Talin as a mechanosensitive signaling hub
https://reasonandscience.catsboard.com/t3175-talin-as-a-mechanosensitive-signaling-hub
A Chat with Ben Goult about the Talin Code
https://www.youtube.com/watch?v=yG8AiBc-trY
Ben Goult: The Mechanical Basis of Memory
https://www.youtube.com/watch?v=C0A8bTehUls&t=5s
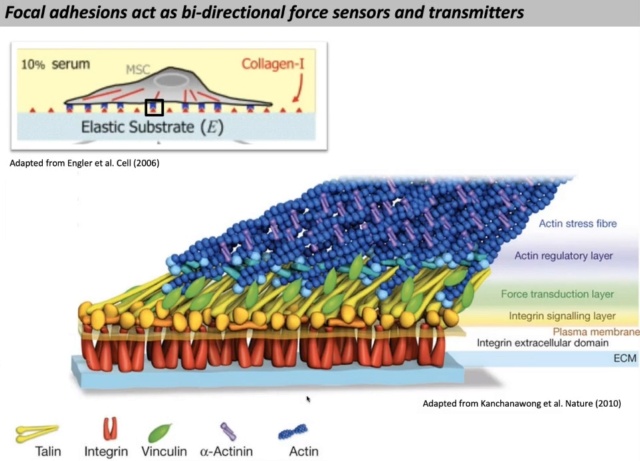
03:53 And then what talin also does is it couples these integrins to the actin cytoskeleton and this is really important you need to be able to couple these attachments to the side skeleton to enable it to withstand forces and if you've cut or removed several ones of these linkages this whole thing will fall apart and your cell will round up and die
My comment: That is evidence of an irreducibly complex system, where all players must be in place in an interlocked, interdependent fashion for the system to work, or nothing goes. That raises the question of how the system could have evolved in the first place if all has to be in place.
04:41 What's really interesting to us and hints again at the complexity which were looking at is that these three proteins integrate alien actin and been killin as a fourth this core assembly of these adhesions is responsible for all the adhesive structures that the cells form or not all but many many of them
Question: .Which of these proteins are not necessary for the cell to survive? Talin, Integrin, Vinculin, alpha-actinin, Actin?
Krishna Chinthalapudi The interaction of talin with the cell membrane is essential for integrin activation and focal adhesion formation April 11, 2018
Vertebrate cell growth, division, locomotion, morphogenesis, and development rely on the dynamic interactions of cells with extracellular matrix components via cell surface complexes termed focal adhesions that are composed of heterodimeric αβ integrin receptors, associated signaling molecules, and the large cytoskeletal protein talin. Talin activation and binding to β-integrin requires interactions with lipids. Here we report that talin binding to the cell membrane seems necessary for integrin activation and focal adhesion formation.
https://www.pnas.org/content/115/41/10339
My comment: It seems that cell adhesion, and signaling, are two essential processes, that have to be instantiated in an integrated fashion. That means, two problems and necessities have to be solved, and implemented at once. That requires the establishment of a sophisticated information and communication system, and the right engineering architecture with multiple players working in a joint venture.
05:28 Our central hypothesis is that talin adopts numerous different conformational States which runs cellular programs to enable these different structures to form
Question: How is the information encoded ?
Benjamin T. Goult Talin as a mechanosensitive signaling hub September 25 2018 [url=One of the most incomprehensible things that are hard to fathom is that God loves us. Despite our sins and shortcomings.]5[/url]
Cell adhesion to the ECM is fundamental to multicellular life. Deletion of major integrins or ECM proteins impairs the development and survival of multicellular organisms.
My comment: That means, that the origin of integrins is a fundamental linked to the quest of how multicellularity emerged.
From an evolutionary perspective, support for the transition from unicellular (single cell) to multicellular organisms requires the emergence of several novel biochemical systems. 1 Such systems include:
- pathways that transform cells from generalized to specialized forms during growth and development;
- mechanisms for the migration of cells relative to each other during growth and development;
- structures that support cell-cell adhesions;
- and mechanisms for cell-cell communication.
- All of these systems have to be in place and operate in an integrated fashion to support multicellularity.
1. Despite a thorough search, no material causes have been discovered that demonstrate the power to produce large amounts of specified information, irreducible, and interdependent biological systems.
2. Intelligent causes have demonstrated the power to produce large amounts of specified information, irreducible and interdependent systems of all sorts.
3. Intelligent design constitutes the best, most causally adequate, explanation for the information and irreducible complexity in the cell, and interdependence of proteins, organelles, and body parts, and even of animals and plants, aka moths and flowers, for example.
Integrin-mediated adhesions sense the mechanical features of the matrix, including stiffness, texture, and externally applied strains, transducing these forces into biological signals 2
Talin plays a central role in cell adhesion, first by converting integrins to high-affinity states (“activation”) and by coupling integrins to the cytoskeleton. Indeed, deletion of talin results in developmental defects in multiple organisms that resemble total loss of integrins
My comment: Here we have another protein that performs essential functions in multicellular organisms. Which rises the question: How could it have evolved, if its essential function only becomes essential once it is integrated in the system? Some might come up with the mullerian two step proposal: Add a part. Make it necessary. That is the heart of Mullerian two-step process. The problem however is always explaining how biological information or function is built up in the first place. 3
Talin is a large (270 kD) multidomain cytosolic protein
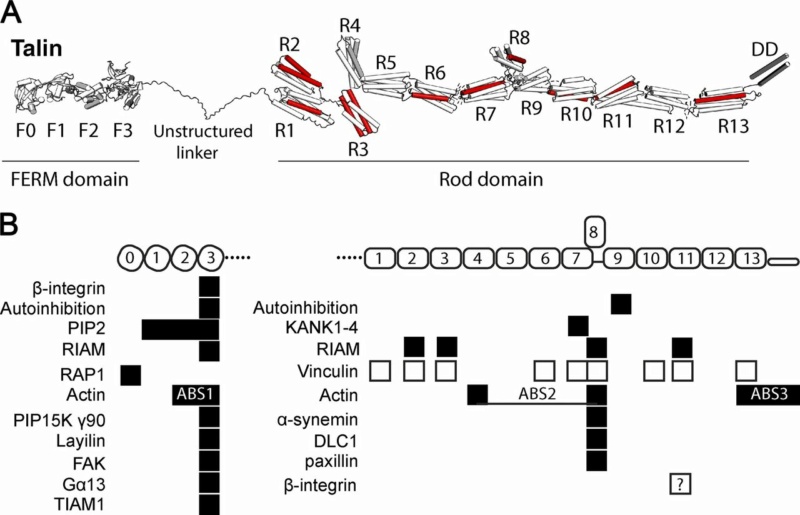
Talin domain organization and interactions.
(A) Talin contains an N-terminal FERM domain (F0–F3) connected via an 80-aa unstructured linker to the 13 talin rod domains R1–R13. 9 of the 13 rod domains contain VBSs (red).
(B) Major binding sites for folded talin domains (black boxes) and unfolded domains (white boxes).
Talin links integrins to F-actin in part via binding of its N-terminal FERM domain to integrin cytoplasmic domains as well as via two sites in its C-terminal flexible rod domain that bind F-actin.
David A. Calderwood Talins and kindlins: partners in integrin-mediated adhesion 17 July 2013 4
Integrin receptors provide a dynamic, tightly regulated link between the extracellular matrix (or cellular counter-receptors) and intracellular cytoskeletal and signalling networks, enabling cells to sense and respond to their chemical and physical environment. Talins and kindlins, two families of FERM–domain proteins, bind the cytoplasmic tail of integrins, recruit cytoskeletal and signalling proteins involved in mechanotransduction and synergize to activate integrin binding to extracellular ligands. Talin mediates integrin activation. RIAM recruits talin to the plasma membrane, whereas vinculin stabilizes talin in cell– matrix junctions.
My comment: Binding,synergizing to activate, mediating, activating, recruiting, stabilizing are all goal-oriented processes. Why and how would/could unguided evolutionary pressures that are neither goal-oriented, nor having foresight, produce such complex mechanistic, and information-based processes, that show clearly that a higher-order/goal/complexity is being achieved, namely producing the interaction of various cells together?
Cell proliferation, cell migration, tissue morphogenesis and homeostasis all depend on cell–cell and cell–extracellular matrix (ECM) interactions, and the integrin family of adhesion receptors has essential roles in these processes.
My comment: Essential means that the processes and events could/would not occur without integrin adhesion receptors. That makes them irreducible.
These binding events are followed by the application of tension from actomyosin that acts on the talin rod, triggering recruitment of a second actin-binding protein, vinculin . This mechanism thus strengthens adhesions under tension, a form of mechanosensitivity.
Despite the presence of the same core components, adhesion complexes are strikingly diverse. Highly dynamic transient adhesions enable cell migration; dynamic and proteolytic (enzymes that break down protein) adhesions mediate invasion; stable adhesions promote tissue organization; and specialized myotendinous junctions transmit very high forces for animal movements. Around the central core of integrin, talin, and actin, numerous additional proteins show selective recruitment that vary widely between adhesion types, likely reflecting distinct effector, signaling, and mechanosensing functions.
The purpose of this perspective is to summarize existing knowledge and propose a new view of talin function. It is therefore split into two sections: the first section reviews recent data on talin and the emerging functional implications, and the second, more speculative section proposes that there exists a “talin code” of force-dependent interactions with signaling proteins and cytoskeletal components, which exhibits some internal hierarchy. This view, where talin serves as a flexible mechanosensitive signaling hub (MSH), has the potential to explain diverse responses of cells to distinct mechanical stimuli on different time scales.
Talin interactions and functions
The multiple domains with numerous force-sensitive binding sites in talin, coupled with their linear arrangement in the path of force transmission (Fig. above), provides opportunities for enormous functional flexibility.
Adhesion dynamics
Regulated adhesion complex assembly and disassembly are vital for cell spreading and migration. In cells freshly plated on ECM or in rapidly migrating cells, activated integrins form small clusters under the lamellipodia. These small adhesions can either rapidly disassemble or can connect to larger actin templates and mature into slightly larger, more stable structures called focal complexes, which themselves either disassemble or further mature into much larger adhesive structures including focal adhesions (FAs; e.g., in contractile cells on rigid substrates), podosomes (in activated cells on soft substrates), or very stable adhesions as in myocytes or myofibroblasts (in highly contractile cells on rigid substrates). In all cases, talin is a major player in force-dependent adhesion growth and stabilization. Indeed, cardiac and skeletal muscle express an alternatively spliced β1-integrin (β1D) with increased affinity for talin, a key event in exertion of ultra-high forces.
Force transmission between actin and integrins at the leading edges of migrating or spreading cells is mediated by a unique, dynamic mechanism in which talin plays a central role. Actin polymerizes at cell edges and flows rearwards, pushed by the force generated by polymerization and/or pulled by myosin motors further back in the cell. This retrograde flow of actin couples to integrins via talin, thereby exerting traction force on the matrix or substrate for spreading, migration, or contraction. This force transfer must occur through highly dynamic bonds that exhibit variable coupling efficiency: the so-called FA clutch.
Talin conformation and mechanotransduction
There are two talin isoforms, talins 1 and 2, that are 76% identical and have identical domain structures. Both contain an N-terminal FERM domain-containing four globular segments (F0 to F3), a disordered linker region, and a C-terminal rod with 13 four- and five-helix bundles (R1 to R13) terminating in a single α-helix that mediates homodimerization (dimerization domain [DD]; Fig. above). With the exception of R8, which is positioned outside the force transmission pathway as will be discussed, the rod domains are arranged linearly like beads on a string, transmitting tension along the talin rod. At least 9 of the 13 rod domains contain cryptic vinculin-binding sites (VBSs) that are exposed when unfolded by mechanical force, allowing vinculin binding and adhesion reinforcement. Talin is autoinhibited by an interaction between the head (F3) and R9, which must be released for actin and integrin-binding and recruitment to FAs. Interaction of talin with negatively charged phosphatidylinositol 4,5-bisphosphate of the plasma membrane inner leaflet also contributes to talin activation and membrane association.
Force-dependent switching of binding partners to the R3 rod domain defines a mechanochemical switch
The best-studied talin activation pathway depends on the small GTPase Rap1, whose effector Rap1-GTP–interacting adaptor molecule (RIAM) binds directly to talin via a high-affinity RIAM binding site in R2–R3 (note that talin R8 and R11 also contain RIAM-binding sites and that Rap1 has been shown to bind directly to talin F0. Rap1/RIAM recruits talin to the plasma membrane and antagonizes talin autoinhibition to promote integrin and actin binding. Importantly, RIAM engages folded talin R3 (Fig. below).
