My book has been published on Amazon
The RNA-DNA Nexus: Unveiling Fine-Tuning in Life's Molecular Machinery at the Atomic Scale and the Paradigm of Intelligent Design
In Paperback. (Here you can also look inside, and see also the Table of Contents:)
https://www.amazon.com/dp/B0CCCNBPVR
and in Kindle format:
https://www.amazon.com/dp/B0CCQJX4HC
Delve into the captivating journey of "The RNA-DNA Nexus," where the mysteries of ribonucleotide and deoxyribonucleotide biosynthesis are unraveled. This groundbreaking book explores life's atomic-scale precision, revealing the handiwork of a superintelligent designer. Whether you seek to solidify your beliefs or embark on an exploration of life's wonders, this book invites you to discover the profound intricacies inside living cells.
Witness the extraordinary complexity of cellular machinery, with enzymes orchestrating biochemical processes with impeccable precision. From the precise placement of atoms to the intricacies of bonding angles, life's sophistication at the atomic level will leave you breathless.
Uncover the foundation of life as we explore the complex biosynthesis pathways of RNA and DNA—the building blocks that shape biological processes. Ribonucleotides play a key role in transmitting genetic instructions and coordinating cellular functions. Meanwhile, deoxyribonucleotides safeguard genetic continuity, ensuring the faithful transmission of life's blueprint.
"The RNA-DNA Nexus" dares to question the conventional view of life's origins, presenting compelling evidence for intelligent design. It navigates through the current scientific knowledge, pondering the enigmatic bridge between prebiotic molecules and living cells. Prepare to be captivated, enlightened, and inspired by the awe-inspiring complexity that underlies life's beginnings.
This enthralling journey will challenge your understanding of life, providing profound insights into the elegance of creation and the purpose behind our existence. From intricate protein folding to gene regulation, from metabolic pathways to organism adaptability—each revelation will deepen your appreciation for life's astonishing architecture.
Join the expedition as the author unravels the complexities of life's origins, offering factual and captivating insights. A thought-provoking exploration awaits, inviting you to contemplate the fingerprint of a divine craftsman—a creator of boundless intelligence and love, shaping life with purpose.
"The RNA-DNA Nexus" delivers a transformative journey that unveils the wonders of cellular life. Step into a world where facts and fascination entwine, and where the symphony of life's mysteries unfolds before your eyes. Prepare to embark on an extraordinary voyage of discovery—a testament to the astonishing complexity and ingenuity of life's grand design.
Meu novo livro foi publicado na Amazon,
"O Nexo RNA- DNA: Revelando o ajuste fino na maquinaria molecular da vida na escala atômica e o paradigma do Design Inteligente
e é disponível no formato Kindle: https://www.amazon.com/dp/B0CD86JWP5
Caso prefira o texto em formato PDF, pode entrar em contato comigo pelo messenger
Mergulhe na cativante jornada de "O Nexo RNA- DNA", onde os mistérios da biossíntese de ribonucleotídeos e desoxirribonucleotídeos são desvendados. Este livro explora a precisão em escala atômica da vida, revelando a obra de um designer superinteligente. Se você procura solidificar suas crenças ou embarcar em uma exploração das maravilhas da vida, este livro o convida a descobrir as profundas complexidades dentro das células vivas.
Testemunhe a extraordinária complexidade da maquinaria celular, com enzimas orquestrando processos bioquímicos com precisão impecável. Do posicionamento preciso dos átomos às complexidades dos ângulos de ligação, a sofisticação da vida no nível atômico o deixará maravilhado.
Descubra a base da vida enquanto exploramos as complexas vias de biossíntese de RNA e DNA - os blocos de construção que moldam os processos biológicos. Os ribonucleotídeos desempenham um papel fundamental na transmissão de instruções genéticas e na coordenação das funções celulares. Enquanto isso, os desoxirribonucleotídeos protegem a continuidade genética, garantindo a transmissão fiel do projeto da vida.
"O Nexo RNA- DNA" ousa questionar a visão convencional das origens da vida, apresentando evidências convincentes para o design inteligente. Navega pelo conhecimento científico atual, refletindo sobre a enigmática ponte entre as moléculas prebióticas e as células vivas. Prepare-se para ser cativado, iluminado e inspirado pela complexidade inspiradora subjacente ao início da vida.
Esta fascinante jornada desafiará sua compreensão da vida, fornecendo percepções profundas sobre a elegância da criação e o propósito por trás de nossa existência. Do intrincado dobramento de proteínas à regulação de genes, das vias metabólicas à adaptabilidade do organismo - cada revelação aprofundará sua apreciação pela surpreendente arquitetura da vida.
Junte-se à expedição enquanto o autor desvenda as complexidades das origens da vida, oferecendo percepções factuais e cativantes. Uma exploração instigante aguarda, convidando você a contemplar a impressão digital de um artesão divino - um criador de inteligência e amor ilimitados, moldando a vida com propósito.
"O Nexo RNA- DNA" oferece uma jornada transformadora que revela as maravilhas da vida celular. Entre em um mundo onde fatos e fascínio se entrelaçam, e onde a sinfonia dos mistérios da vida se desenrola diante de seus olhos. Prepare-se para embarcar em uma extraordinária viagem de descoberta - uma prova da surpreendente complexidade e engenhosidade do grande projeto da vida.
