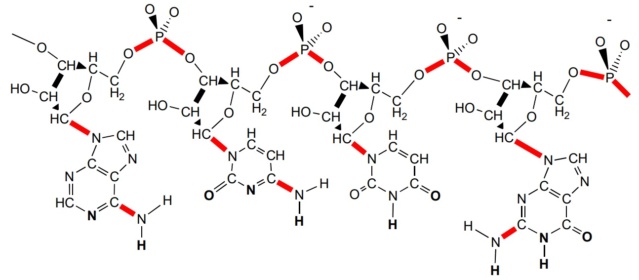
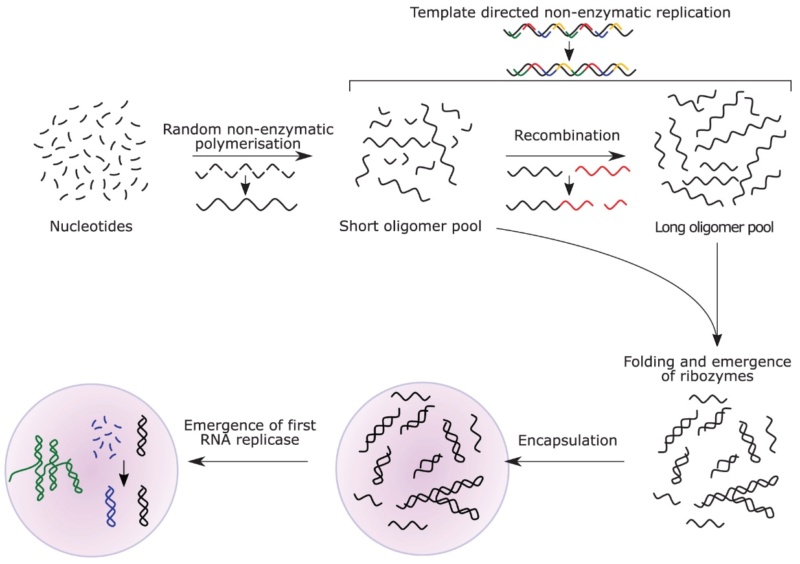
Index Page: On the origin of Cell factories by the means of an intelligent designer
https://reasonandscience.catsboard.com/t2809-on-the-origin-of-cell-factories-by-the-means-of-an-intelligent-designer
Chapter 1
Introduction
God of the gaps
Limited causal alternatives do not justify to claim of
A God, or no God. That's the question
What's the Mechanism of Intelligent Design?
Is the
Did God create Ex-nihilo?
Intelligence vs no intelligence
Genesis or Darwin?
Naturalists hijack science by imposing philosophical naturalism
Consensus in science
What is life?
The constraint of philosophical naturalism, and consensus science, leads to bullshit science
Paley's watchmaker argument 2.0
Chapter 2
Living Cells are chemical factories
Argument from analogy
Cells are factories in a literal sense, or just as a metaphor?
Cell Metabolism as a production line system
Following would give a good sci-fi movie.
Cells are full of robotic assembly lines: evolved, or created?
Cells superb manufacturing concepts and incredible performance evidences intelligent design
Does the fact that cells self-replicate refute the claim that cells are factories?
The origin of cell factories
Difficulties in top-down approaches: Could life have started simple?
The origin of cell factories
Difficulties in top-down approaches: Could life have started simple?
LUCA, the last universal common ancestor
Nobody knows what LUCA and FUCA looked like
Viruses
Giant Viruses
What are the oldest life forms?
Timeline of the earliest evidence of life
Are the first life forms traced back to submarine vents?
Maybe Cyanobacteria?
Gloeobacter violaceus, a basal cyanobacteria
What does science know about a supposed last bacterial common ancestor (LBCA)?
The first bacterial lineages to diverge were most similar to modern Clostridia
But, after all, how simple can we go, and what is the best model candidate to study the origin of life?
Spontaneous generation of life
Chapter 3
The bottom-up approach
What are the possible mechanisms & causes to explain the origin of life?
Evolution
Physical necessity
Unguided random accidental events
Frozen accidents
Emergent properties:
Time: the naturalist's friend?
Abiogenesis research is a failure
Life requires 1. Matter, 2. Energy, and 3. Information.
There was no prebiotic selection to get the basic building blocks of life
Undesired contamination, and mixtures
Biomolecules decompose and degrade. They do not complexify
Chapter 4
The earth, and the atmosphere, just right for life
Essential elements and building blocks for the origin of life
Energy cycles, how did they
Carbon, essential for life
A finely tuned Carbon-cycle - is essential for life
Origin of carbon fixation.
