What are the odds to have a functional interactome for the smallest known living cell?
https://reasonandscience.catsboard.com/t3120-what-are-the-odds-to-have-a-functional-interactome-for-the-smallest-known-living-cell
1. All organisms have a genome. Some an epigenome and all life has a proteome, a metabolome, and less known, as well, an interactome, which defines all cellular interactions, amongst it, all protein-protein interactions of a cell.
2. Protein-protein interactions (PPIs) are one of the most important components of biological networks. Proteins, in order to perform their functions, often need to be interlinked in an interdependent manner with other proteins to form production line-like associations to produce the various molecules, building blocks, co-factors, and proteins of the cell. Without the right interdependent linkages, there would be no life on earth.
3. Besides explaining the origin of a minimal protein set on early earth ( the smallest proteome of a free-living cell is 1350 proteins by Pelagibacter ubique) it must also be explained how they got interlinked together.
4. The odds to connect all 1350 proteins in the right, functional order ( supposing that all outcomes would be equally likely. In other words, every connection would have an equal chance of being chosen. The odds of an event occurring are equal to the ratio of favorable outcomes to unfavorable outcomes) would be 4.1431^3641. That is an unimaginably large number. ( with 3641 zeroes! ) There are 10^22 stars in the knowable universe !! This is the odds on top of the odds to have a functional proteome, which is in the case of Pelagibacter, again with 1350 proteins, average 300 Amino Acids size: 10^722000 ) It should be evidently clear, by the astronomical odds, that having a functional interactome of the currently known smallest life form, Pelagibacter, on top of a functional proteome, is in the real of the ABSOLUTELY impossible !!
Peter Tompa: The Levinthal paradox of the interactome 2011
The inability of the interactome to self-assemble de novo imposes limits on efforts to create artificial cells and organisms, that is, synthetic biology. In particular, the stunning experiment of “creating” a viable bacterial cell by transplanting a synthetic chromosome into a host stripped of its own genetic material36 has been heralded as the generation of a synthetic cell43 (although not by the paper's authors). Such an interpretation is a misnomer, rather like stuffing a foreign engine into a Ford and declaring it to be a novel design.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3302650/
All genes of an organism are called a genome. The information systems beyond genes are called epigenetics, and as derivation, we have the epigenome. And the set of proteins in a cell is called a proteome. All the players in metabolic reactions are called metabolomes. And all interactions in the cell are called interactome. All these subjects merge into a higher-level category called -omics. 2 Organelles, cells, tissues, organs, and organisms, populations, and ecology, planet earth, the planetary and solar system work as integrated systems. They work together as a cooperative and organized system.
The interactome network of protein-protein interactions captures the structure of molecular machinery and gives rise to a bewildering degree of life complexity. A critical property of interactomes is the resilience to protein failures as the breakdown of proteins affects the exchange of any biological information between proteins in a cell and may lead to cell death or disease. 6
A paper in Nature reported that: Present technology already enables analysis of the complete protein inventory of biological systems, including cell-type-specific proteomes of mammalian organs. One outcome of in-depth proteomics studies has unraveled that the identity of cells and tissues seems to be determined primarily by the abundance at which they express their constituent proteins, and perhaps by the manner in which the proteins are organized in the proteome, rather than the presence or absence of certain proteins.
‘Interactomics’ is an interdisciplinary field of biology and bioinformatics refers to the study of both the interactions among various proteins and other molecules within a cell 1 The totality of protein-protein interactions taking place within a cell, organism, or a specific biological context, is known as Interactome.
The interactome is defined as the number of possible interactions between the proteins found in a cell.
Protein-protein interactions (PPIs) are one of the most important components of biological networks.5
The origins of life -- the 'protein interaction world' hypothesis: protein interactions were the first form of self-reproducing life and nucleic acids evolved later as memory molecules 4
The 'protein interaction world' (PIW) hypothesis of the origins of life assumes that life emerged as a self-reproducing and expanding system of protein interactions. In mainstream molecular biology, 'replication' refers to the material copying of molecules such as nucleic acids. However, PIW is conceptualized as an abstract communication system constituted by the interactions between proteins, in which 'replication' happens at the level of self-reproduction of these interactions between proteins.