In my previous literary endeavor, "On the Origin of Life and Virus World by Means of an Intelligent Designer," I embarked on a journey to illuminate the true nature of cells. These remarkable entities, I proposed, are not mere metaphors for human-made factories; they are veritable chemical factories in their own right. Within the pages of this book, we embark on a captivating journey into the realm of ribonucleotide and deoxyribonucleotide biosynthesis. In this enthralling exploration, we will uncover the intricate pathways through which these essential building blocks of life are meticulously crafted. Ribonucleotides and deoxyribonucleotides take center stage in the intricate theater of cellular life, playing pivotal roles that underpin the very essence of biological processes. These molecules, with their distinct structures and functions, are the foundation upon which the dance of life unfolds. Ribonucleotides are key components of RNA, the versatile messenger that carries genetic information and orchestrates a multitude of vital cellular functions. Through the synthesis of RNA, ribonucleotides serve for the transmission of genetic instructions, dictating the production of proteins and enabling the intricate coordination of biological processes. RNA's flexibility allows it to adapt and respond rapidly to changing cellular demands. It acts as a bridge between DNA and protein synthesis, translating the genetic code into functional proteins. Moreover, RNA molecules themselves can possess enzymatic activity, catalyzing essential reactions within the cell. This dual role, as both a messenger and an active participant in cellular metabolism, positions ribonucleotides at the heart of cellular communication and regulation. In the grand scheme of cellular life, deoxyribonucleotides assume an even more fundamental role as the building blocks of DNA, the repository of genetic information. DNA serves as the blueprint for life, housing the instructions that shape an organism's development, traits, and potential. The double helix structure of DNA allows for remarkable stability and fidelity in the transmission of genetic information from one generation to the next. Deoxyribonucleotides, through their precise assembly and pairing, contribute to the integrity and accuracy of DNA replication, ensuring the faithful transmission of genetic material. Furthermore, amazingly, cells have intricate systems in place to detect and correct errors or damage in DNA. Deoxyribonucleotides contain and perpetuate the genetic code, safeguarding the continuity of life across generations. Ribonucleotides and deoxyribonucleotides, with their distinct functions and interplay, form the backbone of cellular life. Together, they facilitate the transmission, expression, and perpetuation of genetic information, shaping the intricate symphony of biological processes. Without these essential molecules, the cellular stage would remain incomplete, lacking the script that guides the performance of life itself.
Science magazines and popular books might provide us with superficial glimpses into the fascinating world of biosynthesis in the molecular world, but they often fail to convey a truly holistic picture of the depth and complexity of these pathways. This book endeavors to bridge that gap, shedding light on the intricacies of these processes and bringing them closer to the reader's grasp. By doing so, I hope to ignite a sense of awe and appreciation for the remarkable complexity that underlies the very fabric of life, and it unmistakenly points to an awe-inspiring creator with capabilities of intelligence, rarely appreciated, nor even imagined.
Louis Pasteur, the renowned French chemist, and microbiologist, is often quoted saying: "Too little science leads away from God, while too much science leads back to Him. " This quote is often attributed to Pasteur, but there is no evidence that he ever said it. The earliest mention of the quote is in a 1924 book by the French writer Paul Dottin. Dottin attributed the quote to Pasteur, but he did not provide any source for it. Nonetheless, i think it is pertinent to mention it here, since this is precisely what this book is about.
For far too long, the quest for the origin of life has been obscured by the shadows of a grand ideological battle between theists and atheists. The focus of attention has often gravitated towards the theory of evolution, the interconnections between humans and apes, and other such contentious topics. It is only in recent years that this quest has emerged from the sidelines and taken center stage, thanks to the proliferation of popular science magazines, books, and the ever-engaging realm of YouTube debates. What becomes apparent, however, is the prevalence of widespread ignorance and oversimplified awareness when it comes to comprehending the sheer complexity that underlies the very essence of life. The journey towards life's actualization requires a meticulous interplay of intricate mechanisms, poised in perfect harmony, and this book strives to shed light on the true state of affairs.
In my previous book, I addressed how 70 years of abiogenesis research did not come to achieve the goal of elucidating how the prebiotic earth could have provided the basic building blocks of life, nucleotides, amino acids, phospholipids, and carbohydrates. Many hypotheses have been elaborated and developed in the past. Some presentations implied that these hypotheses were more certain than they actually are. For example, the textbook "Biology" by Campbell and Reece states that the Miller-Urey experiment "provided strong support for the idea that life could have arisen spontaneously on the early Earth." The Miller-Urey experiment was a landmark experiment in the field of abiogenesis, but it did not provide strong support for the idea that life could have arisen spontaneously on early Earth. The experiment showed that it was possible to synthesize some of the basic building blocks of life from inorganic molecules under conditions that were thought to be similar to those of the early Earth. However, the experiment did not show by any means how these building blocks could have spontaneously assembled into living cells.
In her book "Symbiosis in Cell Evolution," published in 1967, Lynn Margulis wrote: To go from a bacterium to people is less of a step than to go from a mixture of amino acids to a bacterium. She addressed the significant challenges involved in bridging the gap between a mixture of basic building blocks and the complexity of living cells. She did highlight several key reasons why this transition is highly difficult to conceptualize by natural, unguided means.
Firstly, the synthesis of the basic building blocks of life requires complex metabolic pathways. These pathways involve the precise coordination of numerous enzymes and the optimization of chemical reactions. It is challenging to conceive how such intricate pathways could have spontaneously arisen from a simple mixture of building blocks. Secondly, the conditions necessary for life to exist involve a delicate balance of various molecules, pH levels, temperature, and other factors. The early Earth's environment was much harsher than it is today, making it difficult to imagine how the required conditions for life could have been sustained. Additionally, the replication of genetic information is vital for life's continuity. The first cells would have needed a pre-existing template to replicate their DNA and pass it on to offspring. This requirement adds another layer of complexity to the emergence of life from chaotic prebiotic molecules. Margulis presented the idea that the gap between a mixture of basic building blocks and a living cell is formidable and, in her opinion, insurmountable. While scientists aim to understand the origin of life and make progress in bridging this gap, the feasibility, based on existing scientific knowledge today is questionable. The gap between a mixture of chaotic prebiotic molecules and a living cell is a daunting one, and is, as it is my opinion, insurmountable. Scientists claim of making progress in understanding the origins of life, and that it would be possible that scientific discoveries will one day be able to bridge this gap. But is this hope warranted, based on what science already does know? In addition, there is also the fact that life uses complex information systems to store and transmit genetic information. These information systems are based on DNA and RNA, which are molecules that are incredibly complex and difficult to synthesize. While other books have focussed on the problem of the origin of genetic information and the genetic code, this book aims to address the problem of the hardware. The metabolic systems, that synthesize RNA and DNA, and the quest for their origins. Within the pages that follow, we embark on an intellectual voyage that seeks to unveil the awe-inspiring intricacies of these metabolic pathways, that gave rise to RNA and DNA. What we find, upon embarking on this bewildering journey, is that life, in its nascent form, will remain an enigma shrouded in mystery - unless, we permit the footsteps of an intelligent designer into the door.