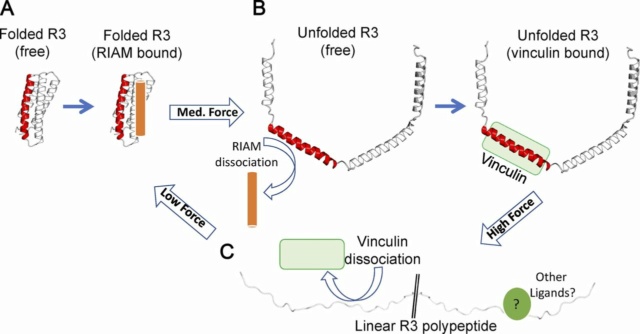
Talin rod domains as mechanochemical switches. (A–C) Each talin rod domain can adopt a number of conformations under different force regimes. (A) Folded bundle at low force. (B) An unfolded string of helices at forces above the mechanical threshold. (C) A fully unfolded polypeptide at high forces. Force-induced domain unfolding leads to a switch in the ligand-binding profile of that domain. Complete unfolding at high force will result in a linear polypeptide unable to bind folded-domain ligands or helix-binding ligands. While no ligands for this form have been identified so far, many proteins bind linear peptide motifs.
R3 is the least stable of the 13 talin rod domains due to a cluster of four threonines in the central hydrophobic core of the four-helix bundle. Consequently, R3 opens under the relatively low force of 5 pN, converting the folded four-helix bundle into a string of helices that could be further unfolded into a disordered conformation. This transition exposes two high-affinity VBSs, which in the absence of force, were cryptic, buried inside the core of the folded R3. Therefore, the force-dependent unfolding of R3 results in exposure of VBS-recruiting vinculin while simultaneously disrupting RIAM binding, severing the link to Rap1 signaling. This switch in ligands is mirrored in cells, where an integrin–talin–RIAM complex at the tip of cell protrusions (“sticky fingers”) is replaced by an integrin–talin–vinculin complex in mature, high-tension FAs, which are devoid of RIAM. This exchange of RIAM, bound to talin in the absence of force for vinculin in the presence of forces, thus defines a mechanochemical switch (Fig. above, A and B).
Structural basis for talin rod interactions
To date, there have been no comprehensive proteomic analyses of the talin interactome; however, structural studies on ligands that bind talin rod domains have begun to reveal interesting themes. Deleted in liver cancer 1 (DLC1), a RhoGAP and tumor suppressor binds the talin R8 domain via an α-helical leucine–aspartic acid (LD) motif. Elucidation of the structure of the DLC1–talin complex revealed a helix-addition mechanism with an amphipathic LD helix of DLC1 packed between two adjacent helices on the surface of the R8 four-helix bundle, effectively converting it to a five-helix bundle. These findings led to the realization that the talin-binding sequence (TBS) in RIAM is also an LD motif as well as the identification of paxillin, which has several LD motifs, as a novel talin ligand. R8 is structurally similar to the FAK FA targeting (FAT) domain, a four-helix bundle that also binds LD motifs. Another class of LD motif-containing proteins, the KANKs (kidney ankyrin repeat-containing), bind to talin R7, a five-helix bundle (Bouchet et al., 2016; Sun et al., 2016b), and mediate connections of microtubules to adhesion complexes. Five-helix-bundle LD motif recognition is also shown with RIAM TBS1 binding the R11 five-helix bundle (Goult et al., 2013). Helix addition thus appears to be a general mechanism for binding to talin rod domain helix bundles in their folded state.
Timo Baade Clustering of integrin β cytoplasmic domains triggers nascent adhesion formation and reveals a protozoan origin of the integrin-talin interaction 05 April 2019 6
The transition from unicellular organisms to multicellular animals (metazoans) coincides with the emergence of dedicated cell adhesion molecules such as cadherins and integrins1. Since their discovery in the mid-80s, it became clear that integrins are not only essential for coupling of the cytoskeleton to the extracellular matrix upon cell attachment, but that they also contribute to a multitude of basic cellular processes including cell survival, proliferation, differentiation, and cell migration.
Integrins and integrin-dependent cell-matrix adhesions are essential for a number of physiological processes. Integrin function is tightly regulated via binding of cytoplasmic proteins to integrin intracellular domains. Yet, the complexity of cell-matrix adhesions in mammals, with more than 150 core adhesome proteins, complicates the analysis of integrin-associated protein complexes. Interestingly, the evolutionary origin of integrins dates back before the transition from unicellular life to complex multicellular animals. Though unicellular relatives of metazoa have a less complex adhesome, nothing is known about the initial steps of integrin activation and adhesion complex assembly in protozoa.
One of the proteins able to initiate integrin inside-out activation is talin. With its N-terminal FERM domain, talin binds to the NPXY/F motif in the membrane proximal part of the integrin β cytoplasmic tail, thereby pushing apart the two integrin subunits followed by the unfolding of their ectodomains. Upon talin binding to the integrin β subunit, additional cytoplasmic proteins are recruited to integrin-initiated focal adhesion (FA) sites.
All β-subunits share the highly conserved membrane proximal NPXY/F and the lesser conserved membrane distal NXXY motif
Clustering of the human integrin β1 tail led to the recruitment of talin, kindlin, and paxillin, and mutation of the known talin binding site abolished recruitment of this protein.
Question: Is the ability of talin to bind to integrin not essential for function? Is that not evidence, that the amino acid sequence of the binding site of talin is only functional if correct right from the beginning? If that is so, how could it be the result of stepwise evolution over long periods of time?
Alexandre R Gingras The structure of the C-terminal actin-binding domain of talin 2008 Jan 23 7
Talin is a large dimeric protein that couples integrins to cytoskeletal actin. Here, we report the structure of the C-terminal actin-binding domain of talin, the core of which is a five-helix bundle linked to a C-terminal helix responsible for dimerization.
Talin is one of a number of cytoskeletal proteins, including α-actinin, filamin, tensin and ILK, implicated in linking members of the integrin family of αβ-heterodimeric cell adhesion molecules to filamentous actin (F-actin). Talin (2541 amino acids) is composed of a globular head (residues 1–400), containing a FERM domain, connected to a flexible rod (residues 482–2541) by a short linker sequence containing a calpain-II cleavage site . The FERM F3 subdomain contains a binding site for the β-integrin cytoplasmic domain, which recognises both the NPxY motif and membrane-proximal sequences within the β-integrin cytodomain
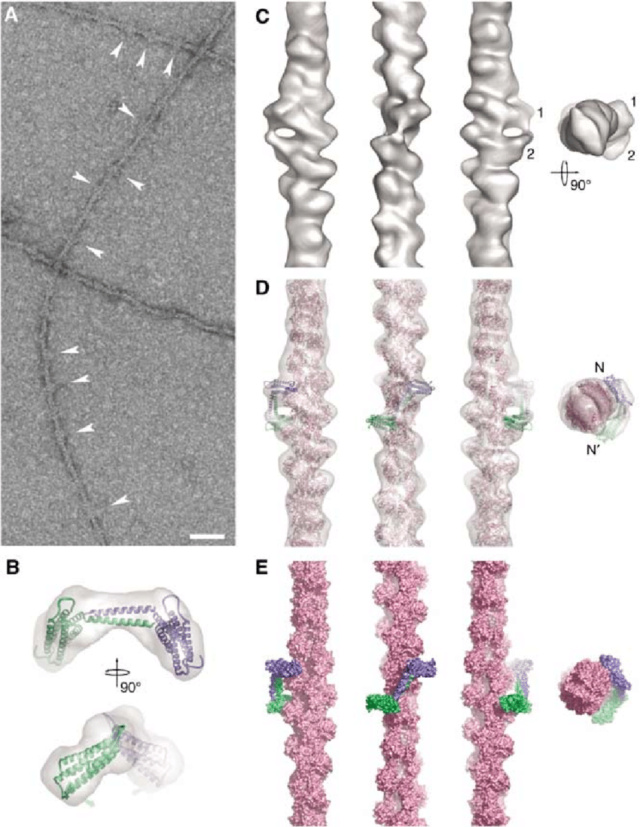
The C-terminal actin-binding site in talin binds to the sides of actin filaments.
(A) The talin fragment binds to the side of actin filaments at specific sites (arrowheads). This binding does not follow helical symmetry. The scale bar represents 50 nm.
(B) Two orthogonal views of the dimer model (monomers in blue and green cartoon representation) and the envelope derived by SAXS (transparent grey). The small grey arrows indicate the direction of the twist that can be used to improve the fit of the SAXS model into the 3D reconstruction.
(C) Surface representation of the 3D reconstruction of F-actin decorated with the talin C-terminal domain. The three views perpendicular to the filament axis are related by successive 901 anticlockwise rotations around the axis. The pointed end of the filament is to the top of the figure for these views. The rightmost view is along the filament axis from the pointed end towards the barbed end. The two connected densities are indicated (1 and 2) (D) Docked atomic models of F-actin (pink) and a dimer of the talin C-terminal domain (monomers in blue and green) inside the 3D reconstruction (transparent grey). Views as in (C).
(E) Molecular surface of the docked models. Views and colours as in (D).
MitaliDas Mechanisms of talin-dependent integrin signaling and crosstalk February 2014 8
Cells undergo dynamic remodeling of the cytoskeleton during adhesion and migration on various extracellular matrix (ECM) substrates in response to physiological and pathological cues. The major mediators of such cellular responses are the heterodimeric adhesion receptors, the integrins. Extracellular or intracellular signals emanating from different signaling cascades cause inside-out signaling of integrins via talin, a cystokeletal protein that links integrins to the actin cytoskeleton. Various integrin subfamilies communicate with each other and growth factor receptors under diverse cellular contexts to facilitate or inhibit various integrin-mediated functions. Since talin is an essential mediator of integrin activation, much of the integrin crosstalk would therefore be influenced by talin.
The communication of extracellular matrix (ECM) with intracellular cytoskeleton is crucial for regulating cell adhesion, cell shape change and cell migration. Such communication depends heavily on integrins, a large family of noncovalent heterodimeric (α/β) adhesion receptors [65], [68]. Integrins function by engaging ECM ligands through their large extracellular domains and actin-binding proteins through their short cytoplasmic tails (CT), thereby linking ECM with the cytoskeleton. The effects of talin on integrin function are broad. It transduces signals across integrins in both the inside-out and outside-in directions and it also influences the organization of the actin network and the composition of focal adhesions.
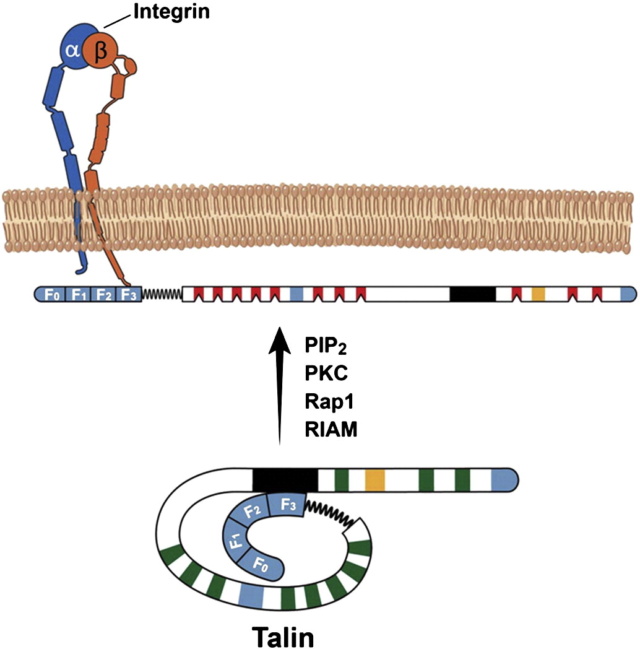
Structure of talin. Talin can be subdivided into head (talin-H) and rod (talin-R) regions. Talin-H is comprised of F0, F1, F2 and F3 domains while the talin-R has ~ 11 vinculin- and 3 actin-binding sites (in blue). The vinculin binding sites are dormant (in green) and are likely mechanoactivated (in red). PIP2, PKC, Rap1 and or RIAM can relieve the autoinhibitory effect of talin-R (1654–2344aa; black region) on F3 domain, promoting talin binding to β CT. The secondary integrin-binding site is in orange.
Benjamin T. Goult The Mechanical Basis of Memory – the MeshCODE Theory 25 February 2021 9
One of the major unsolved mysteries of biological science concerns the question of where and in what form information is stored in the brain. I propose that memory is stored in the brain in a mechanically encoded binary format written into the conformations of proteins found in the cell-extracellular matrix (ECM) adhesions that organise each and every synapse.
My comment: I believe that memory is a fearture of the mind, which is not physical. I do not see how for example the imagination of an event, for example walking on the street to school, could be transformed/encoded in physical form, since we are talking here about a dynamical image, that unfolds in our imagination. If i close my eyes, and imagine that event, it simply appears in my mind, but that imagination seems not to be bound somehow to the physical.
The MeshCODE framework outlined here represents a unifying theory of data storage in animals, providing read-write storage of both dynamic and persistent information in a binary format. Mechanosensitive proteins that contain force-dependent switches can store information persistently, which can be written or updated using small changes in mechanical force. These mechanosensitive proteins, such as talin, scaffold each synapse, creating a meshwork of switches that together form a code, the so-called MeshCODE. Large signalling complexes assemble on these scaffolds as a function of the switch patterns and these complexes would both stabilise the patterns and coordinate synaptic regulators to dynamically tune synaptic activity. Synaptic transmission and action potential spike trains would operate the cytoskeletal machinery to write and update the synaptic MeshCODEs, thereby propagating this coding throughout the organism. Based on established biophysical principles, such a mechanical basis for memory would provide a physical location for data storage in the brain, with the binary patterns, encoded in the information-storing mechanosensitive molecules in the synaptic scaffolds, and the complexes that form on them, representing the physical location of engrams. Furthermore, the conversion and storage of sensory and temporal inputs into a binary format would constitute an addressable read-write memory system, supporting the view of the mind as an organic supercomputer.
