Virus Capsids: A Work of art and marvellous engineering pointing to design
https://reasonandscience.catsboard.com/t3254-virus-capsids-a-work-of-art-and-marvellous-engineering-pointing-to-design
EKATERINA E. HELDWEIN (2018): Viral capsids are marvels of biological engineering. They are sturdy enough to withstand pressure exerted by the tightly packed genomes inside yet can come apart or loosen easily to release the viral genome once the virus penetrates the cell. They are also great examples of genetic economy. Because of the limited coding capacity of viruses, capsids are built by using a few proteins over and over. As a result, capsids are symmetrical. Capsids of many viruses, such as herpesviruses, have icosahedral symmetry, which means nearly spherical soccer ball–like particles with 20 triangular faces and 12 vertices. 3
Hong Zhou (2015): Chainmail is a system formed by concatenated rings. Chainmail in the form of interlocking rings of metal was used by medieval knights to protect their bodies from external forces in battle. The term protein chainmail was coined to explain how capsid proteins of HK97 form large complexes that behave abnormally under biochemical analyses. Viruses likely originated from ancient cells through encapsulating cellular plasmids or genome fragments by cellular protein 1 ( Really?!!)
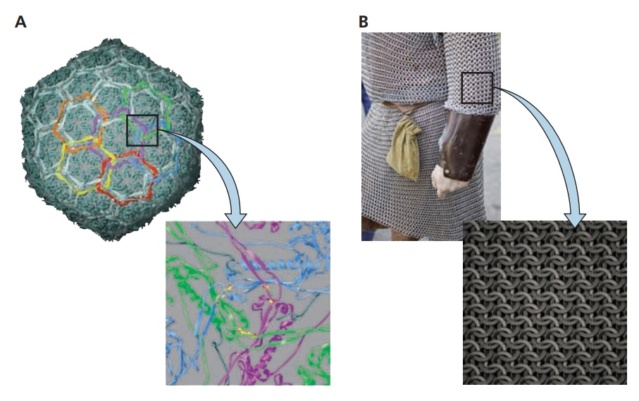
Chain mail in the bacteriophage HK97 capsid. (A) The exterior of the HK97 capsid is shown at the top, with structural units of the Gp5 protein in cyan. The segments of subunits that are cross-linked into rings are colored the same, to illustrate the formation of catenated rings of subunits. The cross-linking is shown in the more detailed view below, down a quasithreefold axis with three pairs of cross-linked subunits. The K-N isopeptide bonds are shown in yellow. The cross-linked monomers (shown in blue) loop over a second pair of covalently joined subunits (green), which in turn cross over a third pair (magenta).
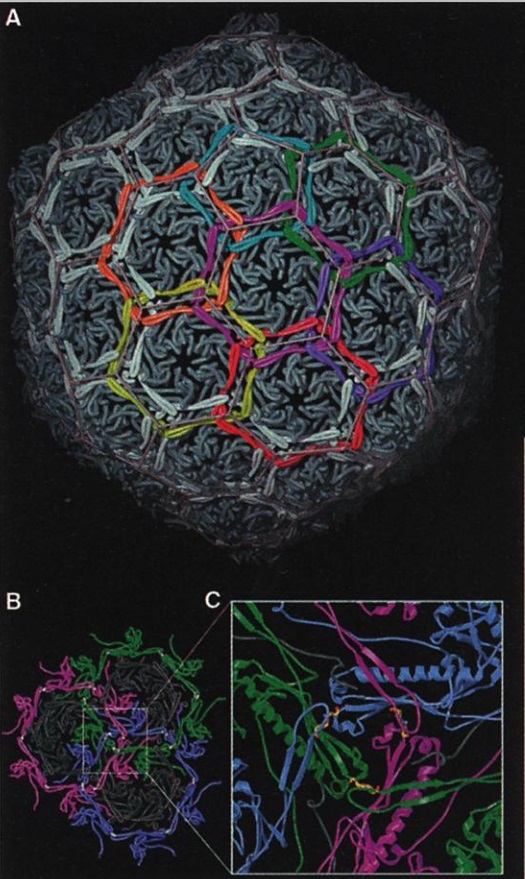
Capsid chainmail
(A) The subunits that are cross-linked into rings are colored identically, highlighting the catenated circle topology (only the region between the cross-linking residues is colored). Of these interlinked rings, 72 form the capsid, locking it into protein catenaries.
