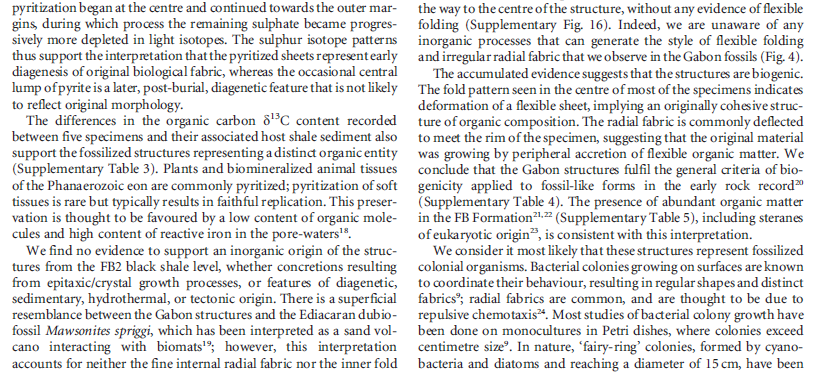
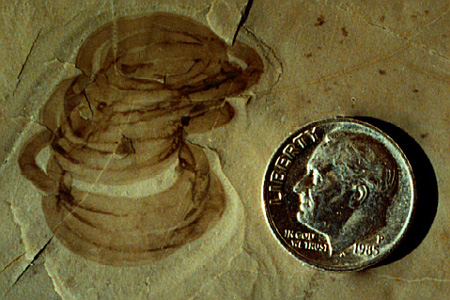
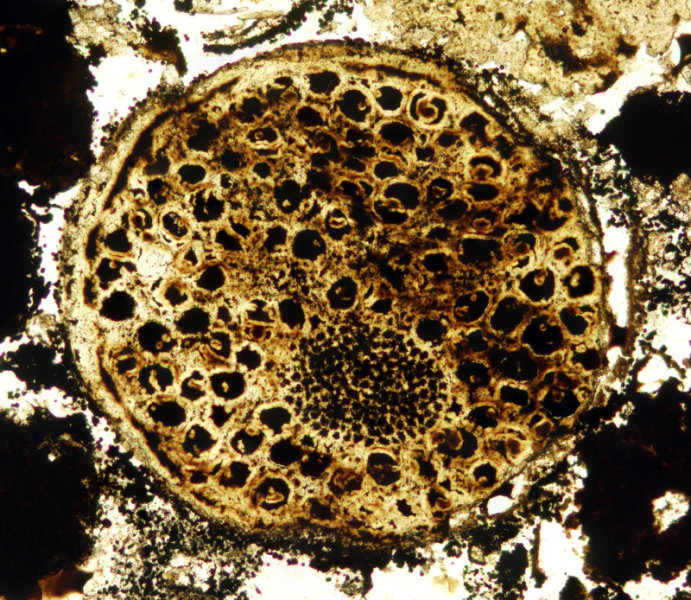
https://reasonandscience.catsboard.com/t2010-unicellular-and-multicellular-organisms-are-best-explained-through-design
One of the central problems of biology is the emergence of complexity. How did complex multicellular organisms emerge, and how is genetic information translated to a spatially organized body plan? The hallmark of a developing multicellular organism is the fact that decisions regarding cell fate are not taken at the level of the individual cell. To allow the building of complex organs with intricate patterns of cellular specialization, such decisions are taken at the population level in a cell-nonautonomous manner. A breakthrough in the elucidation of the genetic control of such intercellular communication in developmental patterning events was made by Nüsslein-Volhard and Wieschaus. 8
Proponents of evolution claim like a mantra, that microevolution leads to macroevolution, and no barrier exists which hinders the transition from one to the other, which last not least explains our biodiversity today
The emergence of multicellularity was supposedly, a major evolutionary leap. Indeed, most biologists consider it one of the most significant transitions in the evolutionary history of Earth’s inhabitants. “How a single cell made the leap to a complex organism is, however, one of life’s great mysteries.”
My comment: Macroevolutionary scenarios and changes include major transitions, that is from LUCA, the last common universal ancestor, to the congregation to yield the first prokaryotic cells, the associations of prokaryotic cells to create eukaryotic cells with organelles such as chloroplasts and mitochondria, and the establishment of cooperative societies composed of discrete multi-cellular individuals. Or in other words: The current hierarchical organization of life reflects a series of transitions in the units of evolution, such as from genes to chromosomes, from prokaryotic to eukaryotic cells, from unicellular to multicellular individuals, and from multi-cellular organisms to societies. Each of these steps requires the overcome of huge hurdles and increase of complexity, which can only be appreciated by the ones, that have to spend time to educate themselves, and gained insight of the extraordinarily complex and manifold mechanisms involved. The emergence of multi-cellularity was ostensibly a major evolutionary leap.