I would like to propose here a unifying theory of rewritable data storage in animals. This theory is based around the realisation that mechanosensitive proteins, which contain force-dependent binary switches, can store information persistently in a binary format, with the information stored in each molecule able to be written and/or updated via small changes in mechanical force. The protein talin contains 13 of these switches, and, as I argue here, it is my assertion that talin is the memory molecule of animals.
My comment: Maybe, if we talk about memory, it can be, that the Talin proteins memorizes states of affairs of the outside world, like environmental conditions, food & nutrition supply and availablity, to respond in a fast manner to the the different conditions constantly imposed by the environment. But that, then, would have nothing to do with the memory, or imagination of past events.
These mechanosensitive proteins scaffold each and every synapse and have been considered mainly structural. However, these synaptic scaffolds also represent a meshwork of binary switches that I propose form a code, the so-called MeshCODE. The appreciation of such a network of switches and the machinery that controls them leads to a new hypothesis for the way the brain might be functioning.
The MeshCODE array of mechanical switches1 would be operated by the cytoskeletal machinery, with synaptic signalling triggering the cytoskeleton to push and pull on these switches to constantly alter and update the coding in each neuron (Figure 1).
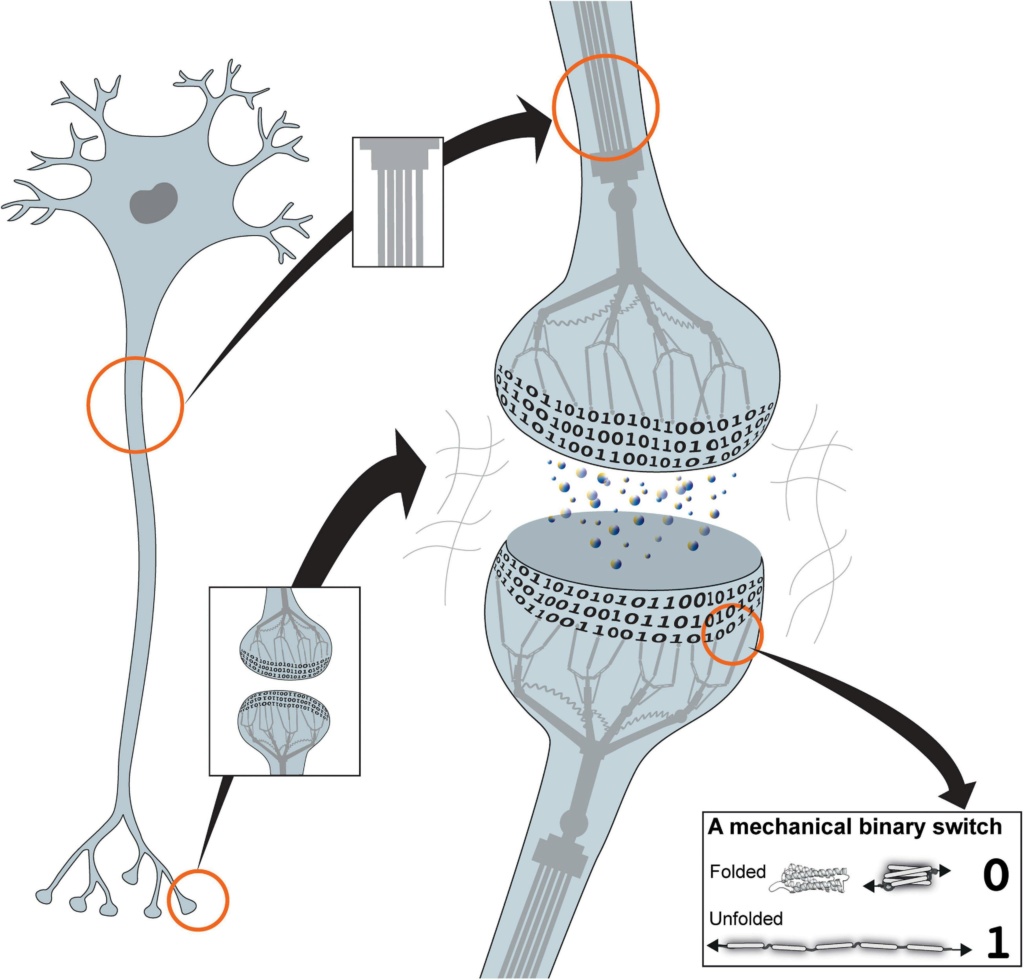
Figure 1. The mechanical cell.
Cartoon of a neuron showing the binary coding that results from the hundreds of mechanical switches built into each synapse. The cytoskeleton is represented as a mechanical machine that operates these switches in response to neuronal activity, thus altering and updating the coding. Inset: example of a mechanical binary switch. These protein domains can be reversibly switched between two different states, folded “0” and unfolded “1.”
Together, this mechanical layer would integrate with the chemical and ionic signalling layers to provide the basis for a new mechanism underlying information-processing and storage. In this scenario, electrochemical signalling between neurons would coordinate a network of trillions of mechanically operated switches, each able to store one bit of data, with action potential spike trains serving as the means to enter new information into this calculation. This mechanical coding would run continuously in every neuron, extending into every cell in the organism, ultimately amounting to a machine code coordinating the entire organism.
The concept of the mechanical computer described herein represents a new hypothesis for how the brain might be performing computation and postulates an addressable read-write memory mechanism. Such a memory mechanism would facilitate both the storage of data in an indexed, hierarchical structure and provide a basis for how information-processing machinery could call on this data as required. This novel concept for biochemical data storage and organic computation has similarities with computers, both old and new. These similarities span from the earliest mechanical computing machines through to solid state disks (SSDs) and the complex subroutines that enable complex computation.
The Cell as an Organic Calculating Machine
The original concept of a digital programmable computer is attributed to Charles Babbage, whose “Analytical Engine” mechanical computer was first described in 1837. Early computers were built using mechanical components, complex machines of levers and gears that were used to perform calculations by turning gears and incrementing counters and ultimately output displays. The inputs initiate the calculation and the levers and gears crunch the numbers, and push and pull until the calculation is complete and the output returned. The programs that the Analytical Engine uses take the form of punched cards, in which the pattern of holes on the card run different parts of the machine. By changing the pattern of holes on the card, a different program will run to give a different calculation and obtain a different output.
There are considerable similarities between a mechanical computer and a cell. Each cell contains a series of levers, pulleys and gears in the form of its cytoskeleton. The cytoskeleton is an incredibly complex, dynamic network formed of three major classes of filaments; actin, microtubules, and intermediate filaments. These filaments can assemble and disassemble rapidly and robustly in response to cellular signals, and the interplay between them is complex. The cytoskeleton can be used to generate forces, with motor proteins pushing and pulling on actin and microtubule filaments, exerting forces on to specific targets. These filaments can also serve as railroads to transport cargos to precise locations in the cell. There are hundreds of cytoskeletal regulators that control these networks with the precise linkages, filaments, and adapters determined by the programme that cell is running.
Almost all cells in our bodies rely on cell-adhesion molecules, that adhere the cell to adjacent cells and/or the surrounding meshwork of proteins called the extracellular matrix (ECM). The adhesions to the ECM, mediated by the integrin family of ECM receptors, serve as information-processing centres, able to feel the surrounding environment and instruct the cell how to function.
My comment: While a big deal of the information stored in DNA is used by the cell to make things, like how to synthesize amino acid chains in the right order to get functional proteins, there is an entire class of proteins that process information to instruct the cell how to function, which is an entirely different function. This is a distinction that goes through all biology. The living needs the know-how to construct, and to operate things, both at once, or no deal.
The cytoskeleton is wired up to the cells adhesions, to the nucleus and to all the organelles in a highly ordered (but dynamic) manner.
My comment: Once again we can observe that one component bears only function in an interdependent fashion together with other players. It is an interlocked machine-like operation with many elementary parts contributing to the functional order.
Once a cell is established in its environment, the cytoskeleton is maintained under tension but is not constantly generating large forces or battling against itself. This homeostasis is achieved when all the forces and tensional restraints are balanced in the system, at which point it is said to be under tensional integrity or “tensegrity”. In a muscle, the actin filaments and myosin motors are highly organised to enable the generation of forces and motion. In a similar fashion, the cytoskeleton in the brain is ordered which I suggest connects all the synaptic adhesions together via dynamic mechanical linkages.
Talin
The major linkage between the integrin-ECM connections and the cytoskeleton is the protein talin. Talin is perfectly positioned to respond to changes in forces, both from outside and inside the cell, and has emerged as a master mechanosensor in that it can sense these forces and convert them into biological signals. Each adhesive structure contains hundreds of talin molecules all connected to the integrins, each other, and the cytoskeleton through direct and indirect connections to actin, creating a complex array of talin molecules (Figure A below).
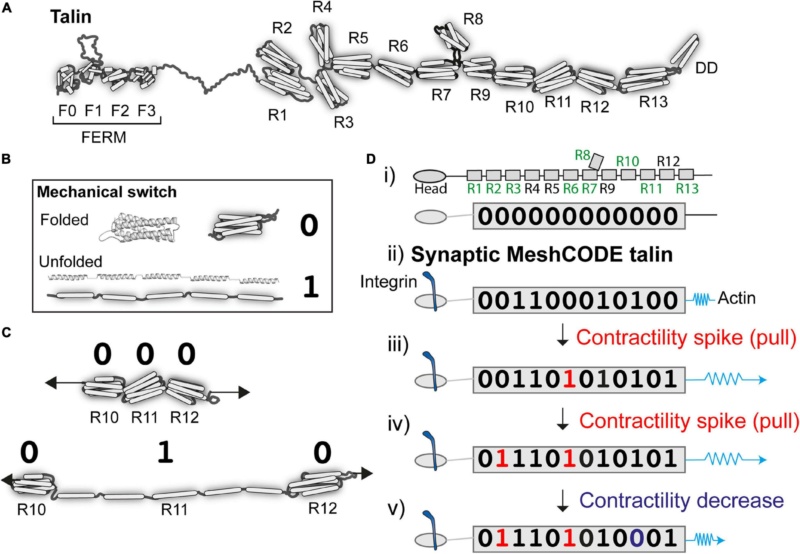
Talin as a memory molecule.
(A) Talin is comprised of an N-terminal FERM domain that binds to integrin, connected to 13 helical bundles, the talin rod domains R1-R13.
(B) A mechanical binary switch. One talin helical bundle is shown. Under tension each bundle can exist in two thermodynamically stable states, folded “0” and unfolded “1” and can be switched back and forth between these states via mechanical force.
(C) Cartoon of three rod domains going from 000 to 010 in response to one contractility spike by the cytoskeletal force-generation machinery.
(D) A talin molecule as a binary string, the nine vinculin binding sites are labelled in green (i) in the absence of force the 13 rod domains are all in the folded “0” state; (ii) upon adhesion formation each talin is tethered between the integrin:ECM and F-actin, and adopts a specific switch pattern; two contractility spikes (shown as a blue spring) result in two of the switches switching (iii and iv), whereas a decrease in contractility resets one switch back to 0 (v). The exact order of switching and the resulting switch state will depend on the system.
Question: Figure 4 defines which of the 13 bundles is switched on, aka unfolds. If all 13 switches are unfold, the proteins gets 13 times the length of each bundle unfolded long, correct ?
Onto this integrin-talin-actin core complex, numerous additional proteins assemble forming distinct, tissue specific signalling complexes.
There are two talin isoforms, talin-1 and talin-2 expressed in the brain; both have the same domain structures but subtly different mechanical properties. Each talin molecule contains 13 force-dependent binary switches ( Figure 2A). These talin domains, can be reversibly switched between two thermodynamically stable states, “folded” and “unfolded,” using mechanical force (Figure 2B). One of the key discoveries that leads to the MeshCODE theory is that the switches in talin exhibit mechanical hysteresis meaning that, provided the molecule is maintained in a mechanical linkage between the integrin-ECM and the actin cytoskeleton, both the folded and unfolded states of each domain are thermodynamically stable (Figure 2B). This imparts a novel function on talin in molecular memory as each talin molecule can be imprinted with a persistent pattern of switch states as a specific outcome of the forces that have acted upon it (Figure 2C). The conformational state of each switch determines which signalling molecules are recruited, thereby providing different instructions to the cell as a function of force, enabling talin to serve as a Mechanosensitive Signalling Hub. As the environment changes, such as when a cell migrates, these switches detect these changes enabling the cell to respond appropriately.