The PIW hypothesis is based on experimentally validated, plausible assumptions about the emergence of early peptide/protein interaction systems in the prebiotic Earth environment
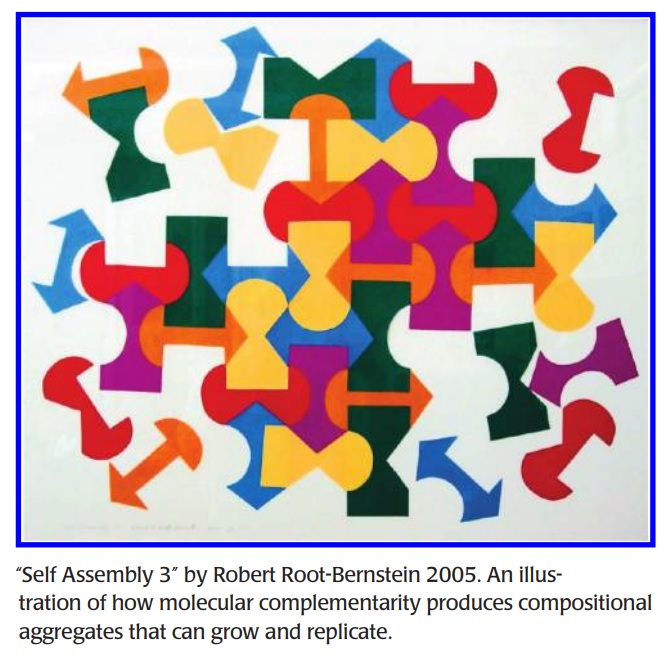
Modular Hierarchy-Based Theory of the Chemical Origins of Life Based on Molecular Complementarity AUGUST 16, 2011 7
Molecular complementarity is ubiquitous in living systems but nowhere else in nature. Why? Molecular complementarity provides means of selecting, concentrating, stabilizing, and producing buffered, homeostatic modules from which hierarchically organized systems can emerge in the most efficient conceivable manner, capturing a maximum of entropy information and providing mechanisms (compositional and linear replication) for chemical “inheritance”. Where to from here? My research has convinced me that the reductionist philosophy of science and hypothesis-driven experimentation have severe limitations. Since emergent properties cannot
be predicted, combinations and mixtures must be explored simply to see what happens.
If living systems are completely integrated molecularly complementary systems, then might the immune system itself have evolved to protect the integrity of that integrated system from noncomplementary components that threaten it?
My comment: If the immune system is vital for life, how could life have evolved without it?
Let us suppose that the first living organism was the size of the smallest currently known life form, Pelagibacter unique, which has about 1350 proteins.
Pelagibacter Ubique is the best candidate to investigate Origin of life scenarios. Here is why
https://reasonandscience.catsboard.com/t3090-pelagibacter-ubique-is-the-best-candidate-to-investigate-origin-of-life-scenarios-here-is-why
What would be the odds to connect them in the right, functional order? Let us suppose that all outcomes would be equally likely. In other words, every connection would have an equal chance of being chosen. The odds of an event occurring are equal to the ratio of favorable outcomes to unfavorable outcomes.
In our case, the odds would be: ( using the Combinations Calculator – Calculate nCr: https://www.inchcalculator.com/combinations-calculator/ )
4.1431^3641. That is an unimaginably large number. ( with 3641 zeroes! ) There are 10^22 stars in the knowable universe !!
This is the odds on top of the odds to have a functional proteome, which is in the case of Pelagibacter, again with 1350 proteins, average 300 Amino Acids size: 10^722000 )
Conclusion: It should be evidently clear, by the astronomical odds, that having a functional interactome of the currently known smallest life form, Pelagibacter, on top of a functional proteome, is in the real of the ABSOLUTELY impossible !!