(B) View down a quasi-threefold axis from the capsid exterior. Three hexamers (black), each with a surrounding meta-hexamer ring of covalently bonded subunits (magenta, green, and blue).
(C) Enlargedview around the quasi-threefold axis, with three pairs of cross-linked subunits; isopeptide bonds are highlighted. Two cross-linked monomers (blue) loop over a second pair (green), which, in turn, crosses over a third pair (magenta).
Hong Zhou (2015): First discovered in bacteriophage HK97, biological chainmail is a highly stable system formed by concatenated protein rings. Each subunit of the ring contains the HK97-like fold, which is characterized by its submarine-like shape with a 5-stranded β sheet in the axial (A) domain, spine helix in the peripheral (P) domain, and an extended (E) loop. HK97 capsid consists of covalently linked copies of just one HK97-like fold protein and represents the most effective strategy to form highly stable chainmail needed for dsDNA genome encapsidation. Recently, near-atomic resolution structures enabled by cryo electron microscopy (cryoEM) have revealed a range of other, more complex variants of this strategy for constructing dsDNA viruses. The first strategy, exemplified by P22-like phages, is the attachment of an insertional (I) domain to the core 5- stranded β sheet of the HK97-like fold. The atomic models of the Bordetella phage BPP-1 showcases an alternative topology of the classic HK97 topology of the HK97-like fold, as well as the second strategy for constructing stable capsids, where an auxiliary jellyroll protein dimer serves to cement the non-covalent chainmail formed by capsid protein subunits. The third strategy, found in lambda-like phages, uses auxiliary protein trimers to stabilize the underlying noncovalent chainmail near the 3-fold axis. Herpesviruses represent highly complex viruses that use a combination of these strategies, resulting in four-level hierarchical organization including a noncovalent chainmail formed by the HK97-like fold domain found in the floor region. A thorough understanding of these structures should help unlock the enigma of the emergence and evolution of dsDNA viruses and inform bioengineering efforts based on these viruses. https://pubmed.ncbi.nlm.nih.gov/29177192/
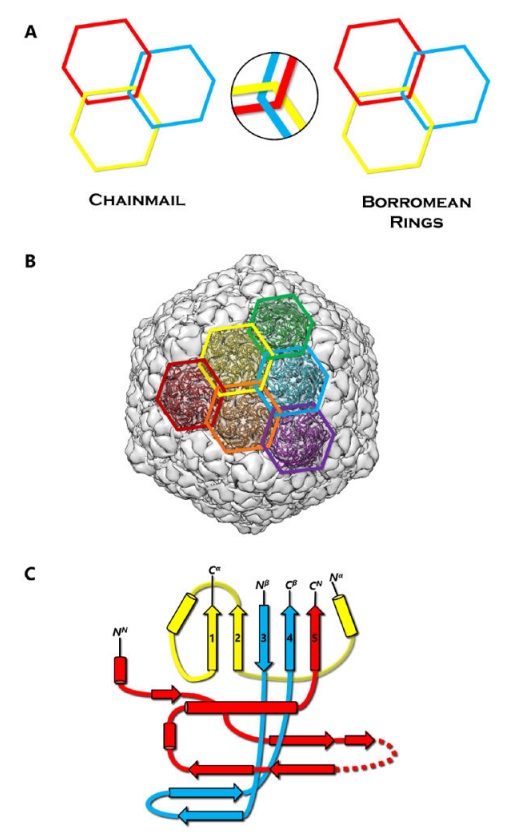
Protein Chainmail and the HK97-like fold.
