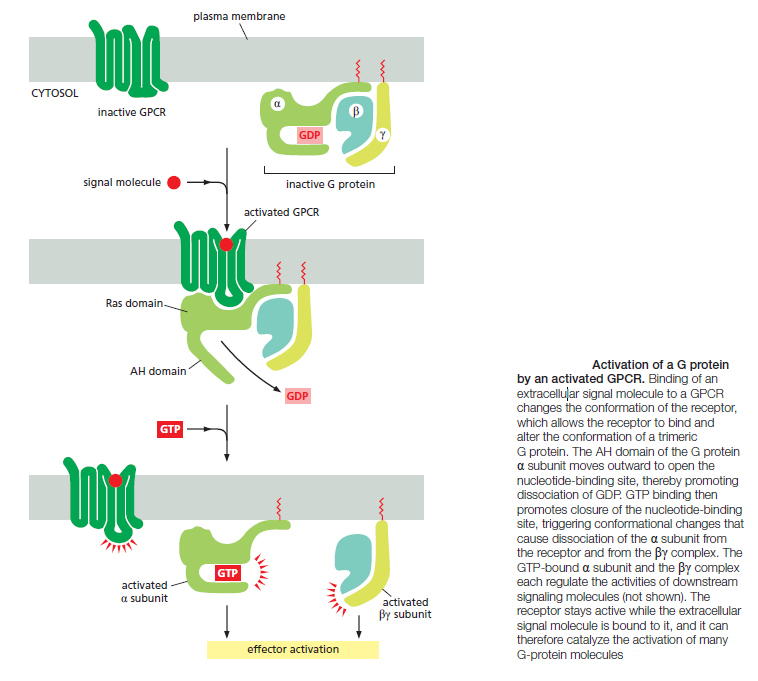
The α subunit is a GTPase and becomes inactive when it hydrolyzes its bound GTP to GDP. The time required for GTP hydrolysis is usually short because the GTPase activity is greatly enhanced by the binding of the α subunit to a second protein, which can be either the target protein or a specific regulator of G protein signaling (RGS). RGS proteins act as α-subunit-specific GTPase-activating proteins (GAPs), and they help shut off G-protein-mediated responses in all eukaryotes. There are about 25 RGS proteins encoded in the
human genome, each of which interacts with a particular set of G proteins.
Some G Proteins Regulate the Production of Cyclic AMP
Cyclic AMP (cAMP) acts as a second messenger in some signaling pathways. An extracellular signal can increase cAMP concentration more than twentyfold in seconds. Such a rapid response requires balancing a rapid synthesis of the molecule with its rapid breakdown or removal. Cyclic AMP is synthesized from ATP by an enzyme called adenylyl cyclase, and it is rapidly and continuously destroyed by cyclic AMP phosphodiesterasesAdenylyl cyclase is a large, multipass transmembrane protein with its catalytic domain on the cytosolic side of the plasma membrane. There are at least eight isoforms in mammals, most of which are regulated by both G proteins and Ca2+. Many extracellular signals work by increasing cAMP concentrations inside the cell. These signals activate GPCRs that are coupled to a stimulatory G protein (Gs). The activated α subunit of Gs binds and thereby activates adenylyl cyclase. Other extracellular signals, acting through different GPCRs, reduce cAMP levels by activating an inhibitory G protein (Gi), which then inhibits adenylyl cyclase. Both Gs and Gi are targets for medically important bacterial toxins. Cholera toxin, which is produced by the bacterium that causes cholera, is an enzyme that catalyzes the transfer of ADP ribose from intracellular NAD+ to the α subunit of Gs. This ADP ribosylation alters the α subunit so that it can no longer hydrolyze its bound GTP, causing it to remain in an active state that stimulates adenylyl cyclase indefinitely. The resulting prolonged elevation in cAMP concentration within intestinal epithelial cells causes a large efflux of Cl– and water into the gut, thereby causing the severe diarrhea that characterizes cholera. Pertussis toxin, which is made by the bacterium that causes pertussis (whooping cough), catalyzes the ADP ribosylation of the α subunit of Gi, preventing the protein from interacting with receptors; as a result, the G protein remains in the inactive GDP-bound state and is unable to regulate its target proteins. These two toxins are widely used in experiments to determine whether a cell’s GPCR-dependent response to a signal is mediated by Gs or by Gi.