1. https://www.biocode.ltd/post/interactomics?fbclid=IwAR2Eb2uPCBMjkIriIDR9Xvx4W8fr1CoW4gdTRnPCAuLtg8AWBJRkorl29H8
2. https://evolutionnews.org/2016/10/imagine_60_mill/
3. https://www.nature.com/articles/nature19949
4. https://pubmed.ncbi.nlm.nih.gov/15694682/#:~:text=Abstract,expanding%20system%20of%20protein%20interactions.&text=Densely%20concentrated%20peptide%20interaction%20systems,by%20lipid%20bi-layer%20membranes
5. https://www.karger.com/Article/Abstract/107601
6. https://snap.stanford.edu/tree-of-life/
7. https://sci-hub.ren/10.1021/ar200209k
8. https://courses.lumenlearning.com/math4libarts/chapter/calculating-the-odds-of-an-event/
https://reasonandscience.catsboard.com/t3120-what-are-the-odds-to-have-a-functional-interactome-for-the-smallest-known-living-cell
1. All organisms have a genome. Some an epigenome and all life has a proteome, a metabolome, and less known, as well, an interactome, which defines all cellular interactions, amongst it, all protein-protein interactions of a cell.
2. Protein-protein interactions (PPIs) are one of the most important components of biological networks. Proteins, in order to perform their functions, often need to be interlinked in an interdependent manner with other proteins to form production line-like associations to produce the various molecules, building blocks, co-factors, and proteins of the cell. Without the right interdependent linkages, there would be no life on earth.
3. Besides explaining the origin of a minimal protein set on early earth ( the smallest proteome of a free-living cell is 1350 proteins by Pelagibacter ubique) it must also be explained how they got interlinked together.
4. The odds to connect all 1350 proteins in the right, functional order ( supposing that all outcomes would be equally likely. In other words, every connection would have an equal chance of being chosen. The odds of an event occurring are equal to the ratio of favorable outcomes to unfavorable outcomes) would be 4.1431^3641. That is an unimaginably large number. ( with 3641 zeroes! ) There are 10^22 stars in the knowable universe !! This is the odds on top of the odds to have a functional proteome, which is in the case of Pelagibacter, again with 1350 proteins, average 300 Amino Acids size: 10^722000 ) It should be evidently clear, by the astronomical odds, that having a functional interactome of the currently known smallest life form, Pelagibacter, on top of a functional proteome, is in the real of the ABSOLUTELY impossible !!
Peter Tompa: The Levinthal paradox of the interactome 2011
The inability of the interactome to self-assemble de novo imposes limits on efforts to create artificial cells and organisms, that is, synthetic biology. In particular, the stunning experiment of “creating” a viable bacterial cell by transplanting a synthetic chromosome into a host stripped of its own genetic material36 has been heralded as the generation of a synthetic cell43 (although not by the paper's authors). Such an interpretation is a misnomer, rather like stuffing a foreign engine into a Ford and declaring it to be a novel design.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3302650/
All genes of an organism are called a genome. The information systems beyond genes are called epigenetics, and as derivation, we have the epigenome. And the set of proteins in a cell is called a proteome. All the players in metabolic reactions are called metabolomes. And all interactions in the cell are called interactome. All these subjects merge into a higher-level category called -omics. 2 Organelles, cells, tissues, organs, and organisms, populations, and ecology, planet earth, the planetary and solar system work as integrated systems. They work together as a cooperative and organized system.
The interactome network of protein-protein interactions captures the structure of molecular machinery and gives rise to a bewildering degree of life complexity. A critical property of interactomes is the resilience to protein failures as the breakdown of proteins affects the exchange of any biological information between proteins in a cell and may lead to cell death or disease. 6
A paper in Nature reported that: Present technology already enables analysis of the complete protein inventory of biological systems, including cell-type-specific proteomes of mammalian organs. One outcome of in-depth proteomics studies has unraveled that the identity of cells and tissues seems to be determined primarily by the abundance at which they express their constituent proteins, and perhaps by the manner in which the proteins are organized in the proteome, rather than the presence or absence of certain proteins.
