https://reasonandscience.catsboard.com/t1322-the-amazing-hemoglobin-molecule
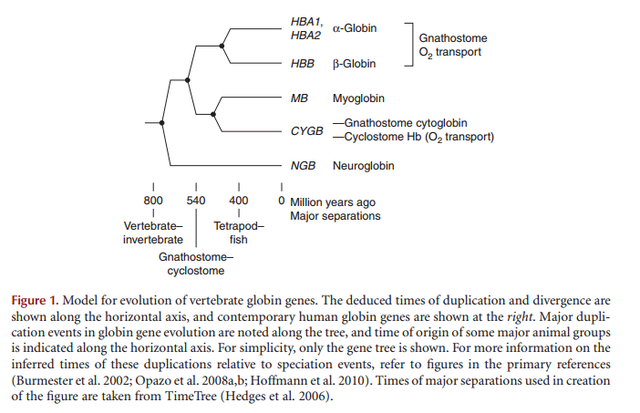
The most ancient heme domain is the globin-like fold and was certainly present in LUCA, the Last Universal Common Ancestor. 2
The argument by hemoglobin
1. The hemoglobin is a protein made of 564 amino acids.
2. The hemoglobin’s three-dimensional structure; the amino acid sequence and the 4 iron atoms in the central region of the hemoglobin are all together enabling the special function of the hemoglobin - the transfer of the oxygen.
3. An alteration of any part of this structure of hemoglobin would cause inability to execute its duty of carrying oxygen.
4. Conclusively, such a structure is a proof of perfect design.
5. Behind a design, there is an intelligent designer.
6. That designer is God.
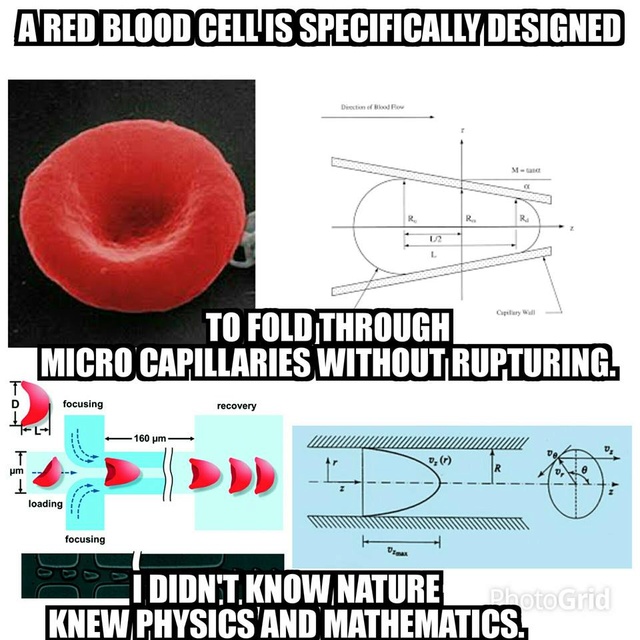
Technological Ingenuity in Red Blood Cells 1
Each mm3 (= 1 ul = 1 microliter) of our blood contains five million red blood cells; so there are 150 million of them in each drop of blood. These highly specialized cells perform functions vital to life.
• Throughout their 120-day lifetime, while circulating through the lungs, they are refueled with oxygen 175,000 times, while simultaneously offloading carbon dioxide, the waste product of oxidation.
• Red blood cells are so tiny that they can squeeze through narrow capillaries to reach every part of the body.
• Our body produces two million new red cells every second and each cell is rich in hemoglobin, a remarkably complex chemical compound.
Hemoglobin is used for transporting oxygen, even during development of the embryo. Up to about the third month of pregnancy, the embryo's oxygen needs are distinctly different from those in the ensuing fetal stage, which are different again from the needs of the infant and adult. All three stages— embryo, fetus, and adult—require the production of chemically different forms of hemoglobin. Shortly before birth, for example, the body's 'factories' start switching to top production mode of the third (adult) type of hemoglobin. These three types of hemoglobin could not have arisen by trial-and-error evolutionary processes because none of the other mutant forms of the hemoglobin molecule could carry enough oxygen and would thus be deadly. Even if the right forms of hemoglobin were to somehow arise to supply the first two stages, but without the genetic coding to produce the third form, the outcome would still be certain death. Each of these three stages of our development requires fundamentally different DNA coding to produce each of the three different hemoglobin molecules.
