The Porphobilinogen deaminase enzyme (PBGD) is involved in the third step of the heme biosynthetic pathway
http://reasonandscience.heavenforum.org/t1986-the-porphobilinogen-deaminase-enzyme-heme-and-chlorophyll-biosynthetic-pathway#3329
https://respondblogs.wordpress.com/2014/07/19/respondblog-new-empirical-evidence-pointing-away-from-darwinian-evolution/
In order to apprechiate the enormous complexity of each step of the biosynthesis pathway of heme ( and subsequently chlorophyll ), i decided to give a closer look at the Porphobilinogen deaminase enzyme. In my view it exemplifies brilliantly why evolution is not a potent cause in biology for macro evolutionary change ( of which i regard the arise of the hemoglobin and chlorophyll molecule ) . One reason is, there would simply not be enough time to evolve such important molecules and proteins , which appeard very early in history of life, its said before the split of prokaryotes and eukaryotes.
http://www.uniprot.org/uniprot/P08397
This enzyme catalyzes the head to tail condensation of four porphobilinogen molecules into the linear hydroxymethylbilane while releasing four ammonia molecules. 2) It assembles the four rings of PBG in a stepwise fashion in which the pyrrole ring A is first bound to the deaminase followed by rings B, C and finally D . The dipyrromethane cofactor ( A cofactor is a non-protein chemical compound that is required for the protein's biological activity. These proteins are commonly enzymes, and cofactors can be considered "helper molecules" that assist in biochemical transformations. ) , which arises from the autocatalytic coupling of two molecules of PBG, is covalently linked to the enzyme. The cofactor functions as a primer to which the four substrate molecules are sequentially attached but is not itself incorporated into the product, hydroxymethylbilane. 8 )
PBGD is highly complex and specified in its structure, using co-factors in order to catalyze the first and second step of the pathway.
Structure and function
Functionally, porphobilinogen deaminase catalyzes the loss of ammonia from the porphobilinogen monomer (deamination) and its subsequent polymerization to a linear tetrapyrrole, which is released as hydroxymethylbilane 3)

The first step is believed to involve an E1 elimination of ammonia from porphobilinogen, generating a carbocation intermediate
The second step : This intermediate is then attacked by the dipyrrole cofactor of porphobilinogen deaminase, which after losing a proton yields a trimer covalently bound to the enzyme .
The third step This intermediate is then open to further reaction with porphobilinogen (1 and 2 repeated three more times)
The fourth step Once a hexamer is formed, hydrolysis allows hydroxymethylbilane to be released, as well as cofactor regeneration
The reaction catalysed by PBGD involves the formation of preuroporphyrinogen, a linear tetrapyrrole (bilane), by the extension of an enzyme-bound DPM (dipyrromethane) cofactor that acts as a reaction primer 4)
Question: how could evolution have achieved the programming of this complex mechanism , and 4 step production of intermediate product, of which any of the 4 steps have any function by its own, and provided no survival advantage ? It had to program into the genome the specification for the production of the co-factor molecule, and its function of extension binding of intermediates during the catalytic reaction at the right place, at the right moment, in the right sequence. Does this not remember the methodology and intelligent planning of a intelligent mind rather than being the product of random mutations, and natural selection ? 1)
Porphobilinogen deaminase, dipyrromethane cofactor binding site (IPR022419)
Short name: Porphobilin_deaminase_cofac_BS
Description
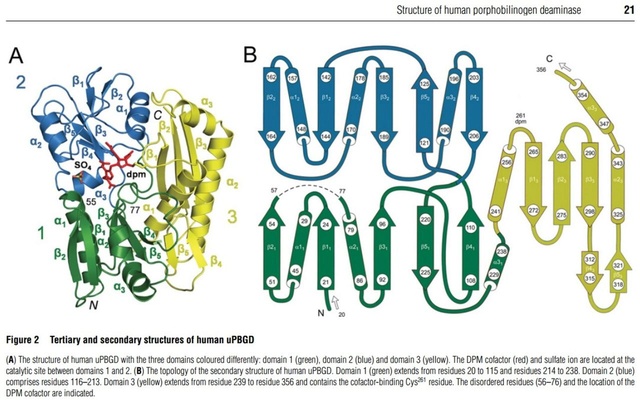
This entry represents the region around a cysteine residues that is conserved in porphobilinogen deaminases from various prokaryotic and eukaryotic sources. The sulphur atom of this cysteine residue has been shown in the Escherichia coli enzyme (gene hemC) to be bound to the dipyrromethane cofactor [PMID: 3196304]. Porphobilinogen deaminase covalently binds a dipyrromethane cofactor to which the PBG subunits are added in a stepwise fashion. Porphobilinogen deaminase has a three-domain structure. Domains 1 (N-terminal) and 2 are duplications with the same structure, resembling the transferrins and periplasmic binding proteins. The dipyrromethane cofactor is covalently linked to domain 3 (C-terminal), but is bound by extensive salt-bridges and hydrogen-bonds within the cleft between domains 1 and 2, at a position corresponding to the binding sites for small-molecule ligands in the analogous proteins [PMID: 1522882]. The enzyme has a single catalytic site, and the flexibility between domains is thought to aid elongation of the polypyrrole product in the active-site cleft of the enzyme. 5)