Question: How could this function be the product of evolutionary pressures ? Was the right state of affairs, that permits the cell to respond appropriately obtained by trial and error? Would each unsuccessful trial not resulted in an "error catastrophe"? How would/could the cell recover from the erroneous result and going back to the previous state of affairs, waiting until another mutation would be selected and tried out, repeating the process as many times as "necessary" until fixing the new function in the organism? In order for that function to be instantiated, would not the whole chain of interacting interdependent molecules, aka. the microtubules, and lectins have to mutate accordingly and in parallel to establish the function? If a myriad of talins and the adjacent interacting proteins are required in the cell to get functionalty, adaptation, plasticity, and different functional states, how could/would single point mutations in the genome lead to a system change in the cell to implement that more advanced, sophisticated structure?
Most current models envisage talin as a rope in a “tug-of-war” between extracellular forces, and those generated by the cells force-generating machinery serving as a “molecular clutch” that enables mechanotransduction and cell migration. Proteins such as vinculin, which bind to nine of the unfolded talin switches, can further stabilise the unfolded state, limiting refolding and reinforcing these connections.
Talin, the Data Molecule of Life
Every animal known to humankind has the same 13 switches in talin, and the high conservation of switch pattern suggests a role that is explicitly dependent on the order of the string of switches. The best way to visualise such a role is to consider the scenario where the extracellular environment is built in such a way that it presents a mechanically stable, predictable environment. In this scenario, the talin switches are no longer required to sense the extracellular environment as that is, to all intents and purposes, constant. Instead, the cell can use its force-generating machinery to operate these mechanical switches and in doing so write data into the adhesions (Figure 2D).
This has profound implications for data storage in animals, as it means that the cell can use the talin switches to store information. Adhesions in controlled, predictable environments can adopt the role of data-storage devices that store information in a manner controlled by the system. I would like to propose that this complex meshwork of switches operates as a code, the MeshCODE outlined here. To simplify the nomenclature of the switches in this view of talin it is easier to regard the “folded” and “unfolded” states as “0” and “1,” respectively, to better reflect that it is data that is being stored in the talin molecule (Figure 2).
Exquisite Calculation in a Single Cell
As a result, the cell is a mechanical computer with all of the architecture necessary for computation. The cytoskeleton serves as the levers and gears that perform the calculation, and the MeshCODE adhesions provide a multifunctional system that can be used to perform calculations and serve as a memory storing the results in the conformations of the switches. An input signal, be it an extracellular signal activating a receptor, a change in physicality, or excitation by a chemical or electrical signal, perturbs the balanced state of the cell, switching the “computer” on, thereby triggering changes in the cytoskeleton as it seeks to return to homeostasis. Key to maintaining the mechanical balance of the cell are the 20 members of the Rho-family of GTPases. These crucial components of the signalling networks coordinate the contractility and behaviour of the actin cytoskeleton and their role in controlling synaptic development and plasticity is established. There are 80 activators [guanine nucleotide exchange factors (GEFs)] and 70 inactivators [GTPase-activating proteins (GAPs)] of the Rho family of proteins, many of which are located at synapses. The balance of these opposing regulators enables the cellular mechanics to be spatially and temporally modulated by a multitude of signalling pathways. Changes in architecture and contractility push and pull on the adhesions which the MeshCODE theory predicts will result in reproducible alterations in the binary switches. When the calculation is complete, homeostasis is restored. At this end point, the conformations of the switches in the adhesions are altered and the array of 0s and 1s reflect the outcome of the calculation. The large adhesion complexes that assemble on the talin scaffold as a function of the altered switch patterns stabilise the patterns and dictate the signalling response.
The appearance of talin at the dawn of multicellularity allowed cells to store information persistently by writing to each talin molecule like a computer writes to a disk. As well as serving as a memory, these switches also coordinate cell signalling and provide a way to control the reading of the genome from the periphery of the cell.
Organic Calculation in the Brain
The brain is a colossal cell signalling machine with a trillion cells all communicating with each other, leveraging the organic calculating power of each cell. Synapses are the perfect system for optimised cell signalling between connected cells, and there are approximately 100 trillion synapses in the brain transmitting signals between neurons, to give rise to brain activity. The specialised architecture of synaptic junctions juxtaposes the presynaptic and postsynaptic neurons to form the synaptic cleft, an arrangement supported by astrocytes which coordinate synaptic maintenance and plasticity, and microglia ( Figure 3A).
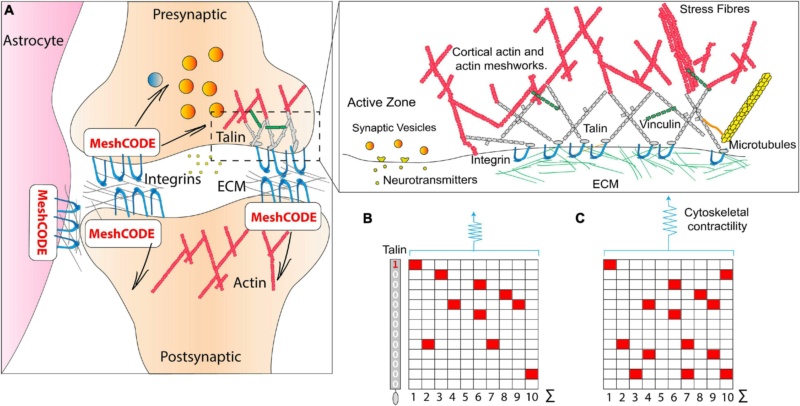
Figure 3. The MeshCODE.
(A) Schematic diagram of a tripartite synaptic junction. The integrin adhesion complexes and MeshCODEs in the presynaptic and postsynaptic zones, and in a neighbouring astrocyte are shown. The synapse is encapsulated by a specialised ECM that protects the mechanical environment of the MeshCODEs. Integrin and talin in one MeshCODE are shown connected to the actin cytoskeleton. (Right) Enlarged view of the highlighted region showing the MeshCODE intricately wired up to the cytoskeleton. Only the integrin-talin-actin adhesion core complex is shown, but up to 250 different proteins decorate this core dependent on the switch patterns of the talin scaffolds. The protein vinculin (green) binds to nine of the unfolded talin switch domains, stabilising them in the unfolded state and reinforcing the cytoskeletal connections.
(B) A schematic of an array of ten talin molecules with the 13 switches arranged vertically. White, 0; red, 1.
(C) Perturbations to the system result in changes in cytoskeletal contractility that alter the pattern of 1s and 0s written in the MeshCODE and its resultant output in a defined way.
Each synapse is scaffolded by adhesions located around the edge of the synapse; these adhesions involve the binding of integrins to ECM components, such as laminin and fibronectin Figure 3A). Integrin adhesion complexes are required for synaptic plasticity and memory, and consolidation of long-term potentiation and the ECM has been linked to learning and memory previously. However, a role for adhesion complexes in the storage of data has not been considered previously.
The brain is soft, which provides the perfect, protective environment for each neuron to tightly control its own mechanical environment independently, building its own ECM niche around each synapse, with the surroundings serving to isolate the neuron from external mechanical forces. The MeshCODE theory proposes that tight regulation of the mechanical environment of each synapse would enable mechanical computation to occur with meticulous precision. The ECM surrounding each synapse is subject to extensive maintenance and remodelling during synaptic function, and it seems reasonable to postulate that this maintenance of the ECM might ensure that the synapses mechanical environment is tightly regulated and predictable.
Therefore, in the MeshCODE framework, these synaptic scaffolds provide the capacity to write data into the synapses themselves in the extensive arrays of binary switches that are located in both the pre-, and postsynaptic side of each synapse as well as in the supporting astrocyte cells (Figure 3A). The capacity to store information in every synapse, with the potential to orchestrate the flow of information through that synapse, makes the synapse a complex computational device.
How Would a Mechanical Code Be Read?
The key to any code is a read-out mechanism and the mechanical code outlined here leads to a number of hypotheses for how such a coding in the brain would be read. This section discusses how MeshCODE driven adhesion signalling might work in the context of a neuron, with the purpose being to demonstrate the feasibility and broad applicability of the theory to well-established neuronal processes.
The contractility-dependent control of integrin-adhesion complexes is a well-studied phenomenon in the cell adhesion field. Adhesion complexes grow as forces are exerted on them, altering their composition and signalling outputs and mechanical forces acting on talin are essential to drive this process. Each synaptic adhesion would serve as a discrete signalling hub, altering its signalling as a function of the mechanical signals received. Following each synaptic signalling event, the neuronal cytoskeleton would contract briefly and precisely generating these mechanical signals and altering the switch patterns at that site. As a result, the code would be read by the ligands that engage the different switch states (Figure 4A) with the read-out being the relative (re)distribution of all the ligands engaging the entire array of switches defining the output.
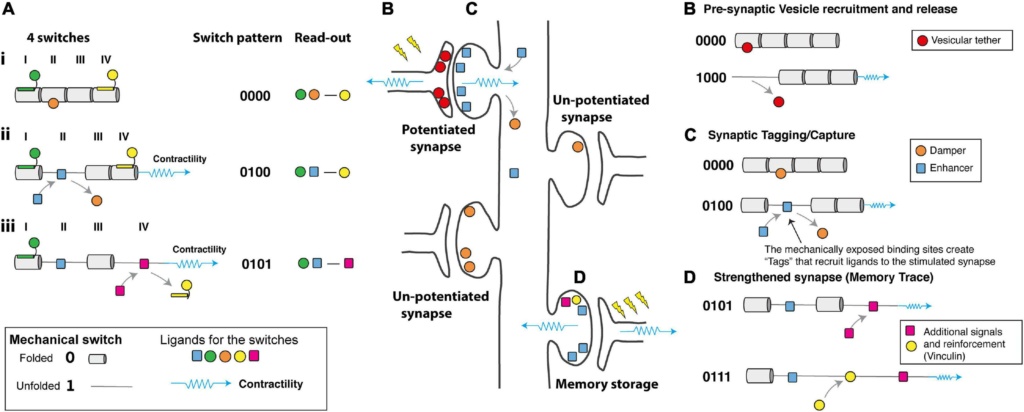
Figure 4. How would a mechanical code be read?
(A) Schematic diagram representing four mechanical switches labelled I–IV, (Left) each switch has the potential to bind different ligands in its folded, 0 and unfolded, 1 states. (Middle) The switch pattern is shown as a binary string. (Right) The ligands decorating the switches present a “read-out” mechanism as the complexes formed will depend on the mechanical coding. (i) at low force the green, orange, and yellow ligands engage the MeshCODE on switches I, II, and IV. (ii) Contractility (shown as a blue spring) switches one domain (here domain II) from the 0 to 1 state which drives a switch in binding partners, displacing the orange ligand and recruiting the blue ligand. The overall signalling complex and read-out is altered. (iii) Further contractility switches a second domain (here domain IV) further altering the coding and the proteins that are recruited to the synaptic scaffolds. Proteins, like vinculin, which bind the unfolded state of a switch lock it in that conformation and limit its ability to refold.
(B–D) A cartoon of a neuron with four synapses in different states showing hypothetical ways a mechanical code might be read. Contractility as a result of synaptic signalling causes alterations to the MeshCODE switch patterns on both sides of a stimulated synapse. (B) in the pre-synaptic terminal these switch patterns might regulate the probability of release of docked vesicles by specific switches controlling this process via a hypothetical interaction with a key regulator of synaptic firing.
(C) Synaptic stimulation triggers contractility in the post-synaptic region that specifically alters the switch patterns in that stimulated synapse. The altered switch patterns create “Tags” that recruit proteins (one is shown as a blue square) specifically to that synapse that enhance long-term potentiation. Switching might also displace proteins that dampen potentiation (orange circle) which then diffuse away.
(D) Future signals through that synapse can trigger additional switch pattern changes and alter the binary coding in that synapse, the protein signals recruited, etc., in a dynamic re/writable way, changing the threshold of each synapse in that neuron. These changes to the synaptic adhesions as they grow and shrink dynamically in response to stimuli form the memory trace.
The location of switches in the pre- and post-synaptic scaffolds, coupled with the ligands available to bind in these different structures, suggests potential mechanical explanations for a number of recognised processes of synaptic regulation including;
(i) dynamical regulation of the probability of vesicular release ( Figure 4B),
(ii) synaptic tagging and capture,
(iii) inverse synaptic tagging,
(iv) synaptic competition Figure 4D),
(v) spike-timing dependent plasticity and
(vi) the capture of temporal information.
Hypothesis: Synapses Are Activated and Deactivated Transiently in Response to Signals
The presence of switches in the synapses raises the possibility that certain MeshCODE patterns might transiently activate, or deactivate, transmission through specific synapses. On the pre-synaptic side, the strength of individual synapses within a single axon are regulated by changes in the probability of vesicular release. Tightly regulated changes in the talin switch conformation at individual synapses might provide a molecular mechanism for this changing probability (Figure 4B). The actin cytoskeleton has been shown to be important for neurotransmitter release, as has the requirement of force generation at these sites, and a role for talin in presynaptic function has also been reported. In the pre-synaptic terminal alterations in certain switch patterns, dynamically exposing and disrupting binding sites in close proximity to the active zone, might regulate the number of docked vesicles, or control the probability of release of docked vesicles via hypothetical interactions with key regulator(s) of synaptic firing (Figure 4B). The probability of each synapse firing would be updated by the mechanical signalling providing a mechanism for synapses to be switched on and off transiently on demand, potentially contributing to the homeostatic synaptic scaling of the neurons output.