Cyclic-AMP-Dependent Protein Kinase (PKA) Mediates Most of the Effects of Cyclic AMP
In most animal cells, cAMP exerts its effects mainly by activating cyclic-AMPdependent protein kinase (PKA). This kinase phosphorylates specific serines or threonines on selected target proteins, including intracellular signaling proteins and effector proteins, thereby regulating their activity. The target proteins differ from one cell type to another, which explains why the effects of cAMP vary so markedly depending on the cell type In the inactive state, PKA consists of a complex of two catalytic subunits and two regulatory subunits. The binding of cAMP to the regulatory subunits alters their conformation, causing them to dissociate from the complex. The released catalytic subunits are thereby activated to phosphorylate specific target proteins
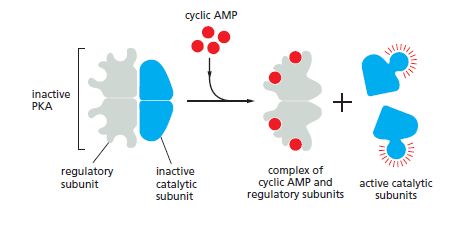
The activation of cyclic- AMP-dependent protein kinase (PKA). The binding of cAMP to the regulatory subunits of the PKA tetramer induces a conformational change, causing these subunits to dissociate from the catalytic subunits, thereby activating the kinase activity of the catalytic subunits. The release of the catalytic subunits requires the binding of more than two cAMP molecules to the regulatory subunits in the tetramer. This requirement greatly sharpens the response of the kinase to changes in cAMP concentration. Mammalian cells have at least two types of PKAs: type I is mainly in the cytosol, whereas type II is bound via its regulatory subunits and special anchoring proteins to the plasma membrane, nuclear membrane, mitochondrial outer membrane, and microtubules. In both types, once the catalytic subunits are freed and active, they can migrate into the nucleus (where they can phosphorylate transcription regulatory proteins), while the regulatory subunits remain in the cytoplasm.
The regulatory subunits of PKA (also called A-kinase) are important for localizing the kinase inside the cell: special A-kinase anchoring proteins (AKAPs) bind both to the regulatory subunits and to a component of the cytoskeleton or a membrane of an organelle, thereby tethering the enzyme complex to a particular subcellular compartment. Some AKAPs also bind other signaling proteins, forming a signaling complex. An AKAP located around the nucleus of heart muscle cells, for example, binds both PKA and a phosphodiesterase that
hydrolyzes cAMP. In unstimulated cells, the phosphodiesterase keeps the local cAMP concentration low, so that the bound PKA is inactive; in stimulated cells, cAMP concentration rapidly rises, overwhelming the phosphodiesterase and activating the PKA. Among the target proteins that PKA phosphorylates and activates in these cells is the adjacent phosphodiesterase, which rapidly lowers the cAMP concentration again. This negative feedback arrangement converts what might otherwise be a prolonged PKA response into a brief, local pulse of PKA activity. Whereas some responses mediated by cAMP occur within seconds others depend on changes in the transcription of specific genes and take
hours to develop fully. In cells that secrete the peptide hormone somatostatin for example, cAMP activates the gene that encodes this hormone. The regulatory region of the somatostatin gene contains a short cis-regulatory sequence, called the cyclic AMP response element (CRE), which is also found in the regulatory region of many other genes activated by cAMP. A specific transcription regulator called CRE-binding (CREB) protein recognizes this sequence. When PKA is activated by cAMP, it phosphorylates CREB on a single serine; phosphorylated CREB then recruits a transcriptional coactivator called CREB-binding protein (CBP), which stimulates the transcription of the target genes
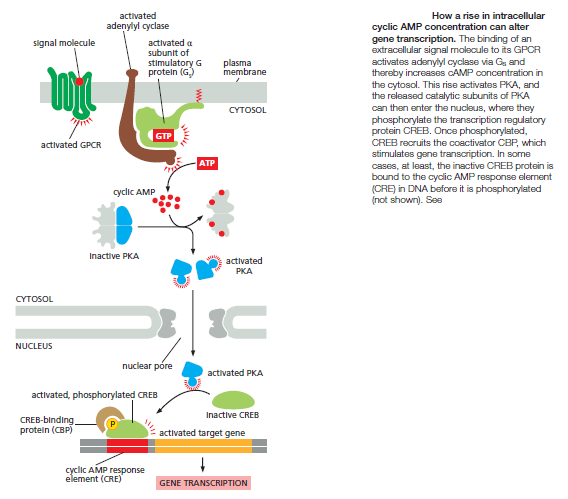
Thus, CREB can transform a short cAMP signal into a long-term change in a cell, a process that, in the brain, is thought to play an important part in some forms of learning and memory.