It is an intricate puzzle, the pieces of which must fit together with extraordinary precision for the grand design to spring forth into existence. The intricate dance of molecular interactions, the delicate choreography of biochemical reactions, and the astonishingly intricate mechanisms that propel life's progression—all of these elements converge to create a symphony of astonishing proportions. It is crucial for us, in order to have an objective picture, to move away from simplistic explanations and explore the profound depths of life's complexity. Only by doing so can we gain a genuine appreciation for the astonishing array of factors that must align harmoniously to kickstart life's extraordinary journey. From the intricate folding of proteins to the intricate regulation of gene expression, from the complex networks of metabolic pathways to the extraordinary adaptability of organisms—each component adds another layer of amazing complexity to the architecture of living systems. Throughout this book, I will unravel these astonishingly complex biological phenomena, uncovering the interconnections that underpin their existence. In doing so, I hope to illuminate the minds of my readers, offering both a factual and captivating exploration of life's origins. So, dear reader, join me on this extraordinary expedition, where facts and fascination intertwine, where knowledge is sought, and where the wonders of life are unveiled before your very eyes. Prepare to be captivated, enlightened, and inspired by the fascinating story that unfolds—one that reveals the remarkable complexity inherent in life's beginnings.
RNA and DNA take the center stage of this book
The biosynthesis pathways of ribonucleotides and deoxyribonucleotides, often overlooked in mainstream literature, are deserving of our special attention and understanding. Within the confines of the cell, an intricate web of molecular reactions and enzymatic activities intertwines, culminating in the precise construction of these vital genetic molecules. As we embark on this journey, we peel back the layers of scientific jargon, aiming to present the material in a relatable and engaging manner. We delve into the intricate mechanisms, providing factual accounts and clear explanations, all while maintaining a fluid and captivating narrative. My aim is to captivate the reader, revealing the awe-inspiring complexity that lies beneath the surface. By unveiling the intricacies of these biosynthesis pathways, we hope to instill a deeper appreciation for the remarkable choreography occurring within the cell. For it is through these pathways that the cell crafts the very essence of life itself. Let us embrace the adventure that awaits.
Within biosynthesis pathways, each protein unveils its own unique marvel of design and sophistication. These proteins, finely crafted with precision and tailored to perform specific tasks, hold hidden treasures waiting to be exposed. As we embark on this journey, we will unravel the intricacies of these remarkable proteins, leaving the reader in awe of their ingenuity. Picture a symphony of proteins, each with its own distinct role, harmoniously working together to create a masterpiece. Like virtuoso performers on a grand stage, these proteins showcase their idiosyncratic qualities and talents. Each protein possesses a finely tuned structure, that allows it to fulfill its designated function with remarkable efficiency. As we delve deeper into the biosynthesis pathways, we encounter proteins that act as catalysts, guiding and facilitating the precise chemical transformations required for nucleotide synthesis. These catalysts, known as enzymes, possess a breathtaking array of mechanisms and active sites, uniquely designed to interact with specific molecules and orchestrate intricate reactions. Consider the elegance of an enzyme's active site, like a lock waiting for the perfect key. This pocket within the protein, precisely sculpted, accommodates the specific shape and properties of its substrate, ensuring a secure fit. It is within this confined space that the magic unfolds, as the enzyme guides and catalyzes the transformation of molecules with astounding specificity. Each protein in the biosynthesis pathway has its own story to tell. Some act as dedicated messengers, shuttling crucial molecules from one step to the next, while others serve as architects, assembling the intricate structures of nucleotides with impeccable precision. As we unravel the hidden treasures within these proteins, we cannot help but marvel at the ingenuity displayed by the designer that shaped them. The remarkable sophistication exhibited by these molecular machines reflects a causal intelligence far beyond our comprehension. It is through their collective efforts that the cell's biosynthesis pathways come to life, culminating in the creation of RNA and DNA. So, let us venture deeper into this captivating world, where proteins reveal their unique marvels of design and sophistication. With each discovery, we draw closer to unraveling the secrets of life, crafted by an intelligence that defies our understanding.
Our story begins with ribonucleotides, the precursors of RNA. Like skilled artisans, the cells carefully construct these molecules to ensure the seamless flow of genetic information. The biosynthesis pathway unfolds within the confines of the cell, where a series of intricate reactions take place. Imagine a bustling workshop, with enzymes as diligent workers. They diligently transform simpler molecules into intricate ribonucleotides. These enzymes, fueled by cellular machinery, perform their tasks with precision and purpose. Step by step, they assemble the nucleotide units, constructing the RNA foundation from scratch. As we delve deeper into the narrative, the spotlight shifts to deoxyribonucleotides, the building blocks of DNA. The biosynthesis pathway for deoxyribonucleotides follows a similar course, yet with subtle nuances that set it apart. Just like before, the cellular factory hums with activity. Enzymes take center stage once again, weaving their magic to transform simpler molecules into complex deoxyribonucleotides. The pathway unfolds with an intricate series of reactions, each step guided by the careful orchestration of enzymes. These reactions occur with astounding accuracy, driven by the cellular machinery's unwavering commitment to ensuring the availability of these vital molecules. It is through this meticulously regulated process that the cell ensures the proper construction and replication of DNA, the blueprint that underpins all forms of life. As we marvel at the intricacies of ribonucleotide and deoxyribonucleotide biosynthesis, we cannot help but recognize the precision and intention woven into the fabric of life. The intelligent design that governs these processes stands as a testament to the intricacy and purpose within the cell. In the pages that follow, we will continue to unravel the secrets of biosynthesis, peering into the inner workings of cellular processes. The captivating journey of ribonucleotides and deoxyribonucleotides will offer us glimpses into the remarkable mechanisms that sustain life itself. So let us forge ahead, guided by curiosity and an insatiable thirst for knowledge.