(A) Concept of chainmail versus Borromean rings. Breaking one ring in a chainmail would not affect the integrity of the whole, unlike the case of Borromean rings.
(B) Protein chainmail of HK97. Chainmail is a structural organization of concatenated rings found in the capsids of icosahedral dsDNA viruses, and allow capsids to withstand internal forces exerted by the dsDNA. The interlocking rings of protein chainmail resemble armor constructed of metal rings worn by medieval knights during times of battle.
(C) Basic building blocks of the HK97-like fold include the N (red), α (yellow), and β (blue) primary elements. Variations of the HK97-like fold can occur when the basic building blocks are connected in different order. Additionally, extra domains can be inserted at the E-loop or at the ends of the building blocks.
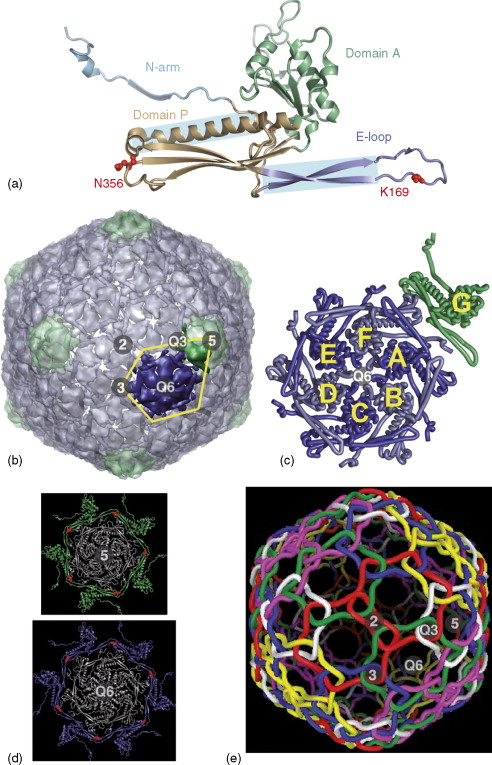
Structure of the T = 7 HK97 particle.
Icosahedral and quasi-symmetry (Q) axes are labeled as in Figure 3. (a) HK97 subunit fold. The small HK97 subunits have mixed α/β structure, forming two domains in the shape of an L that have a continuous hydrophobic core, and placing the two residues involved in cross-linking other subunits (red) on opposite sides of the protein. (b) Surface representation of the entire HK97 capsid viewed down an icosahedral twofold axis, with pentamers colored separately from hexamers. The IAU is outlined in yellow. Unlike the T = 3 and T = 4 structures, none of the hexamers sit on icosahedral axes. (c) Ribbon diagram of the seven subunits in the IAU showing how they align such that the P-domains and E-loops overlap to define the sides of the local capsomers, but place their tips (with the cross-linking residues) outside the capsomer to be accessible to neighboring subunits. (d) The two types of cross-linked subunit ‘circles’ or ‘rings’ in HK97. The surrounding cross-linked subunits (one from each neighboring capsomer) are shown in green (5-circle) or blue (Q6-circle), with the cross-link shown in red. (e) Diagrammatic representation of the interlocking cross-links in HK97. The 5- or Q6-circles, now with subunit and cross-links just represented by tubes, are colored separately showing how they form molecular chainmail by interweaving at the icosahedral and quasi-threefold axes. This molecular chainmail stabilizes the unusually thin 18-Å-wide protein shell (only half the width usually formed by capsids composed of β-barrel subunits), which can be described as a protein balloon. The capsids are also stabilized by other extensive inter-subunit interactions with up to nine other coat proteins. The N-arm, which is not involved in the cross-links, extends 67 Å away from the subunit to contact subunits in neighboring capsomers. (a–d) Courtesy of Dr. Lu Gan. (e) Courtesy of Gabe Lander.