The switch from single-celled organisms to ones made up of many cells have supposedly evolved independently more than two dozen times. Evolution requires more than a mere augmentation of an existing system for co-ordinated multicellularity to evolve; it requires the ex nihilo creation of an entirely new system of organization to coordinate cells appropriately to form a multicellular individual.
There is a level of structure found only in multicellular organisms: intercellular co-ordination. The organism has strategies for arranging and differentiating its cells for survival and reproduction. With this comes a communication network between the cells that regulate the positioning and abundance of each cell type for the benefit of the whole organism. A fundamental part of this organization is cellular differentiation, which is ubiquitous in multicellular organisms. This level cannot be explained by the sum of the parts, cells, and requires coordination from an organizational level above what exists in individual cells. There is a 4-level hierarchy of control in multicellular organisms that constitute a gene regulatory network. This gene regulatory network is essential for the development of the single cell zygote into a full-fledged multicellular individual.
My comment: If evolution and transition from unicellular to multicellular life is exceedingly complex, the chance that it happened once is also exceedingly small. That it happened multiple times separately, becomes even more remotely possible. Convergent evolution of similar traits is evidence against, not for evolution. In order to infer that a proposition is true, these nuances are important to observe. The key is in the details. As Behe states: In order to say that some function is understood, every relevant step in the process must be elucidated. The relevant steps in biological processes occur ultimately at the molecular level, so a satisfactory explanation of a biological phenomenon such as the de novo make of cell communication and cell junction proteins essential for multi-cellular life must include a molecular explanation.
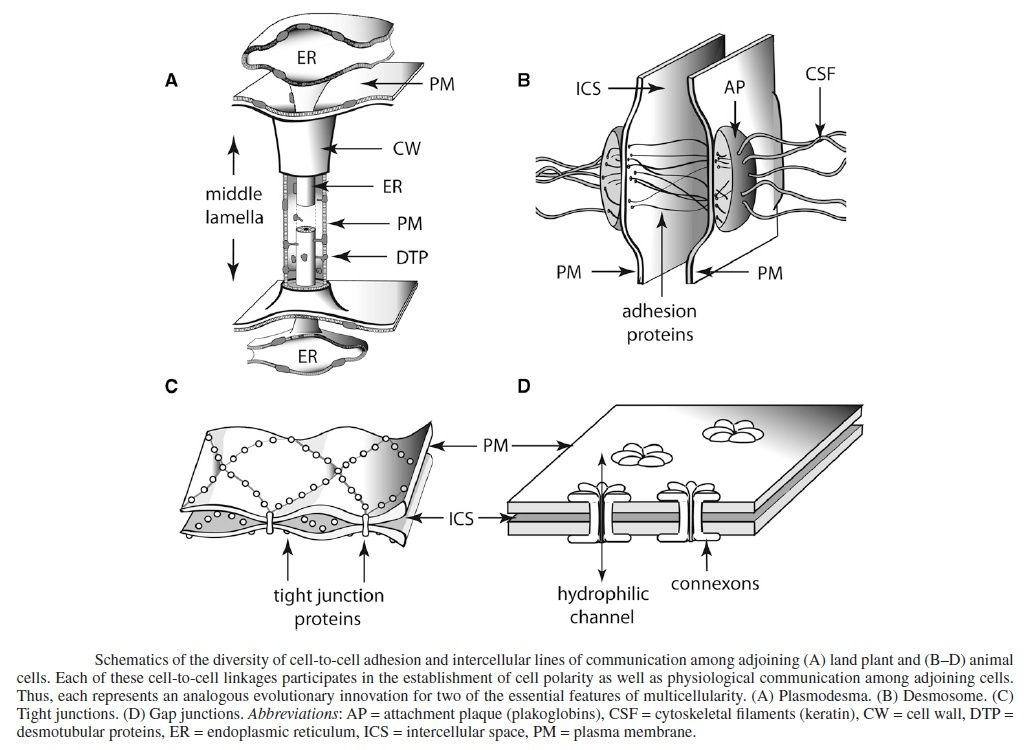
The cells had not only to hold together, but important mechanisms to stick the cells together had to emerge, that is, the ability of individual cells to associate in precise patterns to form tissues, organs, and organ systems requires that individual cells be able to recognize, adhere to, and communicate with each other.