1
The Cell, an amazing chemical factory
Before we embark on a journey through the intricate labyrinth of biosynthetic pathways responsible for the production of ribonucleotides and deoxyribonucleotides, the fundamental building blocks of RNA and DNA, let me elucidate the vital role these metabolic pathways play in the marvelous workings of the chemical cell factory. Imagine the cell as a bustling factory, teeming with activity and ceaseless productivity. Within this microscopic world, countless molecular workers toil relentlessly, executing their specialized tasks to maintain the delicate equilibrium of life. Just as a well-oiled machine requires a constant supply of raw materials, so do cells, that rely on the steady stream of materials that it imports to fuel the remarkable processes that unfold within. These nucleotide monomers serve as the foundation upon which the intricate blueprints of life are encoded. Without ribonucleotides and deoxyribonucleotides, the cell's ability to replicate its DNA, produce proteins, and orchestrate its essential functions would come to a grinding halt. Complex pathways, carefully choreographed by ingenious molecular machinery, guide the transformation of simple starting materials into these vital nucleotide building blocks. Enzymes act as skilled artisans, diligently carrying out each step of the process with precision and finesse. But why are these pathways so important? Well, consider DNA, the master archive of genetic information. It holds the secrets of life, passed down from one generation to the next, encoding the instructions for building and maintaining an entire organism. It is the essential thread that weaves together the tale of life's blueprint, shaping the destiny of each living being. Meanwhile, RNA, the dynamic cousin of DNA, plays a diverse range of roles within the cell. It acts as a messenger, shuttling genetic instructions from the DNA to the protein-making machinery. It also functions as an architect, guiding the assembly of proteins according to the blueprint etched in the DNA. Moreover, certain RNA molecules exhibit enzymatic properties, catalyzing essential chemical reactions within the cell.
To grasp the essence of life, we must delve into the intricate workings of these cellular powerhouses, understanding their ability to reproduce, self-replicate, and engage in a symphony of metabolic reactions. Within the pages of this compelling narrative, I reveal the multifaceted dimensions that render cells truly alive. They possess an unparalleled capacity to transform chemicals through the orchestration of complex enzymatic processes. Like master alchemists, they take up materials, recycle and expel waste products, and grow, develop, and evolve in response to their ever-changing environment. Their complexity is awe-inspiring, comprising millions of essential components such as monomers, polymers, proteins, and cell membranes. Yet, the grandeur of cellular life extends far beyond mere structural intricacies. Cells must maintain a delicate balance, a homeostatic milieu, where stable internal conditions harmoniously coexist. Energy, the lifeblood of these remarkable factories, fuels their cellular functions and empowers their every endeavor. Moreover, cells possess the remarkable ability to perceive and respond to external stimuli, navigating the world around them with unparalleled finesse. At the core of cellular existence lies the artistry of information. Within the recesses of these microscopic realms, intricate data storage, transmission, transcription, translation, and decoding processes unfold. This vast repository of genetic and epigenetic material serves as a blueprint, directing the assembly and operation of the cell factory itself. Replication and transmission of this precious genetic inheritance to future generations, to the daughter cells, stand as paramount pillars of cellular life. Yet, no account of cellular existence would be complete without acknowledging the indispensable mechanism of error detection and repair. In a world where imperfection is inevitable, cells possess the extraordinary ability to identify and rectify errors, ensuring the continuity of life. In our quest to unravel the intricacies of cellular life, let us take a moment to contemplate the fundamental tasks that a basic autonomous cell must accomplish. Its noble purpose: reproduction and the synthesis of essential components from raw materials.
The Information System: In the realm of cellular existence, mere chemicals are incapable of orchestrating the symphony required for reproduction and synthesis. Left to their own devices, they dance chaotically or crystallize into patterns, like delicate snowflakes. Thus, a profound solution emerges – the presence of information. An entity, such as RNA or its counterparts, bears the sacred duty of storing the intricate instructions necessary to guide the assembly process.
The Energy System: Yet, information alone is a dormant force, a whisper awaiting manifestation. The implementation of these instructions demands a wellspring of energy. A system devoid of the capacity to generate or source energy drifts aimlessly or succumbs to the path of least resistance, crystallizing into mundane forms. Hence, a robust system of energy production or sourcing takes center stage, accompanied by intricate subsystems that ensure the proper distribution and management of this vital life force.
The Copy System: Reproduction, the very essence of life's continuum, demands that the device diligently executes the instructions of the information system, drawing upon the reserves of the energy system. This arduous endeavor entails the restoration and reconstruction of critical infrastructure, including the information and energy systems themselves. Such a feat epitomizes the remarkable capabilities of cellular existence.
The Growth System: Without a growth system, the very fabric of this extraordinary device would dwindle with each generation, vanishing into oblivion. To ensure its longevity, a magnificent ensemble of subsystems comes into play. Ingestion of materials from the surrounding world, their meticulous processing, and the artistry of assembly converge to form a formidable chemical factory. Through this intricate process, the device fortifies itself, nurturing growth and vitality.
The Transportation System: Within the bustling corridors of the cell, materials must navigate their destined paths, arriving at their designated locations. A transportation system emerges an intricate network that facilitates the movement of raw materials and products within the cell's inner realm. Moreover, a system arises to manage the influx of raw materials and the egress of waste materials, orchestrating the grand ballet of chemical reactions that sustains the cell's existence.