Further, each set of different DNA coding and its biomachinery that synthesizes the hemoglobin molecules must be switched on and off at the right point in time. Where did such a complex system of information-controlled machinery come from? All conceivable evolutionary explanations fail miserably because any partially completed transitional stage required by evolution would not permit the organism to survive. The whole complex of
information and machinery must be present and functional from the start. This concept of 'irreducible complexity' also applies to the immune system and to the flagellum that many bacteria use to propel themselves. In each case, the organisms 'on the way' to their completed state would not have been able to survive. A more obvious explanation is that this information-controlled machinery was initially complete—something only possible if a wise Creator conceived and made everything fully functional in the beginning.
The heme biosynthesis pathway is irreducible complex.
Heme biosynthesis is a complex pathway with 8 highly specific steps, of which 6 steps are used by specific enzymes uniquely in this pathway.
The pathway must go all the way through, otherwise, heme is not synthesized.
Therefore, the heme biosynthesis pathway is irreducibly complex.
Questions:
What good would there be, if the pathway would go only up to the 7th step? none
What good would there be, if the pathway would go all the way through the 8th step? Heme would be produced, BUT :
What good for survival would there be for Heme by its own, if not fully embedded in the globin proteins? none.
What good would there be for red blood cells without hemoglobin, transporting oxygen to the cells in the body? none, transporting oxygen is essential for the whole process. I conclude therefore that the heme biosynthesis pathway is irreducibly complex, and could not have evolved upon mutation and natural selection.
I mentioned that some enzymes have to be imported into the mitochondrion. These enzymes contain special protein sequences called targeting signals that direct them to the right place. So the next question: is globin targeted to the mitochondrion? No - it is synthesized on ribosomes, attached to the Golgi apparatus in the cytoplasm and it stays there. Some of the haem made in the mitochondrion is used by mitochondrial proteins called cytochromes, but the rest is exported back outside where it can attach to the globin protein. Have a look at these Wikipedia pages: heme and porphyrin, for some more details. Porphyrins, by the way, are intermediates in haem synthesis that also have the tetrapyrrole structure.
Researchers have done experiments in which they synthesized globin protein chains to see at what point the haem attached. It can attach when about 80-90 amino acids have emerged from the ribosome - in other words, it attaches to the "nascent chain" as the protein is being synthesized. One of the mysteries that we don't fully understand is how the hemoglobin assembles itself properly - so as it has 2 alpha chains and 2 beta chains each with a hemoglobin attached.
Question: for what reason would evolution try to assemble the heme to the globin? what survival advantage would there be provided by a globin without the heme? and what advantage of the heme without the globin?
Dr. Kofahl, a chemist, observes:
‘A good example of alleged molecular homology is afforded by the a- and b-hemoglobin molecules of land vertebrates including man. These supposedly are homologous with an ancestral myoglobin molecule similar to human myoglobin. Two a- and two b-hemoglobin associate together to form the marvelous human hemoglobin molecule that carries oxygen and carbon dioxide in our blood. But myoglobin acts as single molecules to transport oxygen in our muscles. Supposedly, the ancient original myoglobin molecules slowly evolved along two paths until the precisely designed a- and b-hemoglobin molecules resulted in that function only when linked together in groups of four to work in the blood in a much different way under very different conditions from myoglobin in the muscle cells. What we have today in modern myoglobin and hemoglobin molecules are marvels of perfect designs for special, highly demanding tasks. Is there any evidence that intermediate, half-evolved molecules could have served useful functions during this imaginary evolutionary change process, or that any creature could survive with them in its blood? There is no such information. Modern vertebrates can tolerate very little variation in these molecules. Thus, the supposed evolutionary history of the allegedly homologous globin molecules is a fantasy, not science.’
https://www.awesomestories.com/asset/view/Breathing-How-Oxygen-Travels-in-the-Body0
http://reasonandscience.heavenforum.org/post?p=1859&mode=editpost
http://creationsmarvels.blogspot.com.br/2013/02/the-amazing-hemoglobin-molecule-miracle.html
“Breathing seems so simple, yet it appears as if this elementary manifestation of life owes its existence to the interplay of many kinds of atoms in a giant molecule of vast complexity.”