Examples of biological processes that show BZ dynamical behavior include glycolysis and aggregation of dispersed cells of the slime mold Dictyostelium discoideum into a slug. But consider the sophisticated components of the aggregation-signaling system of D. discoideum, which include: a cyclic AMP membrane receptor protein that can exist in an active and inactive form; an adenylate cyclase that binds to the active form of the receptor and itself becomes activated; a protein to export cyclic AMP into the extracellular medium; and more (Goldbeter 1996, Part I). All of that complicated machinery is ignored in BZ models--treated as a black box. Oscillations in the cellular concentration of glycolytic intermediates are due in large part to the multi-talented phosphofructokinase (PFK), a tetrameric enzyme that can exist in two conformational states (an active form and a less-active one) and which has binding sites for a dozen different activators and inhibitors (Goldbeter 1996, Part III). Mathematical models of BZ behavior don't explain the origin of the impressive abilities of PFK any more than models of traffic flow explain the origin of brakes or gas pedals. Thus, even if a biological system displays self-organizing behavior, the question of its origin remains. 2
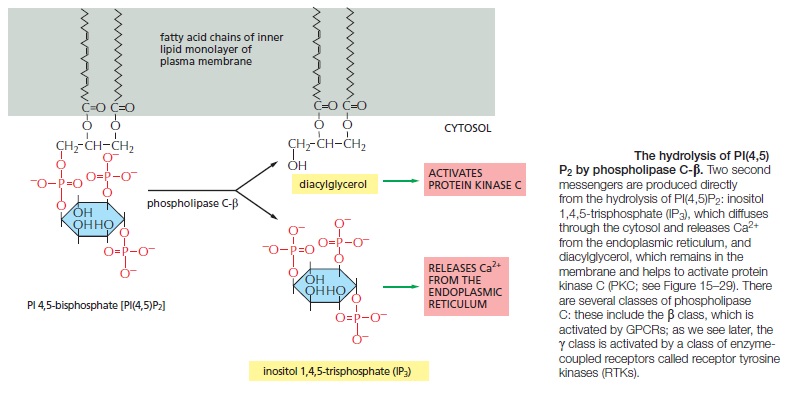
Some G Proteins Signal Via Phospholipids
Many GPCRs exert their effects through G proteins that activate the plasma-membrane bound enzyme phospholipase C-β (PLCβ). The phospholipase acts on a phosphorylatedinositol phospholipid (a phosphoinositide) called phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], which is present in small amounts in the inner half of the plasma membrane lipid bilayer
1) http://www.garlandscience.com/garlandscience_resources/resource_detail.jsf?landing=student&resource_id=9780815344926_CH03_QTM03
2) http://www.arn.org/docs/behe/mb_selforganizationreplytoshanksandjoplin.htm
Last edited by Admin on Sat May 18, 2019 12:40 pm; edited 40 times in total