A growth and a reproduction system: These systems are intricately interconnected and must be synchronized to maintain a harmonious balance. If reproduction outpaces growth, the organism will diminish in size faster than it can replenish itself, ultimately fading away within a few generations. To avert such a fate, a well-designed timing or feedback mechanism becomes indispensable.
Furthermore, effective coordination is essential among all the diverse tasks undertaken by these biological systems. A communication system emerges as a necessity, enabling the synchronization of activities. The reproduction system cannot function optimally without proper coordination with the growth and power systems. Similarly, the power system alone is rendered futile without the harmonious interplay of the growth and reproduction systems. Just as a computer lacks purpose without interconnections between its power supply, CPU, memory, hard drive, video, operating system, and more, the potential of these biological systems can only be realized when all the intricate "circuitry" is in place and the power is activated.
In the wondrous realm of cellular biology, a remarkable truth unfolds before our eyes: cells are not mere abstractions or poetic metaphors. The complexity and sophistication of biological systems, particularly cells, defy the limitations of happenstance. They orchestrate the intricate interplay of molecular networks and regulatory mechanisms. Within the depths of these remarkable systems, we find the essence of life's true marvels. At the heart of cellular existence lies the dance of information. Cells possess the incredible ability to process, store, and utilize genetic information, inscribed within the fabric of their DNA. This information, akin to a sublime software program, guides and directs the cell's every move. It is through the execution of these encoded instructions that cells accomplish extraordinary feats. Immersed in this world of genetic information, we witness the awe-inspiring architecture of life's machinery. The intricate molecular networks and regulatory circuits transform these instructions into tangible manifestations. Proteins, those masterful actors, take the stage, participating in the cell's structure, metabolism, and signaling processes. The orchestration of this symphony requires precise coordination, as multiple components interact harmoniously to bring forth life. Within cells, the storage, transmission, and interpretation of genetic information emerge as feats of molecular brilliance. The complex machinery and intricate cellular processes safeguard the integrity and faithful execution of these instructions. It is through the transcription and translation of the genetic code that cells bring forth the wonder of life, akin to the operation of software programs in the realm of computers. Information is not a mere abstraction; it is the lifeblood that courses through the veins of cellular existence. As we contemplate the awe-inspiring truth of cells as "information-driven chemical factories," we cannot ignore the profound implications that arise. Such intricate complexity, driven by the execution of information-encoded instructions, defies the notion of random, unguided processes.
Cells possess the profound ability to absorb raw materials from their environment and transmute them into an array of essential products crucial for their survival. Proteins, lipids, energy – these are the life-giving elixirs that cells meticulously manufacture through their intricate machinery. Envision, if you will, each cell as a marvel of engineering – an intricate network of interlocking assembly lines, where unfathomably complex protein machines and complexes labor in perfect harmony. The nucleolus, a sprawling factory within eukaryotic cells, the bigger brothers of prokaryotes, emerges as a hub of activity where non-coding RNAs are transcribed, processed, and deftly assembled with proteins to form ribonucleoprotein complexes. The endoplasmic reticulum, a veritable factory floor, assumes the monumental task of synthesizing nearly all of the cell's lipids. And in moments of DNA damage, repair factories materialize, where the cell meticulously brings together and mends its fragile genetic material. Indeed, the protein assemblies within cells echo the artistry of human inventions. These extraordinary structures, akin to sophisticated machines, comprise highly coordinated moving parts. Like masterful clockwork, they engage in precise intermolecular collisions, restricted to a meticulously choreographed set of possibilities. Conformational changes ripple through these assemblies, orchestrated by the energy derived from nucleoside triphosphate hydrolysis and other remarkable sources. It is through these intricate movements that cells function in a polarized fashion, aligning along filaments or nucleic acid strands. They elevate the fidelity of biological reactions and catalyze the formation of complex protein ensembles. To comprehend the true magnitude of cellular complexity, let us make a thought experiment, daring to magnify the size of a cell ten thousand millionfold. In this grand visualization, a cell expands to a radius of 200 miles – a vast expanse, surpassing the very boundaries of human imagination. Even within this sprawling canvas, we find that the required number of buildings to house the factories and machines that cells possess would far surpass the architectural landscape of a metropolis like New York City. As we contemplate the awe-inspiring nature of cellular life, we find ourselves standing in reverence of its grand design. Within the chambers of each cell, chemical transformations and intricate machinery unfurl, painting a portrait of intelligent craftsmanship. The complexity and scale of these remarkable entities surpass our comprehension, beckoning us to embrace the profound truths that lie within. We are presented with a choice – to marvel at the exquisite design woven into the very fabric of cellular existence or to turn a blind eye to the intricacies and dismiss them as mere products of happenstance. I invite you, dear reader, to embark on a journey of discovery, to peer through the lens of reason and contemplation, as we unravel the mysteries that lie within the cellular realm. In doing so, we may find ourselves drawn closer to the undeniable truth of the existence of an intelligent designer, that orchestrated the symphony of life itself.
Premise 1: Unguided events, without the influence of an intelligent agent, has not been demonstrated to assemble complex chemical factories driven by software programs, which in the context of biology, are the instructions encoded in DNA and executed by cellular machinery. Cells are information-driven chemical factories due to their ability to process, store, and utilize genetic information to carry out complex biological processes. They consist of intricate molecular networks and regulatory mechanisms that enable them to perform a wide range of functions necessary for life.
Premise 2: Genetic information is stored in the form of DNA (or RNA) within cells and is transcribed and translated into functional molecules, such as proteins. This genetic code acts as a set of instructions guiding the synthesis of specific proteins, which play key roles in the cell's structure, metabolism, and signaling processes. The storage, transmission, and interpretation of genetic information within cells require complex molecular machinery and cellular processes. Cells utilize sophisticated molecular machinery and biochemical processes to interpret and execute the instructions encoded in the genetic code, similar to how computers use hardware and software to process and execute instructions.