Max F. Perutz, a sharer of the Nobel Prize in 1962 for his studies of the hemoglobin molecule.
Breathing could not keep us alive if it were not for the human hemoglobin molecule, a complex molecular masterpiece of design. The hemoglobin that is inside each of our 30 trillion red blood cells transports the oxygen from the lungs to the tissues throughout the body. Without hemoglobin, we would die almost instantly. But why?
see here :
http://reasonandscience.heavenforum.org/t1439-atp-synthase#2089
Question: How do hemoglobin molecules manage to pick up tiny oxygen molecules at the right time, hold them safely until the right time, and release them at the right time? Several amazing feats of molecular engineering are required. What use would there be to do this " shuttle " service, if cells and their energy requirement were not existing already? Mitochondria, the energy factory in the cell, and ATP synthase rotary motors had to exist already, otherwise, hemoglobin would have no function. So had both not to be in place right from the beginning, and together? how could they have evolved separately?
Tiny Molecular “Taxis”
You might think of each hemoglobin molecule in a cell as a tiny four-door taxi, with room for exactly four “passengers.” This molecular taxi does not require a driver since it is riding inside a red blood cell, which could be described as a traveling container full of these hemoglobin molecules.
Question: what good would blood be for without red blood cells and hemoglobin? no use. (Channichthyidae fish
http://en.wikipedia.org/wiki/Channichthyidae
is an exception because of their low metabolic rates and the high solubility of oxygen in water at the low temperatures of their environment.) What good would blood be for without the veins, and the heart, and the lungs, and the kidneys ? no use. So can we not say these are interdependent? did they not have to be there and fully developed right from the beginning ? each organ, and the blood? how could evolution manage to evolve all the organs and blood at the exact same time ? how could it manage to bring all parts together and interconnect them in the right manner? how could it manage to find all required multiple parts? isn't it rational to conclude that is a task, that evolution isn't able to do?
The journey for a hemoglobin molecule begins when red blood cells arrive at the alveoli of the lungs—the “airport.”
Alveoli are tiny organs that help our body parts get the oxygen that we breathe in and get rid of the carbon dioxide we don't need

As we inhale air into our lungs, the huge crowds of tiny recently arrived oxygen molecules start looking for a ride in a taxi. These molecules quickly diffuse into red blood cells, the “containers.” Oxygen in the lungs passes through the thin-walled blood vessels and into the red blood cells, where it binds to the hemoglobin, turning it into the bright red oxy-hemoglobin. The blood then passes around the body until it reaches cells and tissues which require oxygen to sustain their processes.At this point, the doors of the hemoglobin taxis within each cell are closed. However, it does not take long before a determined oxygen molecule in the bustling crowd squeezes in and takes a seat in a hemoglobin taxi.
Now, something very interesting happens. Inside the red cell, the hemoglobin molecule begins to change its shape. All four “doors” of the hemoglobin taxi begin to open automatically as the first passengers get in, which allows the remaining passengers to hop aboard more easily. This process, called cooperativity, is so efficient that in the time it takes to draw a single breath, 95 percent of the “seats” in all the taxis in a red blood cell are taken. Together the more than one-quarter of a billion hemoglobin molecules in a single red blood cell can carry about a billion oxygen molecules! Soon the red blood cell containing all these taxis is off to deliver its precious supply of oxygen to body tissues that need it.