Questions : how did gene duplication, followed by random mutations and natural selection figure out to produze the PBG deaminase enzyme,, used as far as reported, exclusively in this pathway, so no co-option possible ? - that would produce this complex reaction, ( which is just the third in the whole heme and chlorophyll biosynthesis pathway of total 8 and 17 steps ) consisting in 4 highly coordenated , ordered, sequenced and complex steps, forming a geometrically correct tetrapyrrole, and repeat the first two steps in total 4 times ? and furthermore use co-enzyme molecules ? Could evolution be a feasable mechanism ? How did evolution be capable to produce the right genetic code and informational sequence ? The genetic code uses average over 300 thousand nucleotides in the sequence. How did evolution figure out to program the release at the end of the process of the hydroxymethylbilane enzyme from the reaction at the right time, after the linear hydroxymethylbilane product is catalized, and while releasing four ammonia molecules ? What adaptive value would the intermediate product provide ?
PBGD enzymes possesses several novel features. First, the initial protein translation product is an apoenzyme that has the ability to construct its own DPM cofactor using two of the pyrrole units of preuroporphyrinogen .
Question: How did it achieve the hability to do so ? evolution, or design ?
Secondly, each of the four substrate condensation steps occurs at a single catalytic site and the enzyme is therefore able to translocate the growing polypyrrole chain to vacate the substrate-binding site for the next incoming substrate.
Question: How did it achieve the hability to do this in a coordinated, ordered and systematic way ? evolution, or design
Thirdly, the enzyme can ‘count’ precisely to four and terminate the polymerization reaction by hydrolysis when the hexapyrrole chain (ES4) has been assembled 6)
Question: What does explain this feat better, design or evolution ? lets remember, that evolution has only mutation and natural selection on hand as mechanism to provide new functions . Heme or chlorophyll can only be produced, if the PBGD enzyme if fully developed and exercising its function. How did evolution be able to provide the right instructions in order of the enzyme to count precisely to four and how did it get this feat to be able to instruct to terminate the polimerization reaction at the right moment?
Porphobilinogen deaminase gene structure
The PBGD gene comprises 15 exons encoded within a 10-kilobase (kb) region of chromosome 11q23.3 7)
The porphobilinogen deaminase gene
The gene coding for PBGD is assigned to chromosome 11q24. The size of the gene is 10 kb of which 1.3 kb represents coding sequence. The genomic sequence is divided into 15 exons ranging from 39 and 438 bp and 14 introns ranging from 87 to 2913 bp (Figure 2.4.).
The mRNA of the isoform contains exons 1 and 3 to 15 coding for an enzyme of 361 amino acids
More recently, it has been shown that the hemoglobin gene precedes even the prokaryote/eukaryote split. 9)
Question: If the hemoglobin gene is that ancient, did it have enough time to evolve the complexity of the pathway, all enzymes and co factor molecules , and provide all the diverse instructions demonstrated here ?
The evolution of hemoglobin. 10)
e. It appears that the apparatus that sequesters oxygen in cells, possibly to protect them, is almost identical to the one that, in different contexts, exploits oxygen for its energy-generating potential. At first this apparatus was quite primitive, probably limited to a caged metal atom capable of binding oxygen or tearing away its electrons, which are used in metabolism. But this basic chemical apparatus grew increasingly complex through time and evolution. At some point the metal atom was fixed inside a kind of flat molecular cage called a porphyrin ring, and later that porphyrin ring became embedded in larger organic compounds called proteins. These organic compounds themselves became increasingly varied through time and evolution.
Question: Isnt that typical superfical pseudoscientic blabbering, based on just so guesswork without a shred of evidence to back up the claim ? Where is the detailled explanation about how the extremely complex biosynthesis pathway could have started and evolved ? That explanation is equal to a NO explanation..... just for the guillible without critical thinking. Yes, there is good reason to be skeptic towards that kind of information and assertions and guesswork.