Conclusion: The emergence of complex biological systems, driven by information and executed through intricate molecular processes, is best explained by the involvement of an intelligent agent. The precise coordination, functionality, and storage of vast amounts of genetic information within cells, along with their ability to carry out a wide range of biological functions, are highly complex phenomena that have not been demonstrated to arise solely through random, unguided processes.
B. Alberts (2022): The surface of our planet is populated by living things—organisms—curious, intricately organized chemical factories that take in matter from their surroundings and use these raw materials to generate copies of themselves. Although all cells function as biochemical factories of a broadly similar type, many of the details of their small-molecule transactions differ. All cells operate as biochemical factories, driven by the free energy released in a complicated network of chemical reactions. Each cell can be viewed as a tiny chemical factory, performing many millions of reactions every second. We can view RNA polymerase II in its elongation mode as an RNA factory that not only moves along the DNA synthesizing an RNA molecule but also processes the RNA that it produces. The nucleolus can be thought of as a large factory at which different noncoding RNAs are transcribed, processed, and assembled with proteins to form a large variety of ribonucleoprotein complexes. mRNA production is made more efficient in the nucleus by an aggregation of the many components needed for transcription and pre-mRNA processing, thereby producing a specialized biochemical factory. The extensive ER network serves as a factory for the production of almost all of the cell’s lipids. In response to DNA damage, they rapidly converge on the sites of DNA damage, become activated, and form “repair factories” where many lesions are apparently brought together and repaired. The formation of these factories probably results from many weak interactions between different repair proteins and between repair proteins and damaged DNA. 1
B. Alberts (1998): We can walk and we can talk because the chemistry that makes life possible is much more elaborate and sophisticated than anything we students had ever considered. Proteins make up most of the dry mass of a cell. But instead of a cell dominated by randomly colliding individual protein molecules, we now know that nearly every major process in a cell is carried out by assemblies of 10 or more protein molecules. And, as it carries out its biological functions, each of these protein assemblies interacts with several other large complexes of proteins. Indeed, the entire cell can be viewed as a factory that contains an elaborate network of interlocking assembly lines, each of which is composed of a set of large protein machines. Consider, as an example, the cell cycle–dependent degradation of specific proteins that helps to drive a cell through mitosis. First, a large complex of about 10 proteins, the anaphase-promoting complex (APC), selects out a specific protein for polyubiquitination; this protein is then targeted to the proteasome's 19S cap complex formed from about 20 different subunits; and the cap complex then transfers the targeted protein into the barrel of the large 20S proteasome itself, where it is finally converted to small peptides. Why do we call the large protein assemblies that underlie cell function protein machines? Precisely because, like the machines invented by humans to deal efficiently with the macroscopic world, these protein assemblies contain highly coordinated moving parts. Within each protein assembly, intermolecular collisions are not only restricted to a small set of possibilities, but reaction C depends on reaction B, which in turn depends on reaction A—just as it would in a machine of our common experience. Underlying this highly organized activity are ordered conformational changes in one or more proteins driven by nucleoside triphosphate hydrolysis (or by other sources of energy, such as an ion gradient). Because the conformational changes driven in this way dissipate free energy, they generally proceed only in one direction. An earlier brief review emphasized how the directionality imparted by nucleoside triphosphate hydrolyses allows allosteric proteins to function in three different ways: as motor proteins that move in a polarized fashion along a filament or a nucleic acid strand; as proofreading devices or “clocks” that increase the fidelity of biological reactions by screening out poorly matched partners; and as assembly factors that catalyze the formation of protein complexes and are then recycled. 2
Magnifying a cell ten thousand million times, it would have a radius of 200 miles, about 10 times the size of New York City
Calling a cell a factory is an understatement. Magnifying the cell to a size of 200 miles, it would only contain the required number of buildings, hosting the factories to make the machines that it requires.
New York City has about 900.000 buildings, of which about 40.000 are in Manhattan, of which 7.000 are skyscrapers of high-rise buildings of at least 115 feet (35 m), of which at least 95 are taller than 650 feet (198 m).

As we go deeper into the profound nature of cellular life, a captivating revelation unfolds before our very eyes: cells are not mere abstractions or fleeting concepts. They are, in their essence, even an entire industrial park, brimming with factories and machines that operate with exquisite precision. Imagine, if you will, a sprawling cityscape teeming with buildings, where the factories producing the very machines used within the industrial park far exceed the grandeur of New York City. Each towering structure is a factory in its own right, comparable in size to the majestic skyscrapers of the Twin Towers in the World Trade Center. These towering edifices host the intricate machinery that propels the cellular machinery forward. In the heart of this bustling metropolis, we encounter a marvel of prodigious proportions: the nucleolus. Within its walls, a myriad of noncoding RNAs are transcribed, processed, and artfully assembled with proteins, giving rise to an astonishing array of ribonucleoprotein complexes. It is here that the factory of ribosomes is born – the very machines responsible for protein synthesis, the lifeblood of the cell itself. The ribosome assembly process unfolds in a stepwise, ordered manner, where the synthesis and modification of its components occur in synchrony with their assembly. This intricate dance is facilitated by an ensemble of assembly factors, each one carefully guiding and monitoring the individual steps. From the modification of ribosomal components to the release of assembly factors from intermediates, every aspect of this process is meticulously controlled. To ensure the highest quality of the ribosome population, an array of nucleases stands guard, ready to dismantle assembly intermediates that possess inappropriate structures or constitute kinetic traps. But the wonders within the cellular landscape do not cease there. Behold the mitochondria – the powerhouses of the cell. Within these remarkable organelles, up to 5000 ATP synthase energy turbines tirelessly toil. Consider, that within each human heart muscle cell, up to 8,000 mitochondria reside, housing a staggering number of up to 40 million ATP synthase energy turbines. These mighty machines, devoted to the production of ATP, the cellular energy currency, epitomize the profound intricacies embedded within the cellular realm. As we peer into the depths of cellular existence, the words of eminent scholars echo in our minds. Michael Denton, in his enlightening work "The Miracle of the Cell," paints a vivid picture of this astonishing molecular reality. He draws parallels between the intricate machinations of cells and the cutting-edge technologies of the late twentieth century. Artificial languages and their decoding systems, memory banks for information storage and retrieval, elegant control systems governing automated assembly, error fail-safe mechanisms for quality control – the analogies are striking and undeniable. Envision an object that defies imagination. A colossal automated factory, surpassing the dimensions of a great city like London or New York. Its complexity rivals the vastness of