Question: What came before: the body tissues, or the hemoglobin, blood, veins, heart, lungs etc. ? Body tissues without hemoglobin would have no oxygen supply. Hemoglobin without body tissues to supply with oxygen would have no function... Isn't there an interdependent relationship? Had both not to be there right from the beginning?
But, you might wonder, ‘What keeps oxygen atoms inside the cell from getting out prematurely?’
The answer is that inside each hemoglobin molecule, oxygen molecules attach to waiting atoms of iron. You have probably seen what happens when oxygen and iron get together in the presence of water. The result is usually iron oxide, rust. When iron rusts, the oxygen is locked up permanently in a crystal. So how does the hemoglobin molecule manage to combine and uncombine iron and oxygen in the watery environment of the red blood cell without producing rust?
Taking a Closer Look
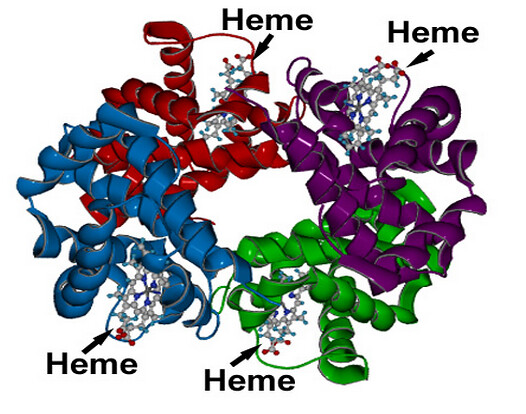
To answer that question, let us take a closer look at the hemoglobin molecule. It is made up of some 10,000 atoms of hydrogen, carbon, nitrogen, sulfur, and oxygen that are carefully assembled around just 4 atoms of iron. Why do four iron atoms need so much support?
First, the four iron atoms are electrically charged and must be carefully controlled. Charged atoms, which are called ions, can do a lot of damage inside cells if they get loose. So each of the four iron ions is secured in the middle of a protective rigid plate.
Question: How did evolution manage to evolve the mechanism to secure the iron ions on the middle of the protective rigid plate ? trial and error ? Why should it do so and try ?
Next, the four plates are carefully fitted into the hemoglobin molecule in such a way that oxygen molecules can get to the iron ions but water molecules cannot get to them. Without water, rust crystals are unable to form.
Question : Isnt that a remarkable feat ? That seems far more likely the engeneering of a super intellect that manages these arrangements at atomic scales, rather than unguided random genetic mutation lucky accident event providing such a highly elaborated mechanism, essential for many life forms.
By itself the iron in the hemoglobin molecule cannot bind and unbind oxygen. Yet, without the four charged iron atoms, the rest of the hemoglobin molecule would be useless. Only when these iron ions are perfectly fitted into the hemoglobin molecule can the transport of oxygen through the bloodstream occur.
Releasing the Oxygen
https://www.youtube.com/watch?v=WXOBJEXxNEo
As a red blood cell leaves the arteries and moves into the tiny capillaries deep in the body tissues, the environment around the red blood cell changes. Now the environment is warmer than in the lungs, and there is less oxygen and more acidity from the carbon dioxide surrounding the cell. These signals tell the hemoglobin molecules, or taxis, inside the cell that it is time to release their precious passengers, oxygen.
When the oxygen molecules get out of the hemoglobin molecule, it changes its shape once more. The change is just enough to “close the doors” and leave the oxygen outside, where it is most needed. Having the doors shut also prevents the hemoglobin from transporting any stray oxygen on the way back to the lungs. Instead, it readily picks up carbon dioxide for the return trip.
Question: How did it " learn " to bring carbon dioxide back to the lungs ?
Soon the deoxygenated red blood cells are back in the lungs, where the hemoglobin molecules will release the carbon dioxide and be recharged with life-sustaining oxygen—a process that is repeated many thousands of times during a red blood cell’s life span of about 120 days.
Clearly, hemoglobin is no ordinary molecule. It is, as stated at the beginning of this article, “a giant molecule of vast complexity.” Surely, this is awe inspiring, brilliant and meticulous microengineering that makes life possible, which demands a adequate explanation of how it came to be.