Origin of ammonia on early earth
The transition to enzymatic fixation of nitrogen
The nitrogen cycle, irreducible interdependence, and the origin of life
Nitrogen levels in the atmosphere must be just right
Oxygen
How could the atmosphere have been aerobic prior the great oxidation event?
From an anaerobic to an aerobic atmosphere
Reactive oxygen species (ROS) & the origin of life
Hydrogen
The faint young sun paradox
Major elements essential for life to start
Calcium
Phosphorus
Chapter 5
Origin of the building blocks of life
Going from prebiotic to biotic synthesis: a major unsolved open question
Amino acids
Origin of the proteinogenic ( protein creating ) amino acids used in life
Extraterrestrial origins
Panspermia
What about the synthesis of amino acids in hydrothermal vents?
The Miller-Urey experiment
Homochirality
Homochirality, its origin a scientifically longstanding unresolved issue
Why only left-handed, and not right-handed amino acids?
From prebiotic to biotic chirality determination
Aspartate Aminotransferase
How were the 20 proteinogenic amino acids selected on early earth?
Optimality of the amino acid set that is used to encode proteins
https://reasonandscience.catsboard.com/t2809-on-the-origin-of-cell-factories-by-the-means-of-an-intelligent-designer
Chapter 1
Introduction
God of the gaps
Limited causal alternatives do not justify to claim of
A God, or no God. That's the question
What's the Mechanism of Intelligent Design?
Is the
Did God create Ex-nihilo?
Intelligence vs no intelligence
Genesis or Darwin?
Naturalists hijack science by imposing philosophical naturalism
Consensus in science
What is life?
The constraint of philosophical naturalism, and consensus science, leads to bullshit science
Paley's watchmaker argument 2.0
Chapter 2
Living Cells are chemical factories
Argument from analogy
Cells are factories in a literal sense, or just as a metaphor?
Cell Metabolism as a production line system
Following would give a good sci-fi movie.
Cells are full of robotic assembly lines: evolved, or created?
Cells superb manufacturing concepts and incredible performance evidences intelligent design
Does the fact that cells self-replicate refute the claim that cells are factories?
The origin of cell factories
Difficulties in top-down approaches: Could life have started simple?
The origin of cell factories
Difficulties in top-down approaches: Could life have started simple?
LUCA, the last universal common ancestor
Nobody knows what LUCA and FUCA looked like
Viruses
Giant Viruses
What are the oldest life forms?
Timeline of the earliest evidence of life
Are the first life forms traced back to submarine vents?
Maybe Cyanobacteria?
Gloeobacter violaceus, a basal cyanobacteria
What does science know about a supposed last bacterial common ancestor (LBCA)?
The first bacterial lineages to diverge were most similar to modern Clostridia
But, after all, how simple can we go, and what is the best model candidate to study the origin of life?
Spontaneous generation of life
Chapter 3
The bottom-up approach
What are the possible mechanisms & causes to explain the origin of life?
Evolution
Physical necessity
Unguided random accidental events
Frozen accidents
Emergent properties:
Time: the naturalist's friend?
Abiogenesis research is a failure
Life requires 1. Matter, 2. Energy, and 3. Information.
There was no prebiotic selection to get the basic building blocks of life
Undesired contamination, and mixtures
Biomolecules decompose and degrade. They do not complexify
Chapter 4
The earth, and the atmosphere, just right for life
Essential elements and building blocks for the origin of life
Energy cycles, how did they
Carbon, essential for life
A finely tuned Carbon-cycle - is essential for life
Origin of carbon fixation.
Origin of ammonia on early earth
The transition to enzymatic fixation of nitrogen
The nitrogen cycle, irreducible interdependence, and the origin of life
Nitrogen levels in the atmosphere must be just right
Oxygen
How could the atmosphere have been aerobic prior the great oxidation event?
From an anaerobic to an aerobic atmosphere
Reactive oxygen species (ROS) & the origin of life
Hydrogen
The faint young sun paradox
Major elements essential for life to start
Calcium
Phosphorus
Chapter 5
Origin of the building blocks of life
Going from prebiotic to biotic synthesis: a major unsolved open question
Amino acids
Origin of the proteinogenic ( protein creating ) amino acids used in life
Extraterrestrial origins
Panspermia
What about the synthesis of amino acids in hydrothermal vents?
The Miller-Urey experiment
Homochirality
Homochirality, its origin a scientifically longstanding unresolved issue
Why only left-handed, and not right-handed amino acids?
From prebiotic to biotic chirality determination
Aspartate Aminotransferase
How were the 20 proteinogenic amino acids selected on early earth?
Optimality of the amino acid set that is used to encode proteins