Hemoglobins Connect Almost All Life on the Planet
We normally think of hemoglobin in the context of our own blood, comprising about 85% of the dry weight of red blood cells and a whopping 35% wet weight. For a protein, this is extremely concentrated. What surprises most people is the ubiquitousness and varied functions of hemoglobins in not just the animal world. Plants use hemoglobins, as do fungi, protists and bacteria. The gene encoding hemoglobin is thus very old, going back to the ancestor common to essentially all life on this planet. In fact, the only group for which hemoglobins have not been found are within the kingdom Archaea, considered to be the living relatives of the most ancient form of life we know about on this planet.
A great deal of speculation has gone into how hemoglobins arose in the first place. Perhaps it initially evolved in order to help protect the cell from the continually rising amounts of the toxin known as oxygen from photosynthetic organisms. Once cells began to use oxygen in respiratory processes, hemoproteins could act as electron-transfer agents or to scavenge oxygen for respiration. Gene duplications with mutation would allow other redox agents (like the cytochrome b molecules) to evolve from the ancestral hemoprotein gene. Once multicellular organisms arose, hemoglobins could then evolve into their current function of oxygen transporers. At an early stage of evolution, it has been proposed that worm-like animals had large polymeric aggreggations of hemoglobin subunits circulating through primitive circulatory systems. Next would be monomeric hemoglobin in blood cells functioning as oxygen storage. 9)
1) http://www.ncbi.nlm.nih.gov/pubmed/3079571
The cofactor is involved in the binding of intermediates during the catalytic reaction but is not incorporated into the product preuroporphyrinogen
2) http://en.wikipedia.org/wiki/Porphobilinogen_deaminase
3) http://scripts.iucr.org/cgi-bin/paper?S0907444912052134
4) http://www.biochemj.org/bj/420/0017/4200017.pdf
5) http://www.ebi.ac.uk/interpro/entry/IPR022419
6) http://www.biochemj.org/bj/420/0017/4200017.pdf
7) http://www.bloodjournal.org/content/bloodjournal/97/3/815.full.pdf?sso-checked=true
8 ) http://ethesis.helsinki.fi/julkaisut/laa/kliin/vk/mustajoki/review.html
9) https://shamelesslyatheist.wordpress.com/2008/10/11/evolution-hemoglobin/
10) http://zoology.wisc.edu/courses/611/Part2/Readings/5The_Evolution_of_Hemoglobin.pdf
Common objection is, that intelligent design , and irreducible complexity, is just argument of ignorance.
My response :
Is irreducible complexity merely an "argument from ignorance?
http://reasonandscience.heavenforum.org/t1983-is-irreducible-complexity-merely-an-argument-from-ignorance
Further readings :
THE AMAZING HEMOGLOBIN MOLECULE
http://reasonandscience.heavenforum.org/t1322-the-amazing-hemoglobin-molecule
http://reasonandscience.heavenforum.org/t1986-the-porphobilinogen-deaminase-enzyme-heme-and-chlorophyll-biosynthetic-pathway#3329
https://respondblogs.wordpress.com/2014/07/19/respondblog-new-empirical-evidence-pointing-away-from-darwinian-evolution/
In order to apprechiate the enormous complexity of each step of the biosynthesis pathway of heme ( and subsequently chlorophyll ), i decided to give a closer look at the Porphobilinogen deaminase enzyme. In my view it exemplifies brilliantly why evolution is not a potent cause in biology for macro evolutionary change ( of which i regard the arise of the hemoglobin and chlorophyll molecule ) . One reason is, there would simply not be enough time to evolve such important molecules and proteins , which appeard very early in history of life, its said before the split of prokaryotes and eukaryotes.

http://www.uniprot.org/uniprot/P08397
This enzyme catalyzes the head to tail condensation of four porphobilinogen molecules into the linear hydroxymethylbilane while releasing four ammonia molecules. 2) It assembles the four rings of PBG in a stepwise fashion in which the pyrrole ring A is first bound to the deaminase followed by rings B, C and finally D . The dipyrromethane cofactor ( A cofactor is a non-protein chemical compound that is required for the protein's biological activity. These proteins are commonly enzymes, and cofactors can be considered "helper molecules" that assist in biochemical transformations. ) , which arises from the autocatalytic coupling of two molecules of PBG, is covalently linked to the enzyme. The cofactor functions as a primer to which the four substrate molecules are sequentially attached but is not itself incorporated into the product, hydroxymethylbilane. 8 )
PBGD is highly complex and specified in its structure, using co-factors in order to catalyze the first and second step of the pathway.