Hypothesis: A Mechanical Basis for Synaptic Tagging?
A well-known, but not fully understood, phenomena is the process of “synaptic tagging and capture,” where following activation of a synapse, that synapse is marked in such a way that proteins are recruited directly to that synapse ( Figure 4C). As a synapse is stimulated, then the MeshCODE theory predicts that the stimulated synapse undergoes localised contractility thereby altering the switch patterns at that site. Certain switch states would be generated that expose novel binding sites in that stimulated synapse only. These “Tags” (Figure 4C) would serve to recruit proteins to that synapse and it is possible that these switches provide the basis for a dynamic form of synaptic tagging. It is already established that alterations in talin conformation cause global relocation of cellular factors. Furthermore, it is also possible that proteins that inactivate synapses, and/or transcriptional regulators, might be bound to the 0 state of the switch which upon the switch unfolding would be released from the newly stimulated synapse to diffuse away (Figure 4C). This displacement of signals from a stimulated synapse would provide a mechanism for “Inverse synaptic tagging”; “inverse tag” proteins would be displaced from stimulated synapses and accumulate at unstimulated synapses where more of that switch is still in the 0 state.
As protein levels in cells are under tight control via transcriptionally regulated programs, these switches would result in the global relocalisation of proteins in the neuron. If proteins are recruited to one synapse following stimulation (Figure 4C), then those same proteins would be depleted from other synapses leading to a basis for “synaptic competition” where the strengthening of one synapse affects the stability of other synapses in that cell. The number of synapses, and the number of scaffold proteins in each synapse will be tightly regulated, and therefore an exact number of mechanical switches will be distributed across each neuron, and throughout the brain. As each switch state can recruit one signalling molecule then as the number of switches in one state increases in a stimulated synapse more of that signalling molecule will be recruited to that site thereby dynamically altering the concentration of molecules at that adhesion/synapse and depleting that molecule from the rest of the neuron and the other unstimulated synapses. This mechanical coordination between synapses, with stimulation of one synapse recruiting and displacing plasticity-related proteins from that site, thereby directly effecting the proteins available to bind at other synapses provides a mechanism for potentiated and depressed synapses to interact in a synergistic manner. This synchronisation would enable the entire neuron to behave as a mechanically coordinated system of synapses where input signals to a synapse can dynamically alter the activity of each synapse in the neuron (and by extension the neurons it is communicating with), providing a way to establish long-term plasticity and orchestrate the flow of information transmitted by the cell.
Overtime this synaptic tagging leads to remodelling of the synapse, driven by the recruitment of newly transcribed proteins to the stimulated site. Remodelling of the actin cytoskeleton occurs at stimulated synapses, which not only enhances that synaptic connection but also strengthens the mechanical linkages mediated through the actin cytoskeleton emanating from that site. Nine of the unfolded switch domains in talin recruit the protein vinculin, which is well known as a means that cells reinforce integrin-talin-actin connections anchoring actin to the adhesion site and also for locking the mechanical switches in the unfolded state.
Hypothesis: Contractility-Dependent Control of Integrin Adhesion Complex Dynamics Is the Way That Neurons Store Information
It might be that the Synaptic tagging observed in synapses is actually a similar process to the contractility-dependent growth of integrin adhesion complexes seen in other cells. The adhesions would grow in stimulated synapses as forces are exerted on them and atrophy in unstimulated synapses, with the cumulative effect that overtime these changes in switch pattern at stimulated synapses would lead to morphological changes in the shape and size of the synapses as they remodel altering the signalling output of each site. This process would be explicitly dependent on the talin switch patterns that dictate which molecules assemble to form each adhesion complex. The engagement of the cytoskeleton with the talin molecules directly, and indirectly through proteins such as vinculin, will alter as the switch patterns change, and these engagements will
(i) help maintain the switch pattern and
(ii) alter where and how future contractility spikes exert tension on talin and the meshwork in general, and
(iii) determine the signalling molecules recruited to that site. It is possible that the patterns of the switch states and the specificity of ligands binding to them, are further regulated by post-translational modification; specific patterns could be “write protected” by locking the domains in those conformations by phosphorylation or other modification.
These synaptic adhesions would be serving as large information-processing centres, with the many proteins recruited to, and interacting on, the patterns of switches stabilising the patterns and encoding/decoding the information. The complexes that assemble on the patterns (Figure 4D) and the signalling outputs that result will vary depending on the expression levels of proteins, and their distribution, in that neuron, but in each case, they will be assembled on the integrin-talin-actin core complex dictated by the mechanical signalling across the whole cell. Furthermore, adhesion complexes also transmit mechanical signals to the nucleus, and can induce modular gene expression patterns in response to both physical and geometric constraints, a process referred to as mechanotransduction. Epigenetic tagging and alterations in gene expression are required for memory system consolidation and it might be that mechanotransduction through these synaptic adhesion complexes contributes to such mechanisms.
Subsequent stimulation of that synapse would result in more contractility changes that further alter the switch patterns in the MeshCODE dependent on the current switch patterns at that site and the cytoskeletal connections that formed. Further contractility might unfold more copies of the same switch domain that was described above (Figure 4C), exposing more “tags” and recruiting more of the ligand that binds to that site further depleting it from other synapses. However, if many copies of that switch are already in the unfolded state, or if the cytoskeletal connections have altered, then additional contractility would trigger different switches to unfold, recruiting and displacing other molecules, causing further remodelling of the adhesion complexes and redistribution of proteins throughout the neuron (Figure 4D).
1. Unicellular and multicellular Organisms are best explained through design
https://reasonandscience.catsboard.com/t2010-unicellular-and-multicellular-organisms-are-best-explained-through-design
2. https://reasonandscience.catsboard.com/t2990-how-does-biological-multicellular-complexity-and-a-spatially-organized-body-plan-emerge#7758
3. https://reasonandscience.catsboard.com/t2254-does-the-mullerian-two-step-proposal-refute-irreducible-complexity
4. https://sci-hub.ren/10.1038/nrm3624
5. https://rupress.org/jcb/article/217/11/3776/120671/Talin-as-a-mechanosensitive-signaling-hubTalin-as
6. https://www.nature.com/articles/s41598-019-42002-6
7. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2168396/
8. https://www.sciencedirect.com/science/article/pii/S0005273613002575
9. https://www.frontiersin.org/articles/10.3389/fnmol.2021.592951/full
https://reasonandscience.catsboard.com/t3175-talin-as-a-mechanosensitive-signaling-hub
A Chat with Ben Goult about the Talin Code
https://www.youtube.com/watch?v=yG8AiBc-trY
Ben Goult: The Mechanical Basis of Memory
https://www.youtube.com/watch?v=C0A8bTehUls&t=5s

03:53 And then what talin also does is it couples these integrins to the actin cytoskeleton and this is really important you need to be able to couple these attachments to the side skeleton to enable it to withstand forces and if you've cut or removed several ones of these linkages this whole thing will fall apart and your cell will round up and die
My comment: That is evidence of an irreducibly complex system, where all players must be in place in an interlocked, interdependent fashion for the system to work, or nothing goes. That raises the question of how the system could have evolved in the first place if all has to be in place.
04:41 What's really interesting to us and hints again at the complexity which were looking at is that these three proteins integrate alien actin and been killin as a fourth this core assembly of these adhesions is responsible for all the adhesive structures that the cells form or not all but many many of them
Question: .Which of these proteins are not necessary for the cell to survive? Talin, Integrin, Vinculin, alpha-actinin, Actin?
Krishna Chinthalapudi The interaction of talin with the cell membrane is essential for integrin activation and focal adhesion formation April 11, 2018
Vertebrate cell growth, division, locomotion, morphogenesis, and development rely on the dynamic interactions of cells with extracellular matrix components via cell surface complexes termed focal adhesions that are composed of heterodimeric αβ integrin receptors, associated signaling molecules, and the large cytoskeletal protein talin. Talin activation and binding to β-integrin requires interactions with lipids. Here we report that talin binding to the cell membrane seems necessary for integrin activation and focal adhesion formation.
https://www.pnas.org/content/115/41/10339
My comment: It seems that cell adhesion, and signaling, are two essential processes, that have to be instantiated in an integrated fashion. That means, two problems and necessities have to be solved, and implemented at once. That requires the establishment of a sophisticated information and communication system, and the right engineering architecture with multiple players working in a joint venture.
05:28 Our central hypothesis is that talin adopts numerous different conformational States which runs cellular programs to enable these different structures to form
Question: How is the information encoded ?
Benjamin T. Goult Talin as a mechanosensitive signaling hub September 25 2018 [url=One of the most incomprehensible things that are hard to fathom is that God loves us. Despite our sins and shortcomings.]5[/url]
Cell adhesion to the ECM is fundamental to multicellular life. Deletion of major integrins or ECM proteins impairs the development and survival of multicellular organisms.
My comment: That means, that the origin of integrins is a fundamental linked to the quest of how multicellularity emerged.
From an evolutionary perspective, support for the transition from unicellular (single cell) to multicellular organisms requires the emergence of several novel biochemical systems. 1 Such systems include:
- pathways that transform cells from generalized to specialized forms during growth and development;
- mechanisms for the migration of cells relative to each other during growth and development;
- structures that support cell-cell adhesions;
- and mechanisms for cell-cell communication.
- All of these systems have to be in place and operate in an integrated fashion to support multicellularity.
1. Despite a thorough search, no material causes have been discovered that demonstrate the power to produce large amounts of specified information, irreducible, and interdependent biological systems.
2. Intelligent causes have demonstrated the power to produce large amounts of specified information, irreducible and interdependent systems of all sorts.
3. Intelligent design constitutes the best, most causally adequate, explanation for the information and irreducible complexity in the cell, and interdependence of proteins, organelles, and body parts, and even of animals and plants, aka moths and flowers, for example.
Integrin-mediated adhesions sense the mechanical features of the matrix, including stiffness, texture, and externally applied strains, transducing these forces into biological signals 2
Talin plays a central role in cell adhesion, first by converting integrins to high-affinity states (“activation”) and by coupling integrins to the cytoskeleton. Indeed, deletion of talin results in developmental defects in multiple organisms that resemble total loss of integrins
My comment: Here we have another protein that performs essential functions in multicellular organisms. Which rises the question: How could it have evolved, if its essential function only becomes essential once it is integrated in the system? Some might come up with the mullerian two step proposal: Add a part. Make it necessary. That is the heart of Mullerian two-step process. The problem however is always explaining how biological information or function is built up in the first place. 3
Talin is a large (270 kD) multidomain cytosolic protein

Talin domain organization and interactions.
(A) Talin contains an N-terminal FERM domain (F0–F3) connected via an 80-aa unstructured linker to the 13 talin rod domains R1–R13. 9 of the 13 rod domains contain VBSs (red).
(B) Major binding sites for folded talin domains (black boxes) and unfolded domains (white boxes).
Talin links integrins to F-actin in part via binding of its N-terminal FERM domain to integrin cytoplasmic domains as well as via two sites in its C-terminal flexible rod domain that bind F-actin.
David A. Calderwood Talins and kindlins: partners in integrin-mediated adhesion 17 July 2013 4
Integrin receptors provide a dynamic, tightly regulated link between the extracellular matrix (or cellular counter-receptors) and intracellular cytoskeletal and signalling networks, enabling cells to sense and respond to their chemical and physical environment. Talins and kindlins, two families of FERM–domain proteins, bind the cytoplasmic tail of integrins, recruit cytoskeletal and signalling proteins involved in mechanotransduction and synergize to activate integrin binding to extracellular ligands. Talin mediates integrin activation. RIAM recruits talin to the plasma membrane, whereas vinculin stabilizes talin in cell– matrix junctions.
My comment: Binding,synergizing to activate, mediating, activating, recruiting, stabilizing are all goal-oriented processes. Why and how would/could unguided evolutionary pressures that are neither goal-oriented, nor having foresight, produce such complex mechanistic, and information-based processes, that show clearly that a higher-order/goal/complexity is being achieved, namely producing the interaction of various cells together?
Cell proliferation, cell migration, tissue morphogenesis and homeostasis all depend on cell–cell and cell–extracellular matrix (ECM) interactions, and the integrin family of adhesion receptors has essential roles in these processes.
My comment: Essential means that the processes and events could/would not occur without integrin adhesion receptors. That makes them irreducible.
These binding events are followed by the application of tension from actomyosin that acts on the talin rod, triggering recruitment of a second actin-binding protein, vinculin . This mechanism thus strengthens adhesions under tension, a form of mechanosensitivity.
Despite the presence of the same core components, adhesion complexes are strikingly diverse. Highly dynamic transient adhesions enable cell migration; dynamic and proteolytic (enzymes that break down protein) adhesions mediate invasion; stable adhesions promote tissue organization; and specialized myotendinous junctions transmit very high forces for animal movements. Around the central core of integrin, talin, and actin, numerous additional proteins show selective recruitment that vary widely between adhesion types, likely reflecting distinct effector, signaling, and mechanosensing functions.