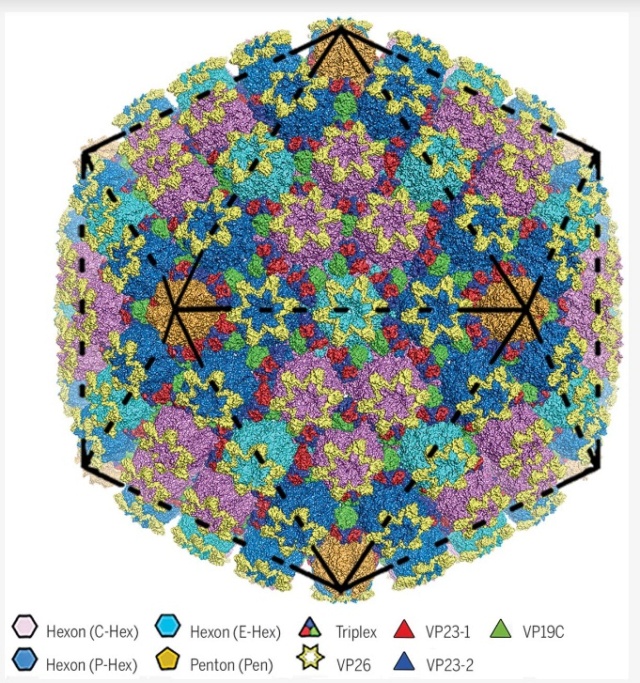
Å structure of HSV-2 B capsid.
Surface representation of HSV-2’s 1250-Å-wide capsid. Black lines represent particle icosahedral facets.
SHUAI YUAN (2018):The herpesvirus virion is genetically and structurally one of the largest and most complex viruses known. It has a T = 16 (triangulation number) icosahedral capsid with a diameter of ~125 nm that not only protects the viral genome physically from damage but also plays an important role in the release of viral genome into the nucleus of the host cell. HSV capsid assembly requires the ordered packing of about 4000 protein subunits into the hexons, pentons, and triplexes that comprise the capsid. 2
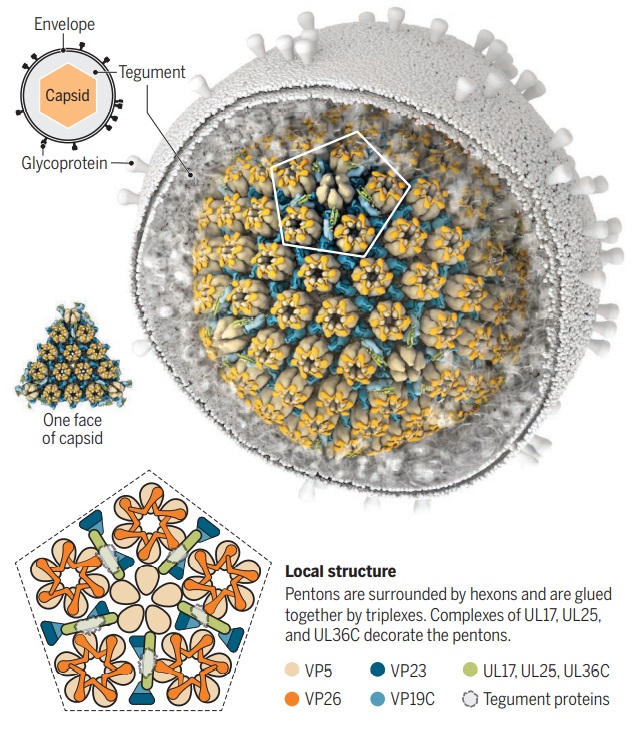
Zooming in on virus structure
The HSV capsid is an icosahedron with 20 faces and 12 vertices. Two high-resolution structures show how ~3000 proteins that form pentons, hexons, and triplexes are arranged within the HSV capsid. One structure also shows how the tegument is anchored to the pentons.