The biosynthesis pathway of hemoglobin is irreducible complex
Hemoglobin is a globular molecule which is made up of four subunits. Each subunit contains heme (an iron-containing porphyrin derivative). Each heme molecule is conjugated to a polypeptide which is called the globin. In each hemoglobin molecule there are 4 chains of polypeptides (2 pairs). In hemoglobin A, which is normal adult human hemoglobin, the two polypeptides are called α chains and the other two, β chains.

How is the heme molecule attached to the globin protein in the synthesis process?
http://www.quora.com/How-is-the-heme-molecule-attached-to-the-globin-protein
The answer is coordinate-covalent bonds, along with other forces.
Hemoglobin is a globular molecule which is made up of four subunits. Each subunit contains heme (an iron-containing porphyrin derivative). Each heme molecule is conjugated to a polypeptide which is called the globin. In each hemoglobin molecule there are 4 chains of polypeptides (2 pairs). In hemoglobin A, which is normal adult human hemoglobin, the two polypeptides are called α chains and the other two, β chains.
http://www.madsci.org/posts/archives/2006-11/1163441912.Bc.r.html
Haem biosynthesis and haemoglobin assembly are very complicated biochemical pathways and scientists still do not understand fully all of the steps.
Synthesis of Hemoglobin
http://www.madsci.org/posts/archives/2006-11/1163441912.Bc.r.html
Hemoglobin (Hb) is synthesized in a complex series of steps in the mitochondria and the cytosol of immature red blood cells, while the globin protein parts are synthesized by ribosomes in the cytosol. Synthesis starts in the mitochondria because they supply a molecule called succinyl-CoA, which is a building block for the haem. However when the haem is partially built, it is moved out into the cytoplasm. Some more reactions occur, then the molecule moves back to the mitochondrion where it is finished and the iron is added.
That seems very complex - why does the synthesis occur in 2 locations? The answer is that it allows the cell to regulate haem synthesis. The mitochondrial enzymes required for haem synthesis have to be imported into the mitochondrion from the cytoplasm, where they are made on ribosomes. If the cell has high levels of haem and doesn't need to make any more, the haem actually inhibits this import process. There is a second level of regulation too - if haem levels are high in the cytoplasm, transcription of the genes for haem synthesis is also inhibited. So by using 2 locations for synthesis, the cell can fine-tune the regulation of haem production. Pretty clever hey?
Question: how did the cell learn to fine tune haem production ?
Chemical steps in the formation of hemoglobin:
2α Ketoglutonic acid(it comes from creb's cycle) + 2 glycine → pyrrole
4 pyrrole → protoporphyrine
porphyrine + Fe+ → heme
4 heme + 4 polypeptide chain(2α + 2β → 1 hemoglobin molecule
http://en.wikipedia.org/wiki/Porphyrin
follwing the pathway :

Globin synthesis
Gene Duplication and the Origin of Novel Biological Information: A Case Study of the Globins
http://www.uncommondescent.com/evolution/gene-duplication-and-the-origin-of-novel-biological-information-a-case-study-of-the-globins/
there are at least five difficulties associated with the evolution of the globins by virtue of gene duplication and divergence. These are:
The question of the adaptive value of proposed intermediates.
Complementary changes involving the regulation of gene expression.
The time constraints associated with finding a selectable function for the duplicated copy.
The fragility problem.
Problems of convergence.
http://sickle.bwh.harvard.edu/hbsynthesis.html
Hemoglobin synthesis requires the coordinated production of heme and globin. Heme is the prosthetic group that mediates reversible binding of oxygen by hemoglobin. Globin is the protein that surrounds and protects the heme molecule.
Question : how could evolution coordenate its production ?
Heme is synthesized in a complex series of steps involving enzymes in the mitochondrion and in the cytosol of the cell
Question: How did these complex series of steps evolve? how and why did the involving enzymes evolve? The enzymes, the mitochondrion, and the cytosol had to be present, in order to synthesize hemoglobin.