The purpose of this perspective is to summarize existing knowledge and propose a new view of talin function. It is therefore split into two sections: the first section reviews recent data on talin and the emerging functional implications, and the second, more speculative section proposes that there exists a “talin code” of force-dependent interactions with signaling proteins and cytoskeletal components, which exhibits some internal hierarchy. This view, where talin serves as a flexible mechanosensitive signaling hub (MSH), has the potential to explain diverse responses of cells to distinct mechanical stimuli on different time scales.
Talin interactions and functions
The multiple domains with numerous force-sensitive binding sites in talin, coupled with their linear arrangement in the path of force transmission (Fig. above), provides opportunities for enormous functional flexibility.
Adhesion dynamics
Regulated adhesion complex assembly and disassembly are vital for cell spreading and migration. In cells freshly plated on ECM or in rapidly migrating cells, activated integrins form small clusters under the lamellipodia. These small adhesions can either rapidly disassemble or can connect to larger actin templates and mature into slightly larger, more stable structures called focal complexes, which themselves either disassemble or further mature into much larger adhesive structures including focal adhesions (FAs; e.g., in contractile cells on rigid substrates), podosomes (in activated cells on soft substrates), or very stable adhesions as in myocytes or myofibroblasts (in highly contractile cells on rigid substrates). In all cases, talin is a major player in force-dependent adhesion growth and stabilization. Indeed, cardiac and skeletal muscle express an alternatively spliced β1-integrin (β1D) with increased affinity for talin, a key event in exertion of ultra-high forces.
Force transmission between actin and integrins at the leading edges of migrating or spreading cells is mediated by a unique, dynamic mechanism in which talin plays a central role. Actin polymerizes at cell edges and flows rearwards, pushed by the force generated by polymerization and/or pulled by myosin motors further back in the cell. This retrograde flow of actin couples to integrins via talin, thereby exerting traction force on the matrix or substrate for spreading, migration, or contraction. This force transfer must occur through highly dynamic bonds that exhibit variable coupling efficiency: the so-called FA clutch.
Talin conformation and mechanotransduction
There are two talin isoforms, talins 1 and 2, that are 76% identical and have identical domain structures. Both contain an N-terminal FERM domain-containing four globular segments (F0 to F3), a disordered linker region, and a C-terminal rod with 13 four- and five-helix bundles (R1 to R13) terminating in a single α-helix that mediates homodimerization (dimerization domain [DD]; Fig. above). With the exception of R8, which is positioned outside the force transmission pathway as will be discussed, the rod domains are arranged linearly like beads on a string, transmitting tension along the talin rod. At least 9 of the 13 rod domains contain cryptic vinculin-binding sites (VBSs) that are exposed when unfolded by mechanical force, allowing vinculin binding and adhesion reinforcement. Talin is autoinhibited by an interaction between the head (F3) and R9, which must be released for actin and integrin-binding and recruitment to FAs. Interaction of talin with negatively charged phosphatidylinositol 4,5-bisphosphate of the plasma membrane inner leaflet also contributes to talin activation and membrane association.
Force-dependent switching of binding partners to the R3 rod domain defines a mechanochemical switch
The best-studied talin activation pathway depends on the small GTPase Rap1, whose effector Rap1-GTP–interacting adaptor molecule (RIAM) binds directly to talin via a high-affinity RIAM binding site in R2–R3 (note that talin R8 and R11 also contain RIAM-binding sites and that Rap1 has been shown to bind directly to talin F0. Rap1/RIAM recruits talin to the plasma membrane and antagonizes talin autoinhibition to promote integrin and actin binding. Importantly, RIAM engages folded talin R3 (Fig. below).

Talin rod domains as mechanochemical switches. (A–C) Each talin rod domain can adopt a number of conformations under different force regimes. (A) Folded bundle at low force. (B) An unfolded string of helices at forces above the mechanical threshold. (C) A fully unfolded polypeptide at high forces. Force-induced domain unfolding leads to a switch in the ligand-binding profile of that domain. Complete unfolding at high force will result in a linear polypeptide unable to bind folded-domain ligands or helix-binding ligands. While no ligands for this form have been identified so far, many proteins bind linear peptide motifs.
R3 is the least stable of the 13 talin rod domains due to a cluster of four threonines in the central hydrophobic core of the four-helix bundle. Consequently, R3 opens under the relatively low force of 5 pN, converting the folded four-helix bundle into a string of helices that could be further unfolded into a disordered conformation. This transition exposes two high-affinity VBSs, which in the absence of force, were cryptic, buried inside the core of the folded R3. Therefore, the force-dependent unfolding of R3 results in exposure of VBS-recruiting vinculin while simultaneously disrupting RIAM binding, severing the link to Rap1 signaling. This switch in ligands is mirrored in cells, where an integrin–talin–RIAM complex at the tip of cell protrusions (“sticky fingers”) is replaced by an integrin–talin–vinculin complex in mature, high-tension FAs, which are devoid of RIAM. This exchange of RIAM, bound to talin in the absence of force for vinculin in the presence of forces, thus defines a mechanochemical switch (Fig. above, A and B).
Structural basis for talin rod interactions
To date, there have been no comprehensive proteomic analyses of the talin interactome; however, structural studies on ligands that bind talin rod domains have begun to reveal interesting themes. Deleted in liver cancer 1 (DLC1), a RhoGAP and tumor suppressor binds the talin R8 domain via an α-helical leucine–aspartic acid (LD) motif. Elucidation of the structure of the DLC1–talin complex revealed a helix-addition mechanism with an amphipathic LD helix of DLC1 packed between two adjacent helices on the surface of the R8 four-helix bundle, effectively converting it to a five-helix bundle. These findings led to the realization that the talin-binding sequence (TBS) in RIAM is also an LD motif as well as the identification of paxillin, which has several LD motifs, as a novel talin ligand. R8 is structurally similar to the FAK FA targeting (FAT) domain, a four-helix bundle that also binds LD motifs. Another class of LD motif-containing proteins, the KANKs (kidney ankyrin repeat-containing), bind to talin R7, a five-helix bundle (Bouchet et al., 2016; Sun et al., 2016b), and mediate connections of microtubules to adhesion complexes. Five-helix-bundle LD motif recognition is also shown with RIAM TBS1 binding the R11 five-helix bundle (Goult et al., 2013). Helix addition thus appears to be a general mechanism for binding to talin rod domain helix bundles in their folded state.
Timo Baade Clustering of integrin β cytoplasmic domains triggers nascent adhesion formation and reveals a protozoan origin of the integrin-talin interaction 05 April 2019 6
The transition from unicellular organisms to multicellular animals (metazoans) coincides with the emergence of dedicated cell adhesion molecules such as cadherins and integrins1. Since their discovery in the mid-80s, it became clear that integrins are not only essential for coupling of the cytoskeleton to the extracellular matrix upon cell attachment, but that they also contribute to a multitude of basic cellular processes including cell survival, proliferation, differentiation, and cell migration.
Integrins and integrin-dependent cell-matrix adhesions are essential for a number of physiological processes. Integrin function is tightly regulated via binding of cytoplasmic proteins to integrin intracellular domains. Yet, the complexity of cell-matrix adhesions in mammals, with more than 150 core adhesome proteins, complicates the analysis of integrin-associated protein complexes. Interestingly, the evolutionary origin of integrins dates back before the transition from unicellular life to complex multicellular animals. Though unicellular relatives of metazoa have a less complex adhesome, nothing is known about the initial steps of integrin activation and adhesion complex assembly in protozoa.
One of the proteins able to initiate integrin inside-out activation is talin. With its N-terminal FERM domain, talin binds to the NPXY/F motif in the membrane proximal part of the integrin β cytoplasmic tail, thereby pushing apart the two integrin subunits followed by the unfolding of their ectodomains. Upon talin binding to the integrin β subunit, additional cytoplasmic proteins are recruited to integrin-initiated focal adhesion (FA) sites.
All β-subunits share the highly conserved membrane proximal NPXY/F and the lesser conserved membrane distal NXXY motif
Clustering of the human integrin β1 tail led to the recruitment of talin, kindlin, and paxillin, and mutation of the known talin binding site abolished recruitment of this protein.
Question: Is the ability of talin to bind to integrin not essential for function? Is that not evidence, that the amino acid sequence of the binding site of talin is only functional if correct right from the beginning? If that is so, how could it be the result of stepwise evolution over long periods of time?
Alexandre R Gingras The structure of the C-terminal actin-binding domain of talin 2008 Jan 23 7
Talin is a large dimeric protein that couples integrins to cytoskeletal actin. Here, we report the structure of the C-terminal actin-binding domain of talin, the core of which is a five-helix bundle linked to a C-terminal helix responsible for dimerization.
Talin is one of a number of cytoskeletal proteins, including α-actinin, filamin, tensin and ILK, implicated in linking members of the integrin family of αβ-heterodimeric cell adhesion molecules to filamentous actin (F-actin). Talin (2541 amino acids) is composed of a globular head (residues 1–400), containing a FERM domain, connected to a flexible rod (residues 482–2541) by a short linker sequence containing a calpain-II cleavage site . The FERM F3 subdomain contains a binding site for the β-integrin cytoplasmic domain, which recognises both the NPxY motif and membrane-proximal sequences within the β-integrin cytodomain

The C-terminal actin-binding site in talin binds to the sides of actin filaments.
(A) The talin fragment binds to the side of actin filaments at specific sites (arrowheads). This binding does not follow helical symmetry. The scale bar represents 50 nm.
(B) Two orthogonal views of the dimer model (monomers in blue and green cartoon representation) and the envelope derived by SAXS (transparent grey). The small grey arrows indicate the direction of the twist that can be used to improve the fit of the SAXS model into the 3D reconstruction.
(C) Surface representation of the 3D reconstruction of F-actin decorated with the talin C-terminal domain. The three views perpendicular to the filament axis are related by successive 901 anticlockwise rotations around the axis. The pointed end of the filament is to the top of the figure for these views. The rightmost view is along the filament axis from the pointed end towards the barbed end. The two connected densities are indicated (1 and 2) (D) Docked atomic models of F-actin (pink) and a dimer of the talin C-terminal domain (monomers in blue and green) inside the 3D reconstruction (transparent grey). Views as in (C).
(E) Molecular surface of the docked models. Views and colours as in (D).
MitaliDas Mechanisms of talin-dependent integrin signaling and crosstalk February 2014 8
Cells undergo dynamic remodeling of the cytoskeleton during adhesion and migration on various extracellular matrix (ECM) substrates in response to physiological and pathological cues. The major mediators of such cellular responses are the heterodimeric adhesion receptors, the integrins. Extracellular or intracellular signals emanating from different signaling cascades cause inside-out signaling of integrins via talin, a cystokeletal protein that links integrins to the actin cytoskeleton. Various integrin subfamilies communicate with each other and growth factor receptors under diverse cellular contexts to facilitate or inhibit various integrin-mediated functions. Since talin is an essential mediator of integrin activation, much of the integrin crosstalk would therefore be influenced by talin.
The communication of extracellular matrix (ECM) with intracellular cytoskeleton is crucial for regulating cell adhesion, cell shape change and cell migration. Such communication depends heavily on integrins, a large family of noncovalent heterodimeric (α/β) adhesion receptors [65], [68]. Integrins function by engaging ECM ligands through their large extracellular domains and actin-binding proteins through their short cytoplasmic tails (CT), thereby linking ECM with the cytoskeleton. The effects of talin on integrin function are broad. It transduces signals across integrins in both the inside-out and outside-in directions and it also influences the organization of the actin network and the composition of focal adhesions.

Structure of talin. Talin can be subdivided into head (talin-H) and rod (talin-R) regions. Talin-H is comprised of F0, F1, F2 and F3 domains while the talin-R has ~ 11 vinculin- and 3 actin-binding sites (in blue). The vinculin binding sites are dormant (in green) and are likely mechanoactivated (in red). PIP2, PKC, Rap1 and or RIAM can relieve the autoinhibitory effect of talin-R (1654–2344aa; black region) on F3 domain, promoting talin binding to β CT. The secondary integrin-binding site is in orange.
Benjamin T. Goult The Mechanical Basis of Memory – the MeshCODE Theory 25 February 2021 9
One of the major unsolved mysteries of biological science concerns the question of where and in what form information is stored in the brain. I propose that memory is stored in the brain in a mechanically encoded binary format written into the conformations of proteins found in the cell-extracellular matrix (ECM) adhesions that organise each and every synapse.
My comment: I believe that memory is a fearture of the mind, which is not physical. I do not see how for example the imagination of an event, for example walking on the street to school, could be transformed/encoded in physical form, since we are talking here about a dynamical image, that unfolds in our imagination. If i close my eyes, and imagine that event, it simply appears in my mind, but that imagination seems not to be bound somehow to the physical.