Of all the social interactions between cells in a multicellular organism, the most fundamental are those that hold the cells together. The apparatus of cell junctions and the extracellular matrix is critical for every aspect of the organization, function, and dynamics of multicellular structures. Animal cells use specialized adhesion receptors to attach to one another. Many of these adhesion proteins are transmembrane proteins, which means the extracellular portion of these proteins can interact with the extracellular portion of similar proteins on the surface of a neighboring cell. Although diagrams of adhesive structures may suggest that they are static once assembled, they are anything but. Cells can dynamically assemble and disassemble adhesions in response to a variety of events. This seems to be an essential requirement for function right from the beginning of multicellularity. Many adhesion proteins are continuously recycled: Protein at the cell surface is internalized by endocytosis, and new protein is deposited at the surface via exocytosis. The molecular machines to exercise these functions, therefore, had to emerge together with adhesion proteins. Furthermore, cell adhesion is coordinated with other major processes, including
1.cell signaling,
2.cell movement,
3.cell proliferation, and
4.cell survival.
We now know that cell-cell adhesion receptors fall into a relatively small number of classes. They include
1.immunoglobulin superfamily (IgSF) proteins,
2.cadherins,
3.selectins, and, in a few cases,
4.integrins
In order to explain multicellularity, its origin must be explained.
https://reasonandscience.catsboard.com/t2187-cell-junctions-and-the-extracellular-matrix
My comment: Thus, the apparatus of cell junctions and the extracellular matrix is critical for every aspect of the organization, function, and dynamics of multicellular structures. The arise of adhesive junctions, tight junctions and gap junctions, and how they emerged is, therefore, a key factor to explain multi-cellular life. The cells of multi-cellular organisms detect and respond to countless internal and extracellular signals that control their growth, division, and differentiation during development, as well as their behavior in adult tissues. At the heart of all these communication systems are regulatory proteins that produce chemical signals, which are sent from one place to another in the body or within a cell, usually being processed along the way and integrated with other signals to provide clear and effective communication. The rise of these communication channels had to arise together with junction mechanisms in order to establish successful multicellular organisms. One feature without the other would not have provided success and advantage of survival.
The ability of cells to receive and act on signals from beyond the plasma membrane is fundamental to life. This conversion of information into a chemical change, signal transduction, is a universal property of living cells. Signal transductions are remarkably specific and exquisitely sensitive. Specificity is achieved by precise molecular complementarity between the signal and receptor molecules.
Question: signal transduction had to be present in the first living cells. How could the specificity of the signal molecule, and the precise fit on its complementary receptor have evolved ? or the Amplification, or the desensitization/adaptation, where the receptor activation triggers a feedback circuit that shuts off the receptor or removes it from the cell surface, once the signal got trough?
Three factors account for the extraordinary sensitivity of signal transducers: the high affinity of receptors for signal molecules, cooperativity (often but not always) in the ligand-receptor interaction, and amplification of the signal by enzyme cascades. The trigger for each system is different, but the general features of signal transduction are common to all: a signal interacts with a receptor; the activated receptor interacts with cellular machinery, producing a second signal or a change in the activity of a cellular protein; the metabolic activity of the target cell undergoes a change; and finally, the transduction event ends. This seems to be an irreducible system, requiring high content of pre-programming and advanced coding.
https://reasonandscience.catsboard.com/t2181-cell-communication-and-signaling-evidence-of-design
Question: how did the high affinity, cooperativity, and amplification have emerged? Is a preestablished convention not necessary, and so a mental process to yield the function? Is trial and error or evolution not a completely incapable mechanism to get this functional information system?
This is an important, essential and fundamental macroevolutionary change, and the explanation of macro-evolution must account for these changes, and provide feasible possible and likely ways through mutation and natural selection. Beside this, a shift on several levels of biological organization had to occur, providing a considerable advantage of survival, considering that for example one of the first cooperative steps required for the evolution of multicellularity in the volvocine algae was the development of the extracellular cell matrix from cell wall components, which can be metabolically costly to produce. But much more is required.
Ann Gauger: New genes and proteins must be invented. The cytoskeleton, Hox genes, desmosomes, cell adhesion molecules, growth factors, microtubules, microfilaments, neurotransmitters, whatever it takes to get cells to stick together, form different shapes, specialize, and communicate must all come from somewhere. Regulatory proteins and RNAs must be made to control the expression in time and space of these new proteins so that they all work together with existing pathways.In fact, in order for development to proceed in any organism, a whole cascade of coordinated genetic and biochemical events is necessary so that cells divide, change shape, migrate, and finally differentiate into many cell types, all in the right sequence at the right time and place. These cascades and the resulting cell divisions, shape changes, etc., are mutually interdependent. Interrupting one disrupts the others.