the cosmos and dwarfs the atomic realm. We step inside, only to be enraptured by a world of supreme technology and bewildering complexity. Endless corridors and conduits branch forth, leading to central memory banks, assembly plants, and processing units. Within the nucleus, a monumental spherical chamber resembling a geodesic dome, we find miles upon miles of coiled DNA chains neatly stacked together in ordered arrays. At every turn, we witness a symphony of control, precision, and adaptation. Millions of openings, like the port holes of a grand spaceship, open and close, allowing the seamless flow of materials in and out of the cell. Robot-like machines surround us, each protein molecule a testament to astounding complexity, consisting of thousands of atoms meticulously arranged in three-dimensional splendor. The life of the cell hinges upon the concerted efforts of countless proteins, each one contributing to the harmonious symphony of cellular existence. The very fabric of our own advanced machines finds its counterpart in the cell. Artificial languages, memory banks, control systems, quality control mechanisms – these analogies permeate the molecular reality of cellular life. The resemblance is so striking that we find ourselves borrowing terminology from the world of late-twentieth-century technology to describe this captivating realm. In our exploration of the cellular world, we witness an awe-inspiring spectacle, where an object resembling an immense automated factory unfurls before our eyes. It transcends our comprehension, surpassing the combined manufacturing activities of humankind on Earth. The most extraordinary aspect? This extraordinary factory has the unparalleled capacity to replicate its entire structure within a matter of hours, a feat that leaves us breathless with wonder. Cells function as intricate chemical factories, adept at acquiring materials from their surroundings, processing them, and generating "finished goods" crucial for their own sustenance and the overall well-being of the larger organism they inhabit. Within the complexity of a cell, specialized receptors serve as entry points ("loading docks") for incoming materials.
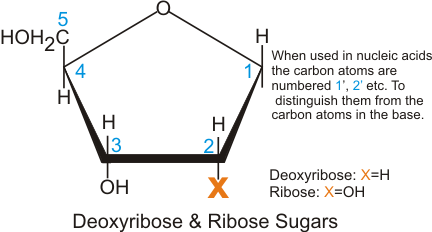
These substances undergo chemical reactions orchestrated by a central information system ("the front office"), and are transported to various locations within the cell ("assembly lines") as the intricate work unfolds. Finally, the transformed products are returned to the larger organism through the same receptors. The cell, a bustling and meticulously organized entity, relies on the harmonious collaboration of its numerous components to ensure its proper functioning. While proteins oversee the operations within these chemical factories, carbohydrates serve as the vital fuel supply for each manufacturing unit. Nucleic acids, specifically DNA and RNA, assume a critical role. These molecules not only carry the genetic blueprint that governs the cell's chemical factories but also serve as the carriers of hereditary information. Carbohydrates, on the other hand, assume the role of energizers for the cell's chemical factories. Serving as the foundational building blocks of carbohydrates, sugars in their small ring-like structures provide the necessary fuel. Similar to any well-organized factory, a cell requires several essential systems to thrive. It necessitates a front office, a repository of information where instructions are formulated and issued to guide the ongoing work. It must possess a physical structure akin to bricks and mortar, where the actual manufacturing occurs, complete with walls and partitions. Additionally, a production system encompassing diverse machines for generating finished goods and a transportation network for the efficient movement of raw materials and final products is imperative. Lastly, an energy plant must be present to provide the necessary power to operate the machinery. Describing cellular factories as comprising walls, partitions, and loading docks aptly captures their architectural makeup. Every living entity, regardless of its size, is composed of one or more cells, each boasting a complex anatomical structure. These "generic" cells harbor a multitude of structures and organelles, which can be likened to miniature chemical factories. The sequence of bases found along the double helix of DNA holds the genetic code—a comprehensive repository of information essential for a cell to reproduce and sustain its chemical factories. It encompasses all the distinctive attributes and idiosyncrasies that contribute to an individual's uniqueness. Soon after, the glucose molecules undergo processing within the cellular chemical factories, ultimately contributing to the formation of cellulose fibers that provide support to every blade of grass. The carbon atom seamlessly integrates into the grass's structural framework, becoming an integral component. In the captivating realm of materials science, a remarkable trend emerges – the miniaturization of complex physical and chemical systems. From the microscale to the nanoscale, scientists have achieved the bottom-up formation of dynamic structures with extraordinary properties. These structures, adorned with an array of molecular species, possess the remarkable ability to convert physical or chemical stimuli into directed motion. They are known as artificial molecular machines (AMMs), often considered molecular counterparts to the macroscopic machines that grace our daily lives – rotors, gears, and cranks that drive the machinery of our world. Yet, the inspiration for these fascinating AMMs often arises not from the realm of human-made machines, but from the intricate world of biology. It is within proteins and multi-protein complexes that we find the source of transformative energy, driving continuous and intricate structural motion. Vision, muscle contraction, and the graceful movement of bacterial flagella stand as awe-inspiring examples of biological responsive systems. These captivating phenomena are made possible by the presence of biological molecular machines (BMMs) such as ATP synthase, ribosomes, and myosin. These BMMs, far more structurally complex than any artificial counterparts created thus far, form an essential part of living systems. Immersed within the cellular landscape, BMMs find themselves embedded or immobilized within intricate scaffolds, such as bilayer lipid membranes or larger protein complexes. Here, within the confines of the cell, their work is continuously synchronized with other machines of similar or different nature. The chemical fuels of ATP or electrochemical gradients propel their functions, while chemical or physical stimuli serve as the controls. The duties entrusted to these remarkable machines encompass a myriad of tasks, from intracellular and transmembrane transport of reagents to the transformation of molecular building blocks into larger, functional structures. In essence, a cell unveils itself as a veritable molecular factory, a realm where diverse components are assembled, transformed, transported, and disassembled in an orchestrated dance of molecular interactions. At the molecular level, the dynamics of these processes are amplified by self-organization, cooperativity, and synchronization. These principles give rise to the living, moving organisms that grace the macroscopic scale. Just as in a grand factory, the performance of the whole system relies upon a modular building concept, periodic alignment, and the synchronization of individual dynamic components across temporal and spatial domains. These organizational principles, irrespective of scale or composition, serve as the bedrock for the design of cooperative dynamic systems. They epitomize the fundamental principles that govern the exquisite harmony and efficiency observed within factories, be they on a microscopic or macroscopic scale. However, it is crucial to recognize the distinctive qualities that set biological systems apart from their man-made counterparts. BMMs and their intricate assemblies possess unparalleled versatility and selectivity, continuously producing a diverse array of complex molecules that remain beyond the reach of any human-engineered system. Their prowess in generating such intricately crafted molecules underscores the remarkable ingenuity woven into the fabric of living organisms.