Structure and function
Functionally, porphobilinogen deaminase catalyzes the loss of ammonia from the porphobilinogen monomer (deamination) and its subsequent polymerization to a linear tetrapyrrole, which is released as hydroxymethylbilane 3)

The first step is believed to involve an E1 elimination of ammonia from porphobilinogen, generating a carbocation intermediate
The second step : This intermediate is then attacked by the dipyrrole cofactor of porphobilinogen deaminase, which after losing a proton yields a trimer covalently bound to the enzyme .
The third step This intermediate is then open to further reaction with porphobilinogen (1 and 2 repeated three more times)
The fourth step Once a hexamer is formed, hydrolysis allows hydroxymethylbilane to be released, as well as cofactor regeneration
The reaction catalysed by PBGD involves the formation of preuroporphyrinogen, a linear tetrapyrrole (bilane), by the extension of an enzyme-bound DPM (dipyrromethane) cofactor that acts as a reaction primer 4)
Question: how could evolution have achieved the programming of this complex mechanism , and 4 step production of intermediate product, of which any of the 4 steps have any function by its own, and provided no survival advantage ? It had to program into the genome the specification for the production of the co-factor molecule, and its function of extension binding of intermediates during the catalytic reaction at the right place, at the right moment, in the right sequence. Does this not remember the methodology and intelligent planning of a intelligent mind rather than being the product of random mutations, and natural selection ? 1)
Porphobilinogen deaminase, dipyrromethane cofactor binding site (IPR022419)
Short name: Porphobilin_deaminase_cofac_BS
Description
This entry represents the region around a cysteine residues that is conserved in porphobilinogen deaminases from various prokaryotic and eukaryotic sources. The sulphur atom of this cysteine residue has been shown in the Escherichia coli enzyme (gene hemC) to be bound to the dipyrromethane cofactor [PMID: 3196304]. Porphobilinogen deaminase covalently binds a dipyrromethane cofactor to which the PBG subunits are added in a stepwise fashion. Porphobilinogen deaminase has a three-domain structure. Domains 1 (N-terminal) and 2 are duplications with the same structure, resembling the transferrins and periplasmic binding proteins. The dipyrromethane cofactor is covalently linked to domain 3 (C-terminal), but is bound by extensive salt-bridges and hydrogen-bonds within the cleft between domains 1 and 2, at a position corresponding to the binding sites for small-molecule ligands in the analogous proteins [PMID: 1522882]. The enzyme has a single catalytic site, and the flexibility between domains is thought to aid elongation of the polypyrrole product in the active-site cleft of the enzyme. 5)
Questions : how did gene duplication, followed by random mutations and natural selection figure out to produze the PBG deaminase enzyme,, used as far as reported, exclusively in this pathway, so no co-option possible ? - that would produce this complex reaction, ( which is just the third in the whole heme and chlorophyll biosynthesis pathway of total 8 and 17 steps ) consisting in 4 highly coordenated , ordered, sequenced and complex steps, forming a geometrically correct tetrapyrrole, and repeat the first two steps in total 4 times ? and furthermore use co-enzyme molecules ? Could evolution be a feasable mechanism ? How did evolution be capable to produce the right genetic code and informational sequence ? The genetic code uses average over 300 thousand nucleotides in the sequence. How did evolution figure out to program the release at the end of the process of the hydroxymethylbilane enzyme from the reaction at the right time, after the linear hydroxymethylbilane product is catalized, and while releasing four ammonia molecules ? What adaptive value would the intermediate product provide ?
PBGD enzymes possesses several novel features. First, the initial protein translation product is an apoenzyme that has the ability to construct its own DPM cofactor using two of the pyrrole units of preuroporphyrinogen .
Question: How did it achieve the hability to do so ? evolution, or design ?
Secondly, each of the four substrate condensation steps occurs at a single catalytic site and the enzyme is therefore able to translocate the growing polypyrrole chain to vacate the substrate-binding site for the next incoming substrate.
Question: How did it achieve the hability to do this in a coordinated, ordered and systematic way ? evolution, or design
Thirdly, the enzyme can ‘count’ precisely to four and terminate the polymerization reaction by hydrolysis when the hexapyrrole chain (ES4) has been assembled 6)
Question: What does explain this feat better, design or evolution ? lets remember, that evolution has only mutation and natural selection on hand as mechanism to provide new functions . Heme or chlorophyll can only be produced, if the PBGD enzyme if fully developed and exercising its function. How did evolution be able to provide the right instructions in order of the enzyme to count precisely to four and how did it get this feat to be able to instruct to terminate the polimerization reaction at the right moment?