The synthesis of heme is a complex process that involves multiple enzymatic steps. The process begins in the mitochondrion with the condensation of succinyl-CoA and glycine to form 5-aminolevulinic acid. A series of steps in the cytoplasm produces coproporphrynogen III, which re-enters the mitochondrion. The final enzymatic steps produce heme.
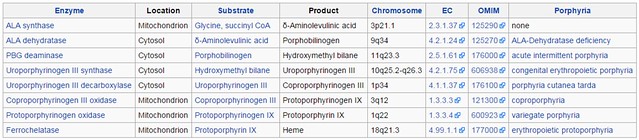
Following the enzymes involved :
ALA synthase
http://en.wikipedia.org/wiki/Aminolevulinic_acid_synthase
ALA synthase (EC 2.3.1.37), or ALAS, catalyzes the synthesis of D-Aminolevulinic acid (ALA) the first common precursor in the biosynthesis of all tetrapyrroles such as hemes, cobalamins and chlorophylls. ( therefore its not unique in this pathway. co-option from other biological systems is theoretically possible )
ALA dehydratase
http://en.wikipedia.org/wiki/Porphobilinogen_synthase
All natural tetrapyrroles, including hemes, chlorophylls and vitamin B12, share porphobilinogen as a common precursor. ( therefore its not unique in this pathway. co-option from other biological systems is theoretically possible )
PBG deaminase
http://en.wikipedia.org/wiki/Porphobilinogen_deaminase
Porphobilinogen deaminase is involved in the third step of the heme and chlorophyll biosynthetic pathway.
Uroporphyrinogen III synthase
http://ghr.nlm.nih.gov/gene/UROS
This enzyme is involved in the production of a molecule called heme. ( therefore its unique in this pathway. No co-option from other biological systems is possible )
Uroporphyrinogen III decarboxylase
http://en.wikipedia.org/wiki/Uroporphyrinogen_III_decarboxylase
This gene encodes the fifth enzyme of the heme biosynthetic pathway. ( therefore its unique in this pathway. No co-option from other biological systems is possible )
Coproporphyrinogen III oxidase
http://en.wikipedia.org/wiki/Coproporphyrinogen_III_oxidase
It is an enzyme involved in the sixth step of porphyrin metabolism it catalyzes the oxidative decarboxylation of coproporphyrinogen III to proto-porphyrinogen IX in the haem and chlorophyll biosynthetic pathways. ( therefore its just used in one other pathway ( chlorophyll )
Protoporphyrinogen oxidase
http://en.wikipedia.org/wiki/Protoporphyrinogen_oxidase
Protoporphyrinogen oxidase (EC 1.3.3.4) is an enzyme that is responsible for the seventh step in the biosynthesis of protoporphyrin IX. This porphyrin is the precursor to hemoglobin, the oxygen carrier in animals, and chlorophyll, the dye in plants. ( therefore its unique in this pathway. No co-option from other biological systems is possible )
Ferrochelatase Mitochondrion
http://en.wikipedia.org/wiki/Ferrochelatase
Ferrochelatase (FECH, protoheme ferrolyase) is an enzyme that catalyzes the terminal (eighth) step in the biosynthesis of heme, converting protoporphyrin IX into heme. ( therefore its unique in this pathway. No co-option from other biological systems is possible )
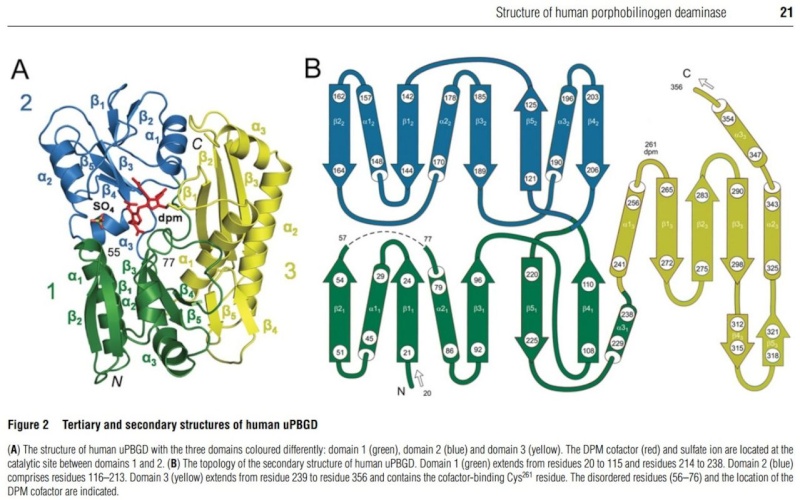
The PBG deaminase enzyme
http://en.wikipedia.org/wiki/Porphobilinogen_deaminase
Porphobilinogen deaminase is involved in the third step of the heme biosynthetic pathway. No other use is mentioned, therefore we can assume its uniquely used in this pathway.