1. Hong Zhou: Protein chainmail variants in dsDNA viruses 2015
2. SHUAI YUAN: Cryo-EM structure of a herpesvirus capsid at 3.1 Å 6 Apr 2018
3. EKATERINA E. HELDWEIN: Up close with herpesviruses 6 Apr 2018
https://reasonandscience.catsboard.com/t3254-virus-capsids-a-work-of-art-and-marvellous-engineering-pointing-to-design
EKATERINA E. HELDWEIN (2018): Viral capsids are marvels of biological engineering. They are sturdy enough to withstand pressure exerted by the tightly packed genomes inside yet can come apart or loosen easily to release the viral genome once the virus penetrates the cell. They are also great examples of genetic economy. Because of the limited coding capacity of viruses, capsids are built by using a few proteins over and over. As a result, capsids are symmetrical. Capsids of many viruses, such as herpesviruses, have icosahedral symmetry, which means nearly spherical soccer ball–like particles with 20 triangular faces and 12 vertices. 3
Hong Zhou (2015): Chainmail is a system formed by concatenated rings. Chainmail in the form of interlocking rings of metal was used by medieval knights to protect their bodies from external forces in battle. The term protein chainmail was coined to explain how capsid proteins of HK97 form large complexes that behave abnormally under biochemical analyses. Viruses likely originated from ancient cells through encapsulating cellular plasmids or genome fragments by cellular protein 1 ( Really?!!)

Chain mail in the bacteriophage HK97 capsid. (A) The exterior of the HK97 capsid is shown at the top, with structural units of the Gp5 protein in cyan. The segments of subunits that are cross-linked into rings are colored the same, to illustrate the formation of catenated rings of subunits. The cross-linking is shown in the more detailed view below, down a quasithreefold axis with three pairs of cross-linked subunits. The K-N isopeptide bonds are shown in yellow. The cross-linked monomers (shown in blue) loop over a second pair of covalently joined subunits (green), which in turn cross over a third pair (magenta).

Capsid chainmail
(A) The subunits that are cross-linked into rings are colored identically, highlighting the catenated circle topology (only the region between the cross-linking residues is colored). Of these interlinked rings, 72 form the capsid, locking it into protein catenaries.
(B) View down a quasi-threefold axis from the capsid exterior. Three hexamers (black), each with a surrounding meta-hexamer ring of covalently bonded subunits (magenta, green, and blue).
(C) Enlargedview around the quasi-threefold axis, with three pairs of cross-linked subunits; isopeptide bonds are highlighted. Two cross-linked monomers (blue) loop over a second pair (green), which, in turn, crosses over a third pair (magenta).
Hong Zhou (2015): First discovered in bacteriophage HK97, biological chainmail is a highly stable system formed by concatenated protein rings. Each subunit of the ring contains the HK97-like fold, which is characterized by its submarine-like shape with a 5-stranded β sheet in the axial (A) domain, spine helix in the peripheral (P) domain, and an extended (E) loop. HK97 capsid consists of covalently linked copies of just one HK97-like fold protein and represents the most effective strategy to form highly stable chainmail needed for dsDNA genome encapsidation. Recently, near-atomic resolution structures enabled by cryo electron microscopy (cryoEM) have revealed a range of other, more complex variants of this strategy for constructing dsDNA viruses. The first strategy, exemplified by P22-like phages, is the attachment of an insertional (I) domain to the core 5- stranded β sheet of the HK97-like fold. The atomic models of the Bordetella phage BPP-1 showcases an alternative topology of the classic HK97 topology of the HK97-like fold, as well as the second strategy for constructing stable capsids, where an auxiliary jellyroll protein dimer serves to cement the non-covalent chainmail formed by capsid protein subunits. The third strategy, found in lambda-like phages, uses auxiliary protein trimers to stabilize the underlying noncovalent chainmail near the 3-fold axis. Herpesviruses represent highly complex viruses that use a combination of these strategies, resulting in four-level hierarchical organization including a noncovalent chainmail formed by the HK97-like fold domain found in the floor region. A thorough understanding of these structures should help unlock the enigma of the emergence and evolution of dsDNA viruses and inform bioengineering efforts based on these viruses. https://pubmed.ncbi.nlm.nih.gov/29177192/

Protein Chainmail and the HK97-like fold.