Porphobilinogen deaminase gene structure
The PBGD gene comprises 15 exons encoded within a 10-kilobase (kb) region of chromosome 11q23.3 7)
The porphobilinogen deaminase gene
The gene coding for PBGD is assigned to chromosome 11q24. The size of the gene is 10 kb of which 1.3 kb represents coding sequence. The genomic sequence is divided into 15 exons ranging from 39 and 438 bp and 14 introns ranging from 87 to 2913 bp (Figure 2.4.).
The mRNA of the isoform contains exons 1 and 3 to 15 coding for an enzyme of 361 amino acids
More recently, it has been shown that the hemoglobin gene precedes even the prokaryote/eukaryote split. 9)
Question: If the hemoglobin gene is that ancient, did it have enough time to evolve the complexity of the pathway, all enzymes and co factor molecules , and provide all the diverse instructions demonstrated here ?
The evolution of hemoglobin. 10)
e. It appears that the apparatus that sequesters oxygen in cells, possibly to protect them, is almost identical to the one that, in different contexts, exploits oxygen for its energy-generating potential. At first this apparatus was quite primitive, probably limited to a caged metal atom capable of binding oxygen or tearing away its electrons, which are used in metabolism. But this basic chemical apparatus grew increasingly complex through time and evolution. At some point the metal atom was fixed inside a kind of flat molecular cage called a porphyrin ring, and later that porphyrin ring became embedded in larger organic compounds called proteins. These organic compounds themselves became increasingly varied through time and evolution.
Question: Isnt that typical superfical pseudoscientic blabbering, based on just so guesswork without a shred of evidence to back up the claim ? Where is the detailled explanation about how the extremely complex biosynthesis pathway could have started and evolved ? That explanation is equal to a NO explanation..... just for the guillible without critical thinking. Yes, there is good reason to be skeptic towards that kind of information and assertions and guesswork.
Hemoglobins Connect Almost All Life on the Planet
We normally think of hemoglobin in the context of our own blood, comprising about 85% of the dry weight of red blood cells and a whopping 35% wet weight. For a protein, this is extremely concentrated. What surprises most people is the ubiquitousness and varied functions of hemoglobins in not just the animal world. Plants use hemoglobins, as do fungi, protists and bacteria. The gene encoding hemoglobin is thus very old, going back to the ancestor common to essentially all life on this planet. In fact, the only group for which hemoglobins have not been found are within the kingdom Archaea, considered to be the living relatives of the most ancient form of life we know about on this planet.
A great deal of speculation has gone into how hemoglobins arose in the first place. Perhaps it initially evolved in order to help protect the cell from the continually rising amounts of the toxin known as oxygen from photosynthetic organisms. Once cells began to use oxygen in respiratory processes, hemoproteins could act as electron-transfer agents or to scavenge oxygen for respiration. Gene duplications with mutation would allow other redox agents (like the cytochrome b molecules) to evolve from the ancestral hemoprotein gene. Once multicellular organisms arose, hemoglobins could then evolve into their current function of oxygen transporers. At an early stage of evolution, it has been proposed that worm-like animals had large polymeric aggreggations of hemoglobin subunits circulating through primitive circulatory systems. Next would be monomeric hemoglobin in blood cells functioning as oxygen storage. 9)
1) http://www.ncbi.nlm.nih.gov/pubmed/3079571
The cofactor is involved in the binding of intermediates during the catalytic reaction but is not incorporated into the product preuroporphyrinogen
2) http://en.wikipedia.org/wiki/Porphobilinogen_deaminase
3) http://scripts.iucr.org/cgi-bin/paper?S0907444912052134
4) http://www.biochemj.org/bj/420/0017/4200017.pdf
5) http://www.ebi.ac.uk/interpro/entry/IPR022419
6) http://www.biochemj.org/bj/420/0017/4200017.pdf
7) http://www.bloodjournal.org/content/bloodjournal/97/3/815.full.pdf?sso-checked=true
8 ) http://ethesis.helsinki.fi/julkaisut/laa/kliin/vk/mustajoki/review.html
9) https://shamelesslyatheist.wordpress.com/2008/10/11/evolution-hemoglobin/
10) http://zoology.wisc.edu/courses/611/Part2/Readings/5The_Evolution_of_Hemoglobin.pdf
Common objection is, that intelligent design , and irreducible complexity, is just argument of ignorance.
My response :
Is irreducible complexity merely an "argument from ignorance?
http://reasonandscience.heavenforum.org/t1983-is-irreducible-complexity-merely-an-argument-from-ignorance
Further readings :
THE AMAZING HEMOGLOBIN MOLECULE
http://reasonandscience.heavenforum.org/t1322-the-amazing-hemoglobin-molecule