The MeshCODE framework outlined here represents a unifying theory of data storage in animals, providing read-write storage of both dynamic and persistent information in a binary format. Mechanosensitive proteins that contain force-dependent switches can store information persistently, which can be written or updated using small changes in mechanical force. These mechanosensitive proteins, such as talin, scaffold each synapse, creating a meshwork of switches that together form a code, the so-called MeshCODE. Large signalling complexes assemble on these scaffolds as a function of the switch patterns and these complexes would both stabilise the patterns and coordinate synaptic regulators to dynamically tune synaptic activity. Synaptic transmission and action potential spike trains would operate the cytoskeletal machinery to write and update the synaptic MeshCODEs, thereby propagating this coding throughout the organism. Based on established biophysical principles, such a mechanical basis for memory would provide a physical location for data storage in the brain, with the binary patterns, encoded in the information-storing mechanosensitive molecules in the synaptic scaffolds, and the complexes that form on them, representing the physical location of engrams. Furthermore, the conversion and storage of sensory and temporal inputs into a binary format would constitute an addressable read-write memory system, supporting the view of the mind as an organic supercomputer.
I would like to propose here a unifying theory of rewritable data storage in animals. This theory is based around the realisation that mechanosensitive proteins, which contain force-dependent binary switches, can store information persistently in a binary format, with the information stored in each molecule able to be written and/or updated via small changes in mechanical force. The protein talin contains 13 of these switches, and, as I argue here, it is my assertion that talin is the memory molecule of animals.
My comment: Maybe, if we talk about memory, it can be, that the Talin proteins memorizes states of affairs of the outside world, like environmental conditions, food & nutrition supply and availablity, to respond in a fast manner to the the different conditions constantly imposed by the environment. But that, then, would have nothing to do with the memory, or imagination of past events.
These mechanosensitive proteins scaffold each and every synapse and have been considered mainly structural. However, these synaptic scaffolds also represent a meshwork of binary switches that I propose form a code, the so-called MeshCODE. The appreciation of such a network of switches and the machinery that controls them leads to a new hypothesis for the way the brain might be functioning.
The MeshCODE array of mechanical switches1 would be operated by the cytoskeletal machinery, with synaptic signalling triggering the cytoskeleton to push and pull on these switches to constantly alter and update the coding in each neuron (Figure 1).

Figure 1. The mechanical cell.
Cartoon of a neuron showing the binary coding that results from the hundreds of mechanical switches built into each synapse. The cytoskeleton is represented as a mechanical machine that operates these switches in response to neuronal activity, thus altering and updating the coding. Inset: example of a mechanical binary switch. These protein domains can be reversibly switched between two different states, folded “0” and unfolded “1.”
Together, this mechanical layer would integrate with the chemical and ionic signalling layers to provide the basis for a new mechanism underlying information-processing and storage. In this scenario, electrochemical signalling between neurons would coordinate a network of trillions of mechanically operated switches, each able to store one bit of data, with action potential spike trains serving as the means to enter new information into this calculation. This mechanical coding would run continuously in every neuron, extending into every cell in the organism, ultimately amounting to a machine code coordinating the entire organism.
The concept of the mechanical computer described herein represents a new hypothesis for how the brain might be performing computation and postulates an addressable read-write memory mechanism. Such a memory mechanism would facilitate both the storage of data in an indexed, hierarchical structure and provide a basis for how information-processing machinery could call on this data as required. This novel concept for biochemical data storage and organic computation has similarities with computers, both old and new. These similarities span from the earliest mechanical computing machines through to solid state disks (SSDs) and the complex subroutines that enable complex computation.
The Cell as an Organic Calculating Machine
The original concept of a digital programmable computer is attributed to Charles Babbage, whose “Analytical Engine” mechanical computer was first described in 1837. Early computers were built using mechanical components, complex machines of levers and gears that were used to perform calculations by turning gears and incrementing counters and ultimately output displays. The inputs initiate the calculation and the levers and gears crunch the numbers, and push and pull until the calculation is complete and the output returned. The programs that the Analytical Engine uses take the form of punched cards, in which the pattern of holes on the card run different parts of the machine. By changing the pattern of holes on the card, a different program will run to give a different calculation and obtain a different output.
There are considerable similarities between a mechanical computer and a cell. Each cell contains a series of levers, pulleys and gears in the form of its cytoskeleton. The cytoskeleton is an incredibly complex, dynamic network formed of three major classes of filaments; actin, microtubules, and intermediate filaments. These filaments can assemble and disassemble rapidly and robustly in response to cellular signals, and the interplay between them is complex. The cytoskeleton can be used to generate forces, with motor proteins pushing and pulling on actin and microtubule filaments, exerting forces on to specific targets. These filaments can also serve as railroads to transport cargos to precise locations in the cell. There are hundreds of cytoskeletal regulators that control these networks with the precise linkages, filaments, and adapters determined by the programme that cell is running.
Almost all cells in our bodies rely on cell-adhesion molecules, that adhere the cell to adjacent cells and/or the surrounding meshwork of proteins called the extracellular matrix (ECM). The adhesions to the ECM, mediated by the integrin family of ECM receptors, serve as information-processing centres, able to feel the surrounding environment and instruct the cell how to function.
My comment: While a big deal of the information stored in DNA is used by the cell to make things, like how to synthesize amino acid chains in the right order to get functional proteins, there is an entire class of proteins that process information to instruct the cell how to function, which is an entirely different function. This is a distinction that goes through all biology. The living needs the know-how to construct, and to operate things, both at once, or no deal.
The cytoskeleton is wired up to the cells adhesions, to the nucleus and to all the organelles in a highly ordered (but dynamic) manner.
My comment: Once again we can observe that one component bears only function in an interdependent fashion together with other players. It is an interlocked machine-like operation with many elementary parts contributing to the functional order.
Once a cell is established in its environment, the cytoskeleton is maintained under tension but is not constantly generating large forces or battling against itself. This homeostasis is achieved when all the forces and tensional restraints are balanced in the system, at which point it is said to be under tensional integrity or “tensegrity”. In a muscle, the actin filaments and myosin motors are highly organised to enable the generation of forces and motion. In a similar fashion, the cytoskeleton in the brain is ordered which I suggest connects all the synaptic adhesions together via dynamic mechanical linkages.
Talin
The major linkage between the integrin-ECM connections and the cytoskeleton is the protein talin. Talin is perfectly positioned to respond to changes in forces, both from outside and inside the cell, and has emerged as a master mechanosensor in that it can sense these forces and convert them into biological signals. Each adhesive structure contains hundreds of talin molecules all connected to the integrins, each other, and the cytoskeleton through direct and indirect connections to actin, creating a complex array of talin molecules (Figure A below).

Talin as a memory molecule.
(A) Talin is comprised of an N-terminal FERM domain that binds to integrin, connected to 13 helical bundles, the talin rod domains R1-R13.
(B) A mechanical binary switch. One talin helical bundle is shown. Under tension each bundle can exist in two thermodynamically stable states, folded “0” and unfolded “1” and can be switched back and forth between these states via mechanical force.
(C) Cartoon of three rod domains going from 000 to 010 in response to one contractility spike by the cytoskeletal force-generation machinery.
(D) A talin molecule as a binary string, the nine vinculin binding sites are labelled in green (i) in the absence of force the 13 rod domains are all in the folded “0” state; (ii) upon adhesion formation each talin is tethered between the integrin:ECM and F-actin, and adopts a specific switch pattern; two contractility spikes (shown as a blue spring) result in two of the switches switching (iii and iv), whereas a decrease in contractility resets one switch back to 0 (v). The exact order of switching and the resulting switch state will depend on the system.
Question: Figure 4 defines which of the 13 bundles is switched on, aka unfolds. If all 13 switches are unfold, the proteins gets 13 times the length of each bundle unfolded long, correct ?
Onto this integrin-talin-actin core complex, numerous additional proteins assemble forming distinct, tissue specific signalling complexes.
There are two talin isoforms, talin-1 and talin-2 expressed in the brain; both have the same domain structures but subtly different mechanical properties. Each talin molecule contains 13 force-dependent binary switches ( Figure 2A). These talin domains, can be reversibly switched between two thermodynamically stable states, “folded” and “unfolded,” using mechanical force (Figure 2B). One of the key discoveries that leads to the MeshCODE theory is that the switches in talin exhibit mechanical hysteresis meaning that, provided the molecule is maintained in a mechanical linkage between the integrin-ECM and the actin cytoskeleton, both the folded and unfolded states of each domain are thermodynamically stable (Figure 2B). This imparts a novel function on talin in molecular memory as each talin molecule can be imprinted with a persistent pattern of switch states as a specific outcome of the forces that have acted upon it (Figure 2C). The conformational state of each switch determines which signalling molecules are recruited, thereby providing different instructions to the cell as a function of force, enabling talin to serve as a Mechanosensitive Signalling Hub. As the environment changes, such as when a cell migrates, these switches detect these changes enabling the cell to respond appropriately.
Question: How could this function be the product of evolutionary pressures ? Was the right state of affairs, that permits the cell to respond appropriately obtained by trial and error? Would each unsuccessful trial not resulted in an "error catastrophe"? How would/could the cell recover from the erroneous result and going back to the previous state of affairs, waiting until another mutation would be selected and tried out, repeating the process as many times as "necessary" until fixing the new function in the organism? In order for that function to be instantiated, would not the whole chain of interacting interdependent molecules, aka. the microtubules, and lectins have to mutate accordingly and in parallel to establish the function? If a myriad of talins and the adjacent interacting proteins are required in the cell to get functionalty, adaptation, plasticity, and different functional states, how could/would single point mutations in the genome lead to a system change in the cell to implement that more advanced, sophisticated structure?
Most current models envisage talin as a rope in a “tug-of-war” between extracellular forces, and those generated by the cells force-generating machinery serving as a “molecular clutch” that enables mechanotransduction and cell migration. Proteins such as vinculin, which bind to nine of the unfolded talin switches, can further stabilise the unfolded state, limiting refolding and reinforcing these connections.
Talin, the Data Molecule of Life
Every animal known to humankind has the same 13 switches in talin, and the high conservation of switch pattern suggests a role that is explicitly dependent on the order of the string of switches. The best way to visualise such a role is to consider the scenario where the extracellular environment is built in such a way that it presents a mechanically stable, predictable environment. In this scenario, the talin switches are no longer required to sense the extracellular environment as that is, to all intents and purposes, constant. Instead, the cell can use its force-generating machinery to operate these mechanical switches and in doing so write data into the adhesions (Figure 2D).
This has profound implications for data storage in animals, as it means that the cell can use the talin switches to store information. Adhesions in controlled, predictable environments can adopt the role of data-storage devices that store information in a manner controlled by the system. I would like to propose that this complex meshwork of switches operates as a code, the MeshCODE outlined here. To simplify the nomenclature of the switches in this view of talin it is easier to regard the “folded” and “unfolded” states as “0” and “1,” respectively, to better reflect that it is data that is being stored in the talin molecule (Figure 2).
Exquisite Calculation in a Single Cell
As a result, the cell is a mechanical computer with all of the architecture necessary for computation. The cytoskeleton serves as the levers and gears that perform the calculation, and the MeshCODE adhesions provide a multifunctional system that can be used to perform calculations and serve as a memory storing the results in the conformations of the switches. An input signal, be it an extracellular signal activating a receptor, a change in physicality, or excitation by a chemical or electrical signal, perturbs the balanced state of the cell, switching the “computer” on, thereby triggering changes in the cytoskeleton as it seeks to return to homeostasis. Key to maintaining the mechanical balance of the cell are the 20 members of the Rho-family of GTPases. These crucial components of the signalling networks coordinate the contractility and behaviour of the actin cytoskeleton and their role in controlling synaptic development and plasticity is established. There are 80 activators [guanine nucleotide exchange factors (GEFs)] and 70 inactivators [GTPase-activating proteins (GAPs)] of the Rho family of proteins, many of which are located at synapses. The balance of these opposing regulators enables the cellular mechanics to be spatially and temporally modulated by a multitude of signalling pathways. Changes in architecture and contractility push and pull on the adhesions which the MeshCODE theory predicts will result in reproducible alterations in the binary switches. When the calculation is complete, homeostasis is restored. At this end point, the conformations of the switches in the adhesions are altered and the array of 0s and 1s reflect the outcome of the calculation. The large adhesion complexes that assemble on the talin scaffold as a function of the altered switch patterns stabilise the patterns and dictate the signalling response.
The appearance of talin at the dawn of multicellularity allowed cells to store information persistently by writing to each talin molecule like a computer writes to a disk. As well as serving as a memory, these switches also coordinate cell signalling and provide a way to control the reading of the genome from the periphery of the cell.
Organic Calculation in the Brain
The brain is a colossal cell signalling machine with a trillion cells all communicating with each other, leveraging the organic calculating power of each cell. Synapses are the perfect system for optimised cell signalling between connected cells, and there are approximately 100 trillion synapses in the brain transmitting signals between neurons, to give rise to brain activity. The specialised architecture of synaptic junctions juxtaposes the presynaptic and postsynaptic neurons to form the synaptic cleft, an arrangement supported by astrocytes which coordinate synaptic maintenance and plasticity, and microglia ( Figure 3A).