And last not least:
Like engineers carefully blowing up a bridge, cells have intricate, programmed suicide mechanisms. Without apoptosis, all multicellular life would be impossible. Good luck to proponents of evolution to explain how it emerged........
https://reasonandscience.catsboard.com/t2193-apoptosis-cell-s-essential-mechanism-of-programmed-suicide-points-to-design
Transition from from Unicellular to Multicellular Organisms
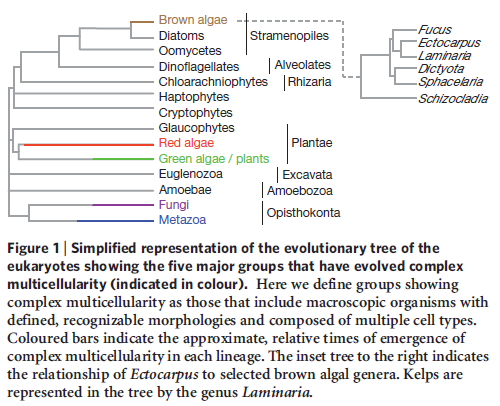
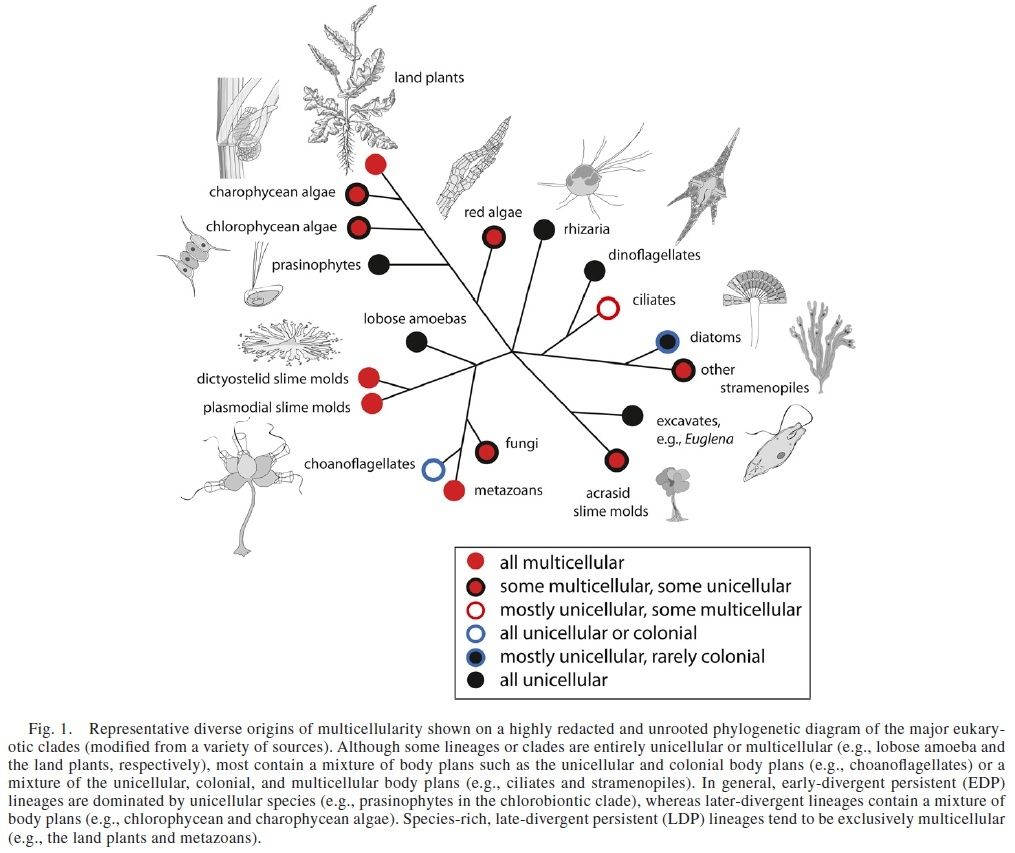
It is now widely accepted, on the basis of structural, biochemical, and molecular evidence, that the five major types of large, complex, multicellular organisms (i.e., red algae, brown algae, land plants, fungi, and animals) arose separately from different types of unicellular ancestors.