(A) Concept of chainmail versus Borromean rings. Breaking one ring in a chainmail would not affect the integrity of the whole, unlike the case of Borromean rings.
(B) Protein chainmail of HK97. Chainmail is a structural organization of concatenated rings found in the capsids of icosahedral dsDNA viruses, and allow capsids to withstand internal forces exerted by the dsDNA. The interlocking rings of protein chainmail resemble armor constructed of metal rings worn by medieval knights during times of battle.
(C) Basic building blocks of the HK97-like fold include the N (red), α (yellow), and β (blue) primary elements. Variations of the HK97-like fold can occur when the basic building blocks are connected in different order. Additionally, extra domains can be inserted at the E-loop or at the ends of the building blocks.

Structure of the T = 7 HK97 particle.
Icosahedral and quasi-symmetry (Q) axes are labeled as in Figure 3. (a) HK97 subunit fold. The small HK97 subunits have mixed α/β structure, forming two domains in the shape of an L that have a continuous hydrophobic core, and placing the two residues involved in cross-linking other subunits (red) on opposite sides of the protein. (b) Surface representation of the entire HK97 capsid viewed down an icosahedral twofold axis, with pentamers colored separately from hexamers. The IAU is outlined in yellow. Unlike the T = 3 and T = 4 structures, none of the hexamers sit on icosahedral axes. (c) Ribbon diagram of the seven subunits in the IAU showing how they align such that the P-domains and E-loops overlap to define the sides of the local capsomers, but place their tips (with the cross-linking residues) outside the capsomer to be accessible to neighboring subunits. (d) The two types of cross-linked subunit ‘circles’ or ‘rings’ in HK97. The surrounding cross-linked subunits (one from each neighboring capsomer) are shown in green (5-circle) or blue (Q6-circle), with the cross-link shown in red. (e) Diagrammatic representation of the interlocking cross-links in HK97. The 5- or Q6-circles, now with subunit and cross-links just represented by tubes, are colored separately showing how they form molecular chainmail by interweaving at the icosahedral and quasi-threefold axes. This molecular chainmail stabilizes the unusually thin 18-Å-wide protein shell (only half the width usually formed by capsids composed of β-barrel subunits), which can be described as a protein balloon. The capsids are also stabilized by other extensive inter-subunit interactions with up to nine other coat proteins. The N-arm, which is not involved in the cross-links, extends 67 Å away from the subunit to contact subunits in neighboring capsomers. (a–d) Courtesy of Dr. Lu Gan. (e) Courtesy of Gabe Lander.

Å structure of HSV-2 B capsid.
Surface representation of HSV-2’s 1250-Å-wide capsid. Black lines represent particle icosahedral facets.
SHUAI YUAN (2018):The herpesvirus virion is genetically and structurally one of the largest and most complex viruses known. It has a T = 16 (triangulation number) icosahedral capsid with a diameter of ~125 nm that not only protects the viral genome physically from damage but also plays an important role in the release of viral genome into the nucleus of the host cell. HSV capsid assembly requires the ordered packing of about 4000 protein subunits into the hexons, pentons, and triplexes that comprise the capsid. 2

Zooming in on virus structure
The HSV capsid is an icosahedron with 20 faces and 12 vertices. Two high-resolution structures show how ~3000 proteins that form pentons, hexons, and triplexes are arranged within the HSV capsid. One structure also shows how the tegument is anchored to the pentons.
1. Hong Zhou: Protein chainmail variants in dsDNA viruses 2015
2. SHUAI YUAN: Cryo-EM structure of a herpesvirus capsid at 3.1 Å 6 Apr 2018
3. EKATERINA E. HELDWEIN: Up close with herpesviruses 6 Apr 2018