Figure 3. The MeshCODE.
(A) Schematic diagram of a tripartite synaptic junction. The integrin adhesion complexes and MeshCODEs in the presynaptic and postsynaptic zones, and in a neighbouring astrocyte are shown. The synapse is encapsulated by a specialised ECM that protects the mechanical environment of the MeshCODEs. Integrin and talin in one MeshCODE are shown connected to the actin cytoskeleton. (Right) Enlarged view of the highlighted region showing the MeshCODE intricately wired up to the cytoskeleton. Only the integrin-talin-actin adhesion core complex is shown, but up to 250 different proteins decorate this core dependent on the switch patterns of the talin scaffolds. The protein vinculin (green) binds to nine of the unfolded talin switch domains, stabilising them in the unfolded state and reinforcing the cytoskeletal connections.
(B) A schematic of an array of ten talin molecules with the 13 switches arranged vertically. White, 0; red, 1.
(C) Perturbations to the system result in changes in cytoskeletal contractility that alter the pattern of 1s and 0s written in the MeshCODE and its resultant output in a defined way.
Each synapse is scaffolded by adhesions located around the edge of the synapse; these adhesions involve the binding of integrins to ECM components, such as laminin and fibronectin Figure 3A). Integrin adhesion complexes are required for synaptic plasticity and memory, and consolidation of long-term potentiation and the ECM has been linked to learning and memory previously. However, a role for adhesion complexes in the storage of data has not been considered previously.
The brain is soft, which provides the perfect, protective environment for each neuron to tightly control its own mechanical environment independently, building its own ECM niche around each synapse, with the surroundings serving to isolate the neuron from external mechanical forces. The MeshCODE theory proposes that tight regulation of the mechanical environment of each synapse would enable mechanical computation to occur with meticulous precision. The ECM surrounding each synapse is subject to extensive maintenance and remodelling during synaptic function, and it seems reasonable to postulate that this maintenance of the ECM might ensure that the synapses mechanical environment is tightly regulated and predictable.
Therefore, in the MeshCODE framework, these synaptic scaffolds provide the capacity to write data into the synapses themselves in the extensive arrays of binary switches that are located in both the pre-, and postsynaptic side of each synapse as well as in the supporting astrocyte cells (Figure 3A). The capacity to store information in every synapse, with the potential to orchestrate the flow of information through that synapse, makes the synapse a complex computational device.
How Would a Mechanical Code Be Read?
The key to any code is a read-out mechanism and the mechanical code outlined here leads to a number of hypotheses for how such a coding in the brain would be read. This section discusses how MeshCODE driven adhesion signalling might work in the context of a neuron, with the purpose being to demonstrate the feasibility and broad applicability of the theory to well-established neuronal processes.
The contractility-dependent control of integrin-adhesion complexes is a well-studied phenomenon in the cell adhesion field. Adhesion complexes grow as forces are exerted on them, altering their composition and signalling outputs and mechanical forces acting on talin are essential to drive this process. Each synaptic adhesion would serve as a discrete signalling hub, altering its signalling as a function of the mechanical signals received. Following each synaptic signalling event, the neuronal cytoskeleton would contract briefly and precisely generating these mechanical signals and altering the switch patterns at that site. As a result, the code would be read by the ligands that engage the different switch states (Figure 4A) with the read-out being the relative (re)distribution of all the ligands engaging the entire array of switches defining the output.

Figure 4. How would a mechanical code be read?
(A) Schematic diagram representing four mechanical switches labelled I–IV, (Left) each switch has the potential to bind different ligands in its folded, 0 and unfolded, 1 states. (Middle) The switch pattern is shown as a binary string. (Right) The ligands decorating the switches present a “read-out” mechanism as the complexes formed will depend on the mechanical coding. (i) at low force the green, orange, and yellow ligands engage the MeshCODE on switches I, II, and IV. (ii) Contractility (shown as a blue spring) switches one domain (here domain II) from the 0 to 1 state which drives a switch in binding partners, displacing the orange ligand and recruiting the blue ligand. The overall signalling complex and read-out is altered. (iii) Further contractility switches a second domain (here domain IV) further altering the coding and the proteins that are recruited to the synaptic scaffolds. Proteins, like vinculin, which bind the unfolded state of a switch lock it in that conformation and limit its ability to refold.
(B–D) A cartoon of a neuron with four synapses in different states showing hypothetical ways a mechanical code might be read. Contractility as a result of synaptic signalling causes alterations to the MeshCODE switch patterns on both sides of a stimulated synapse. (B) in the pre-synaptic terminal these switch patterns might regulate the probability of release of docked vesicles by specific switches controlling this process via a hypothetical interaction with a key regulator of synaptic firing.
(C) Synaptic stimulation triggers contractility in the post-synaptic region that specifically alters the switch patterns in that stimulated synapse. The altered switch patterns create “Tags” that recruit proteins (one is shown as a blue square) specifically to that synapse that enhance long-term potentiation. Switching might also displace proteins that dampen potentiation (orange circle) which then diffuse away.
(D) Future signals through that synapse can trigger additional switch pattern changes and alter the binary coding in that synapse, the protein signals recruited, etc., in a dynamic re/writable way, changing the threshold of each synapse in that neuron. These changes to the synaptic adhesions as they grow and shrink dynamically in response to stimuli form the memory trace.
The location of switches in the pre- and post-synaptic scaffolds, coupled with the ligands available to bind in these different structures, suggests potential mechanical explanations for a number of recognised processes of synaptic regulation including;
(i) dynamical regulation of the probability of vesicular release ( Figure 4B),
(ii) synaptic tagging and capture,
(iii) inverse synaptic tagging,
(iv) synaptic competition Figure 4D),
(v) spike-timing dependent plasticity and
(vi) the capture of temporal information.
Hypothesis: Synapses Are Activated and Deactivated Transiently in Response to Signals
The presence of switches in the synapses raises the possibility that certain MeshCODE patterns might transiently activate, or deactivate, transmission through specific synapses. On the pre-synaptic side, the strength of individual synapses within a single axon are regulated by changes in the probability of vesicular release. Tightly regulated changes in the talin switch conformation at individual synapses might provide a molecular mechanism for this changing probability (Figure 4B). The actin cytoskeleton has been shown to be important for neurotransmitter release, as has the requirement of force generation at these sites, and a role for talin in presynaptic function has also been reported. In the pre-synaptic terminal alterations in certain switch patterns, dynamically exposing and disrupting binding sites in close proximity to the active zone, might regulate the number of docked vesicles, or control the probability of release of docked vesicles via hypothetical interactions with key regulator(s) of synaptic firing (Figure 4B). The probability of each synapse firing would be updated by the mechanical signalling providing a mechanism for synapses to be switched on and off transiently on demand, potentially contributing to the homeostatic synaptic scaling of the neurons output.
Hypothesis: A Mechanical Basis for Synaptic Tagging?
A well-known, but not fully understood, phenomena is the process of “synaptic tagging and capture,” where following activation of a synapse, that synapse is marked in such a way that proteins are recruited directly to that synapse ( Figure 4C). As a synapse is stimulated, then the MeshCODE theory predicts that the stimulated synapse undergoes localised contractility thereby altering the switch patterns at that site. Certain switch states would be generated that expose novel binding sites in that stimulated synapse only. These “Tags” (Figure 4C) would serve to recruit proteins to that synapse and it is possible that these switches provide the basis for a dynamic form of synaptic tagging. It is already established that alterations in talin conformation cause global relocation of cellular factors. Furthermore, it is also possible that proteins that inactivate synapses, and/or transcriptional regulators, might be bound to the 0 state of the switch which upon the switch unfolding would be released from the newly stimulated synapse to diffuse away (Figure 4C). This displacement of signals from a stimulated synapse would provide a mechanism for “Inverse synaptic tagging”; “inverse tag” proteins would be displaced from stimulated synapses and accumulate at unstimulated synapses where more of that switch is still in the 0 state.
As protein levels in cells are under tight control via transcriptionally regulated programs, these switches would result in the global relocalisation of proteins in the neuron. If proteins are recruited to one synapse following stimulation (Figure 4C), then those same proteins would be depleted from other synapses leading to a basis for “synaptic competition” where the strengthening of one synapse affects the stability of other synapses in that cell. The number of synapses, and the number of scaffold proteins in each synapse will be tightly regulated, and therefore an exact number of mechanical switches will be distributed across each neuron, and throughout the brain. As each switch state can recruit one signalling molecule then as the number of switches in one state increases in a stimulated synapse more of that signalling molecule will be recruited to that site thereby dynamically altering the concentration of molecules at that adhesion/synapse and depleting that molecule from the rest of the neuron and the other unstimulated synapses. This mechanical coordination between synapses, with stimulation of one synapse recruiting and displacing plasticity-related proteins from that site, thereby directly effecting the proteins available to bind at other synapses provides a mechanism for potentiated and depressed synapses to interact in a synergistic manner. This synchronisation would enable the entire neuron to behave as a mechanically coordinated system of synapses where input signals to a synapse can dynamically alter the activity of each synapse in the neuron (and by extension the neurons it is communicating with), providing a way to establish long-term plasticity and orchestrate the flow of information transmitted by the cell.
Overtime this synaptic tagging leads to remodelling of the synapse, driven by the recruitment of newly transcribed proteins to the stimulated site. Remodelling of the actin cytoskeleton occurs at stimulated synapses, which not only enhances that synaptic connection but also strengthens the mechanical linkages mediated through the actin cytoskeleton emanating from that site. Nine of the unfolded switch domains in talin recruit the protein vinculin, which is well known as a means that cells reinforce integrin-talin-actin connections anchoring actin to the adhesion site and also for locking the mechanical switches in the unfolded state.
Hypothesis: Contractility-Dependent Control of Integrin Adhesion Complex Dynamics Is the Way That Neurons Store Information
It might be that the Synaptic tagging observed in synapses is actually a similar process to the contractility-dependent growth of integrin adhesion complexes seen in other cells. The adhesions would grow in stimulated synapses as forces are exerted on them and atrophy in unstimulated synapses, with the cumulative effect that overtime these changes in switch pattern at stimulated synapses would lead to morphological changes in the shape and size of the synapses as they remodel altering the signalling output of each site. This process would be explicitly dependent on the talin switch patterns that dictate which molecules assemble to form each adhesion complex. The engagement of the cytoskeleton with the talin molecules directly, and indirectly through proteins such as vinculin, will alter as the switch patterns change, and these engagements will
(i) help maintain the switch pattern and
(ii) alter where and how future contractility spikes exert tension on talin and the meshwork in general, and
(iii) determine the signalling molecules recruited to that site. It is possible that the patterns of the switch states and the specificity of ligands binding to them, are further regulated by post-translational modification; specific patterns could be “write protected” by locking the domains in those conformations by phosphorylation or other modification.
These synaptic adhesions would be serving as large information-processing centres, with the many proteins recruited to, and interacting on, the patterns of switches stabilising the patterns and encoding/decoding the information. The complexes that assemble on the patterns (Figure 4D) and the signalling outputs that result will vary depending on the expression levels of proteins, and their distribution, in that neuron, but in each case, they will be assembled on the integrin-talin-actin core complex dictated by the mechanical signalling across the whole cell. Furthermore, adhesion complexes also transmit mechanical signals to the nucleus, and can induce modular gene expression patterns in response to both physical and geometric constraints, a process referred to as mechanotransduction. Epigenetic tagging and alterations in gene expression are required for memory system consolidation and it might be that mechanotransduction through these synaptic adhesion complexes contributes to such mechanisms.
Subsequent stimulation of that synapse would result in more contractility changes that further alter the switch patterns in the MeshCODE dependent on the current switch patterns at that site and the cytoskeletal connections that formed. Further contractility might unfold more copies of the same switch domain that was described above (Figure 4C), exposing more “tags” and recruiting more of the ligand that binds to that site further depleting it from other synapses. However, if many copies of that switch are already in the unfolded state, or if the cytoskeletal connections have altered, then additional contractility would trigger different switches to unfold, recruiting and displacing other molecules, causing further remodelling of the adhesion complexes and redistribution of proteins throughout the neuron (Figure 4D).
1. Unicellular and multicellular Organisms are best explained through design
https://reasonandscience.catsboard.com/t2010-unicellular-and-multicellular-organisms-are-best-explained-through-design
2. https://reasonandscience.catsboard.com/t2990-how-does-biological-multicellular-complexity-and-a-spatially-organized-body-plan-emerge#7758
3. https://reasonandscience.catsboard.com/t2254-does-the-mullerian-two-step-proposal-refute-irreducible-complexity
4. https://sci-hub.ren/10.1038/nrm3624
5. https://rupress.org/jcb/article/217/11/3776/120671/Talin-as-a-mechanosensitive-signaling-hubTalin-as
6. https://www.nature.com/articles/s41598-019-42002-6
7. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2168396/
8. https://www.sciencedirect.com/science/article/pii/S0005273613002575
9. https://www.frontiersin.org/articles/10.3389/fnmol.2021.592951/full