The evolution of multicellular life from simpler, unicellular microbes was a pivotal moment in the history of biology on Earth and has drastically reshaped the planet’s ecology. However, one mystery about multicellular organisms is why cells did not return back to single-celled life. 2
In the beginning there were supposedly single cells. Today, many millions of years later, most plants, animals, fungi, and algae are composed of multiple cells that work collaboratively as a single being. Despite the various ways these organisms achieved multicellularity, their conglomeration of cells operate cooperatively to consume energy, survive, and reproduce. But how did multicellularity evolve? Did it evolve once or multiple times? How did cells make the transition from life as a solo cell to associating and cooperating with other cells such that they work as a single, cohesive unit? 3
What we usually teach when discussing the origin and evolution of metazoans in geology class is that single-celled eukaryotes (organisms with a nucleus) like choanoplagellates learned how to clump together and chemically communicate to form sponges - thought to be one of the earliest multicellular organisms.4
While evolutionists can provide plenty of guesses about how multicellular organisms could have evolved from unicellular organisms, the fact is evolutionists have no idea how they actually evolved. And if evolutionists have no idea how they evolved, can we really be sure that they did evolve? Evolutionists scoff at such skepticism. It is unwarranted, they say, because evolution is a fact. 6
This transition is probably the easiest to understand.
Several colonial flagellated green algae provide a clue. These species are called colonial because they are made up simply of clusters of independent cells. If a single cell of Gonium, Pandorina, or Eudorina is isolated from the rest of the colony, it will swim away looking quite like a Chlamydomonas cell. Then, as it undergoes mitosis, it will form a new colony with the characteristic number of cells in that colony.
(The figures are not drawn to scale. Their sizes range from Chlamydomonas which is about 10 µm in diameter — little larger than a human red blood cell — to Volvox whose sphere is some 350 µm in diameter — visible to the naked eye.)
The situation in Pleodorina and Volvox is different. In these organisms, some of the cells of the colony (most in Volvox) are not able to live independently. If a nonreproductive cell is isolated from a Volvox colony, it will fail to reproduce itself by mitosis and eventually will die. What has happened? In some way, as yet unclear, Volvox has crossed the line separating simple colonial organisms from truly multicellular ones. Unlike Gonium, Volvox cannot be considered simply a colony of individual cells. It is a single organism whose cells have lost their ability to live independently. If a sufficient number of them become damaged, the entire sphere of cells will die.