This enzyme catalyzes the head to tail condensation of four porphobilinogen molecules into the linear hydroxymethylbilane while releasing four ammonia molecules.
Structure and function
Functionally, porphobilinogen deaminase catalyzes the loss of ammonia from the porphobilinogen monomer (deamination) and its subsequent polymerization to a linear tetrapyrrole, which is released as hydroxymethylbilane:

The first step is believed to involve an E1 elimination of ammonia from porphobilinogen, generating a carbocation intermediate (1).[6]
The second step : This intermediate is then attacked by the dipyrrole cofactor of porphobilinogen deaminase, which after losing a proton yields a trimer covalently bound to the enzyme (2).
The third step This intermediate is then open to further reaction with porphobilinogen (1 and 2 repeated three more times). Once a hexamer is formed, hydrolysis allows hydroxymethylbilane to be released, as well as cofactor regeneration (3)
The fourth step Once a hexamer is formed, hydrolysis allows hydroxymethylbilane to be released, as well as cofactor regeneration
What co-factor are we talking about? let's see.
Porphobilinogen deaminase, dipyrromethane cofactor binding site (IPR022419)
Short name: Porphobilin_deaminase_cofac_BS
http://www.ebi.ac.uk/interpro/entry/IPR022419
Description
This entry represents the region around cysteine residues that is conserved in porphobilinogen deaminases from various prokaryotic and eukaryotic sources. The sulfur atom of this cysteine residue has been shown in the Escherichia coli enzyme (gene hemC) to be bound to the dipyrromethane cofactor [PMID: 3196304]. Porphobilinogen deaminase covalently binds a dipyrromethane cofactor to which the PBG subunits are added in a stepwise fashion. Porphobilinogen deaminase has a three-domain structure. Domains 1 (N-terminal) and 2 are duplications with the same structure, resembling the transferrins and periplasmic binding proteins. The dipyrromethene cofactor is covalently linked to domain 3 (C-terminal), but is bound by extensive salt-bridges and hydrogen-bonds within the cleft between domains 1 and 2, at a position corresponding to the binding sites for small-molecule ligands in the analogous proteins [PMID: 1522882]. The enzyme has a single catalytic site, and the flexibility between domains is thought to aid elongation of the polypyrrole product in the active-site cleft of the enzyme.
Questions: how did gene duplication, followed by random mutations and natural selection figure out to produce the PBG deaminase enzyme,, used as far as science knows, exclusively in this pathway, so no co-option possible? - that would produce this complex reaction, ( which is just the third in the whole pathway of total 8 steps ) consisting in 4 highly coordinated, ordered, sequenced and complex steps, forming a geometrically correct tetrapyrrole, and repeat the first two steps in total 4 times? How did evolution be capable to produce the right genetic code and informational sequence? How did evolution figure out to program the release of the hydroxymethylbilane enzyme at the right time, after the product, the linear hydroxymethylbilane was catalyzed, and while releasing four ammonia molecules?
1. Werner Gitt, Without excuse, page 308
2. http://www.sciencedirect.com.https.sci-hub.hk/science/article/pii/S0005272812010407
Last edited by Admin on Tue Aug 07, 2018 7:35 pm; edited 45 times in total