Within the living cell, a realm of extraordinary sophistication unfolds, showcasing a breathtaking array of nanotechnological wonders. Like a masterful workshop, it harbors an assortment of intricate instruments: pumps, levers, motors, rotors, turbines, propellers, and even scissors, each crafted with unparalleled precision. These minute marvels orchestrate a symphony of activity, operating with remarkable efficiency. At times, they work autonomously, while at others, they collaborate harmoniously with neighboring cells through a vast network of intercellular communication, facilitated by intricate chemical signaling. At the heart of this astonishing cellular orchestra lies the command and control center—the DNA database. Encoded within its molecular structure are the blueprints that govern the cell's functions. It serves as the custodian of instructions, diligently executing them through intermediary molecules. Employing an optimal mathematical code, these instructions seamlessly transform into tangible hardware products, each exquisitely tailored with specific functionality. It is a seamless interplay between software and hardware, orchestrated by the elegant dance of molecules. Yet, let us not forget that this awe-inspiring spectacle represents only a fraction of the complexity of life. In a larger organism, countless cells harmonize their efforts, joining forces to form intricate organs such as eyes, ears, brains, livers, and kidneys. These organs, akin to architectural marvels, exhibit a complexity that defies comprehension. Their structures, as elaborate as their functions, leave us humbled. Consider, for instance, the human brain—an entity teeming with an abundance of cells that surpasses the very number of stars adorning our vast Milky Way galaxy. When we contemplate the sheer magnitude of this orchestration, our minds are engulfed in awe. The cumulative marvels that abound within the realm of life are nothing short of stupefying. It is a testament to a grand design, one that invites us to delve deeper into the mysteries of existence. As we unravel the secrets concealed within these intricate cellular enclaves, we embark on a remarkable journey—a journey that both captivates and confounds us, igniting our curiosity and fueling our wonder.
Computer programs and machines are subsystems of the mind
In the realm of information and function, computer programs and machines serve as remarkable subsystems of the human mind. Unlike natural phenomena governed by the laws of physics and chemistry, these formal systems operate under the strict control of abstract rules. They belong to a distinct category of phenomena, separate from entities like rocks or ponds. Similarly, functional objects possess goal-oriented properties that depend on how a system is utilized, rather than being inherent physical properties. The concept of a machine, in the engineering sense, transcends the realms of physics and chemistry. Unlike the immutable laws of nature, machines are subject to malfunction and breakdown. Even when a machine is destroyed, the laws of physics and chemistry continue to operate unaffected in the remaining parts. The structure of a machine, rooted in engineering principles, effectively harnesses these laws to fulfill its designed purpose. However, the practical principles of design and coordination that imbue a machine with functionality can only be attributed to the mind of the engineer. In the realm of biology, an intriguing interdependence emerges between biological function and sign systems. To ensure the transmission of biological function through time, it must be stored within a time-independent sign system. Only through an abstract sign-based language can the necessary information be stored to construct functional biomolecules. Likewise, the very definition of the genetic code is intricately linked to biological function. This interdependency lies at the core of the origin of life, extending beyond the mere existence of designed organisms we observe today. The visionary thinker John von Neumann believed that life's reproduction was fundamentally based on logic, suggesting the presence of a logical construct supporting this phenomenon. To address the implications of Gödel's incompleteness theorem, von Neumann introduced a blueprint of the machine. His abstract model comprised a Universal Computer and a Universal Constructor. The Universal Constructor, guided by the directions encoded in the Universal Computer, constructs another Universal Constructor. Once complete, the Universal Constructor copies the Universal Computer and passes it down to its descendant. This model of self-replication finds its parallel in life, where the Universal Computer represents the genetic instructions within genes, and the Universal Constructor represents the cell and its machinery. The necessity of symbolic self-reference is a general premise in logic, crucial for replication. When it comes to copying biological bodies, their three-dimensional structure and relationship to the whole play a significant role. Accessing the internal sequence of amino acids (or nucleic acids in the case of ribozymes) necessary for replication would inevitably interfere with their structure and function. Enzymes, for instance, would require inherent self-replication properties or the ability to reconstitute their internal parts from existing components. In this context, it is more pertinent to consider physical and practical possibilities rather than applying logical terms such as "paradox" and "consistent" to biological systems. These constraints highlight the categorical distinction between the machine that reads instructions and the description of the machine. As we delve into the intricate web of information, function, and replication, we encounter the profound implications of intelligent design and the interplay between genetic instructions and cellular machinery. It is within this realm that the mysteries of life's complexity unravel. Exploring the intricacies of these systems invites us to contemplate the remarkable interdependence of information, function, and design—a testament to the ingenuity and purpose that underlie the living world
Last edited by Otangelo on Tue Aug 01, 2023 9:10 am; edited 22 times in total