What has Volvox gained? In giving up their independence, the cells of Volvox have become specialists. No longer does every cell carry out all of life's functions (as in colonial forms); instead certain cells specialize to carry out certain functions while leaving other functions to other specialists. In Volvox this process goes no further than having certain cells specialize for reproduction while others, unable to reproduce themselves, fulfill the needs for photosynthesis and locomotion.
In more complex multicellular organisms, the degree of specialization is carried much further. Each cell has one or two precise functions to carry out. It depends on other cells to carry out all the other functions needed to maintain the life of the organism and thus its own.
The specialization and division of labor among cells is the outcome of their history of differentiation. One of the great problems in biology is how differentiation arises among cells, all of which having arisen by mitosis, share the same genes. Link to a discussion of the solution.
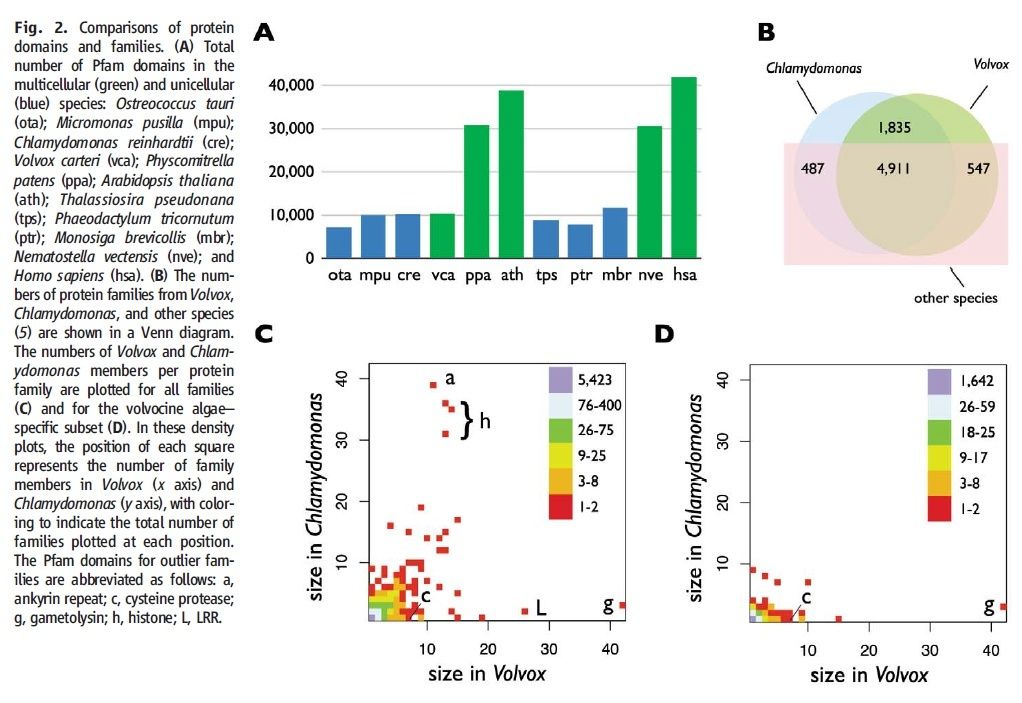
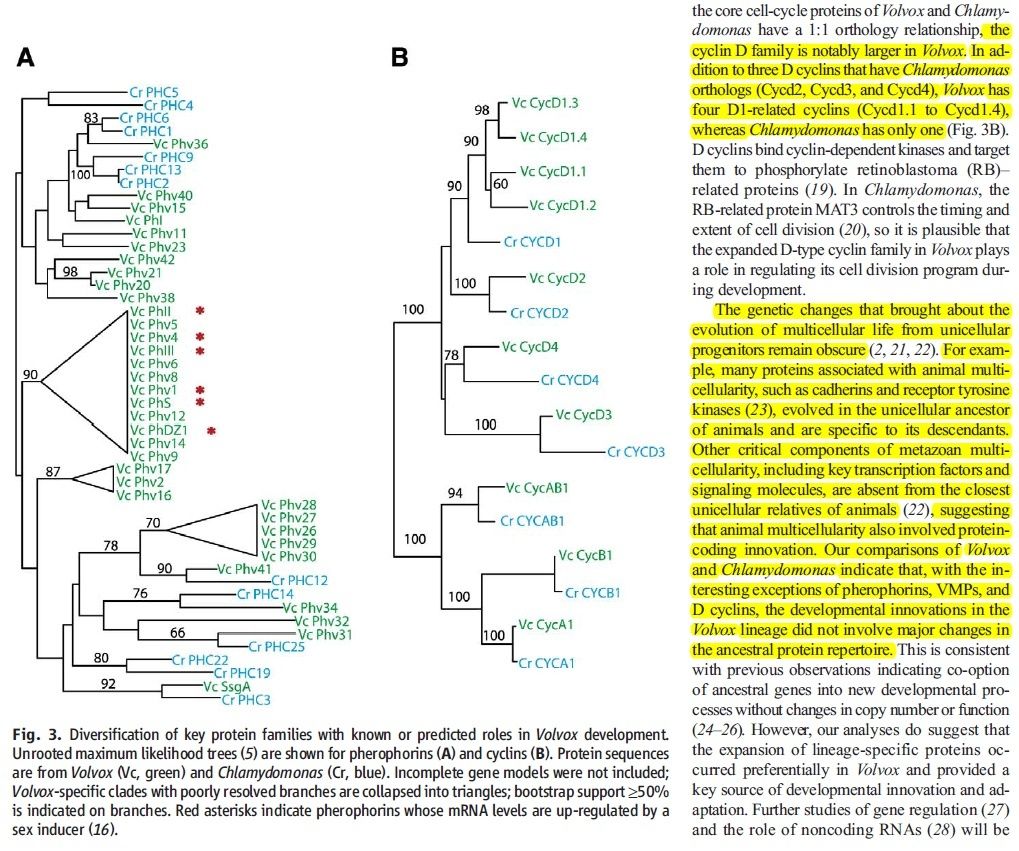
The genomes of both Chlamydomonas and Volvox have been sequenced. Although one is unicellular, the other multicellular, they have not only about the same number of protein-encoding genes (14,516 in Chlamydomonas, 14,520 in Volvox) but most of these are homologous. Volvox has only 58 genes that have no relatives in Chlamydomonas and even fewer unique mRNAs.
At one time, many of us would have expected that a multicellular organism like Volvox with its differentiated cells and complex life cycle would have had many more genes than a single-celled organism like Chlamydomonas. But that turns out not to be the case.
How to explain this apparent paradox? My guess is that just as we have seen in the evolution of animals [Examples], we are seeing here that the evolution of organismic complexity is not so much a matter of the evolution of new genes but rather the evolution of changes in the control elements (promoters and enhancers) that dictate how and where the basic tool kit of eukaryotic genes will be expressed .
The evidence is compelling that all these organisms are close relatives; that is, belong to the same clade. They illustrate how colonial forms could arise from unicellular ones and multicellular forms from colonial ones.