‘Interactomics’ is an interdisciplinary field of biology and bioinformatics refers to the study of both the interactions among various proteins and other molecules within a cell 1 The totality of protein-protein interactions taking place within a cell, organism, or a specific biological context, is known as Interactome.
The interactome is defined as the number of possible interactions between the proteins found in a cell.
Protein-protein interactions (PPIs) are one of the most important components of biological networks.5
The origins of life -- the 'protein interaction world' hypothesis: protein interactions were the first form of self-reproducing life and nucleic acids evolved later as memory molecules 4
The 'protein interaction world' (PIW) hypothesis of the origins of life assumes that life emerged as a self-reproducing and expanding system of protein interactions. In mainstream molecular biology, 'replication' refers to the material copying of molecules such as nucleic acids. However, PIW is conceptualized as an abstract communication system constituted by the interactions between proteins, in which 'replication' happens at the level of self-reproduction of these interactions between proteins.
The PIW hypothesis is based on experimentally validated, plausible assumptions about the emergence of early peptide/protein interaction systems in the prebiotic Earth environment
Modular Hierarchy-Based Theory of the Chemical Origins of Life Based on Molecular Complementarity AUGUST 16, 2011 7
Molecular complementarity is ubiquitous in living systems but nowhere else in nature. Why? Molecular complementarity provides means of selecting, concentrating, stabilizing, and producing buffered, homeostatic modules from which hierarchically organized systems can emerge in the most efficient conceivable manner, capturing a maximum of entropy information and providing mechanisms (compositional and linear replication) for chemical “inheritance”. Where to from here? My research has convinced me that the reductionist philosophy of science and hypothesis-driven experimentation have severe limitations. Since emergent properties cannot
be predicted, combinations and mixtures must be explored simply to see what happens.
If living systems are completely integrated molecularly complementary systems, then might the immune system itself have evolved to protect the integrity of that integrated system from noncomplementary components that threaten it?
My comment: If the immune system is vital for life, how could life have evolved without it?
Let us suppose that the first living organism was the size of the smallest currently known life form, Pelagibacter unique, which has about 1350 proteins.
Pelagibacter Ubique is the best candidate to investigate Origin of life scenarios. Here is why
https://reasonandscience.catsboard.com/t3090-pelagibacter-ubique-is-the-best-candidate-to-investigate-origin-of-life-scenarios-here-is-why
What would be the odds to connect them in the right, functional order? Let us suppose that all outcomes would be equally likely. In other words, every connection would have an equal chance of being chosen. The odds of an event occurring are equal to the ratio of favorable outcomes to unfavorable outcomes.
In our case, the odds would be: ( using the Combinations Calculator – Calculate nCr: https://www.inchcalculator.com/combinations-calculator/ )
4.1431^3641. That is an unimaginably large number. ( with 3641 zeroes! ) There are 10^22 stars in the knowable universe !!
This is the odds on top of the odds to have a functional proteome, which is in the case of Pelagibacter, again with 1350 proteins, average 300 Amino Acids size: 10^722000 )
Conclusion: It should be evidently clear, by the astronomical odds, that having a functional interactome of the currently known smallest life form, Pelagibacter, on top of a functional proteome, is in the real of the ABSOLUTELY impossible !!
1. https://www.biocode.ltd/post/interactomics?fbclid=IwAR2Eb2uPCBMjkIriIDR9Xvx4W8fr1CoW4gdTRnPCAuLtg8AWBJRkorl29H8
2. https://evolutionnews.org/2016/10/imagine_60_mill/
3. https://www.nature.com/articles/nature19949
4. https://pubmed.ncbi.nlm.nih.gov/15694682/#:~:text=Abstract,expanding%20system%20of%20protein%20interactions.&text=Densely%20concentrated%20peptide%20interaction%20systems,by%20lipid%20bi-layer%20membranes
5. https://www.karger.com/Article/Abstract/107601
6. https://snap.stanford.edu/tree-of-life/
7. https://sci-hub.ren/10.1021/ar200209k
8. https://courses.lumenlearning.com/math4libarts/chapter/calculating-the-odds-of-an-event/