The Origin of Multicellularity 5
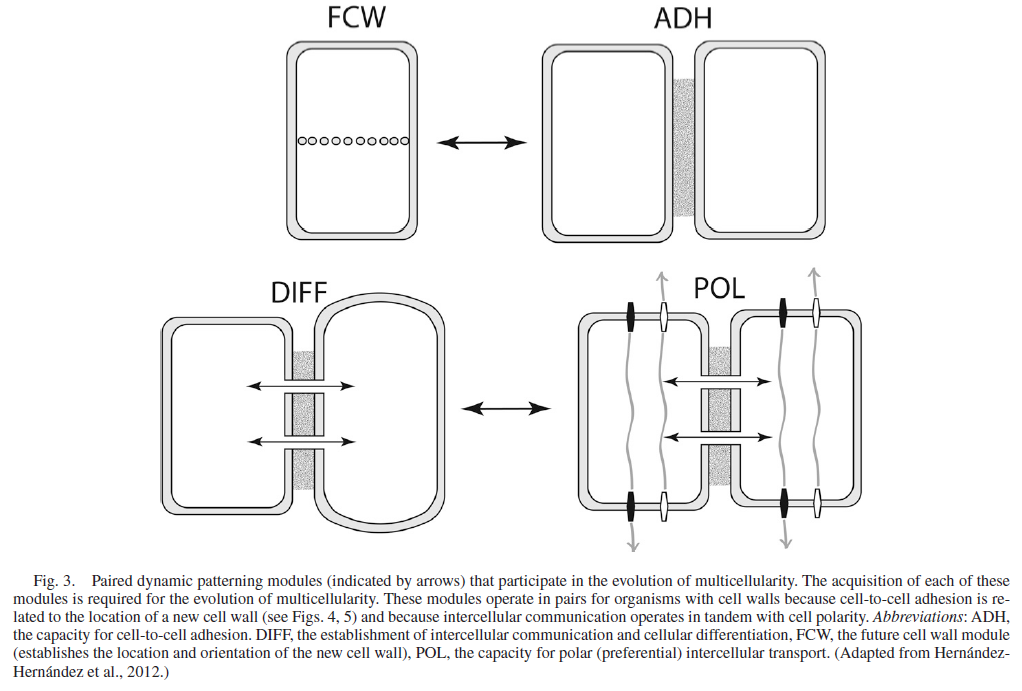
From an evolutionary perspective, support for the transition from unicellular (single cell) to multicellular organisms requires the emergence of several novel biochemical systems. Such systems include:
- pathways that transform cells from generalized to specialized forms during growth and development;
- mechanisms for the migration of cells relative to each other during growth and development;
- structures that support cell-cell adhesions;
- and mechanisms for cell-cell communication.
- All of these systems have to be in place and operate in an integrated fashion to support multicellularity.
The Origin of Multicellularity in Brown Algae
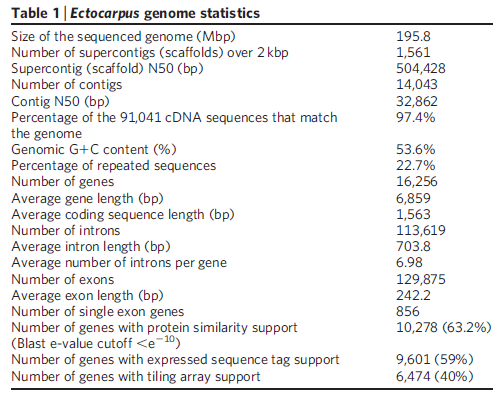
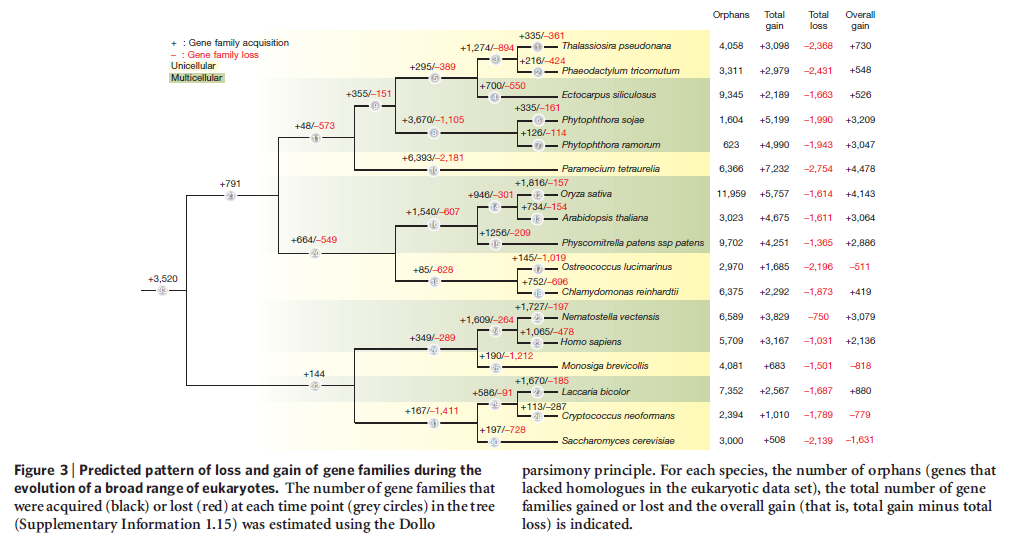
To gain understanding about how multicellularity originated in brown algae, a research consortium sequenced the entire genome (consisting of 214 million genetic letters) of the brown algae Ectocarpus siliculosus. They then compared it to closely related unicellular organisms. The research team noticed that multicellularity’s origin in the brown algae appears to correlate with the appearance of a rich ensemble of genes that encode proteins involved in signal transduction (processes that support cell-cell communication).
They also noted that this same collection of genes also correlates with the advent of multicellularity in plants and animals. According to the evolutionary paradigm, multicellularity arose independently in brown algae, plants, and animals. And the same biochemical systems that make multicellularity possible also appear to have arisen in each group of organisms on three separate occasions.
Implications for the Evolutionary Paradigm
Evolutionary processes are blind and undirected. Their very essence renders evolutionary outcomes unpredictable and nonrepeatable. According to the concept of historical contingency, espoused by late scientist Stephen Jay Gould in his book Wonderful Life, chance governs biological and biochemical evolution at its most fundamental level. Evolutionary pathways consist of a historical sequence of chance genetic changes operated on by natural selection, which also consists of chance components. Thus, if evolutionary events could be “rewound” and “replayed,” the outcome would be dramatically different every time. The inability of evolutionary processes to retrace the same path makes it highly unlikely that the same biological and biochemical designs would appear repeatedly throughout nature among unrelated organisms.
Yet it looks as if evolution has repeated itself, time and time again! Evolutionary biologists note that evolutionary processes frequently seem to converge independently on identical anatomical, physiological, behavioral, and biochemical systems. (As a case on point, go here and here to see two articles I wrote on this phenomenon some time ago.) Evolutionary biologists refer to repeated evolutionary outcomes as convergence.
In my book The Cell’s Design I document over one hundred examples of convergence at the biochemical level. On this basis I argue that if historical contingency truly reflects the nature of evolutionary processes, then the widespread occurrence of a broad range of biochemical systems emerging repeatedly raises significant questions about the validity of evolutionary explanations for life’s origin and diversity.
The repeated origin of multicellularity in brown algae, plants, and animals defies an evolutionary explanation, particularly since it involves the independent origin of virtually the same biochemical systems. But this new insight makes sense if a Creator responsible for the multi-member church was also responsible for assembling multicellular life-forms.
Birds and bats belong to different groups, with birds assigned to the class Aves and bats to the class Mammalia. 4According to the evolutionary paradigm, undirected natural processes yielded the identical outcome (wings, in this case) because the forces of selection channeled evolutionary pathways to the same endpoint.
This explanation doesn’t square up, however. If biological systems are the product of evolution, then the same biological systems should not recur throughout nature. Chance governs biological and biochemical evolution at its most fundamental level. Evolutionary pathways consist of a historical sequence of chance genetic changes operated on by natural selection, which, too, consists of chance components. The consequences are profound. If evolutionary events could be repeated, the outcome would be dramatically different every time. The inability of evolutionary processes to retrace the same path makes it highly unlikely that the same biological and biochemical designs should repeatedly appear throughout nature.
1) http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/A/AbioticSynthesis.html
2) http://www.astrobio.net/news-exclusive/multicellular-life-evolve/
3) http://www.eurekalert.org/pub_releases/2014-01/ajob-foc012414.php
4) http://www.reasons.org/articles/deja-vu-again-part-1-of-2
5) http://www.reasons.org/articles/repeated-origin-of-multicellularity-points-to-a-creator
6) http://darwins-god.blogspot.com.br/2010/03/does-evidence-support-evolution-or-does.html
7) http://physrev.physiology.org/content/87/4/1343
More:
The Multiple Origins of Complex Multicellularity

Last edited by Otangelo on Sun Oct 03, 2021 6:36 am; edited 35 times in total