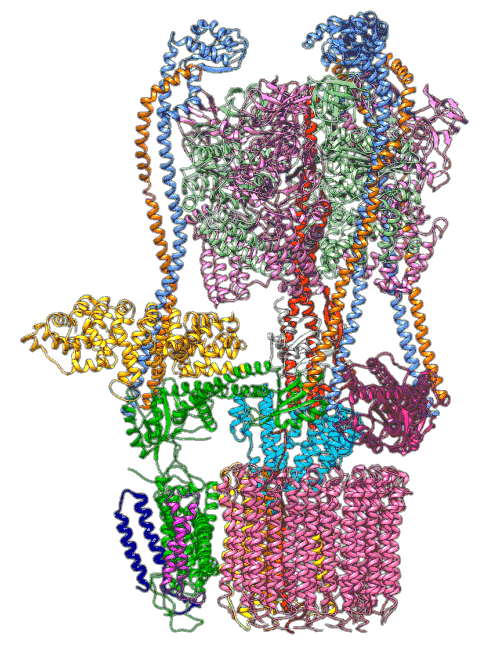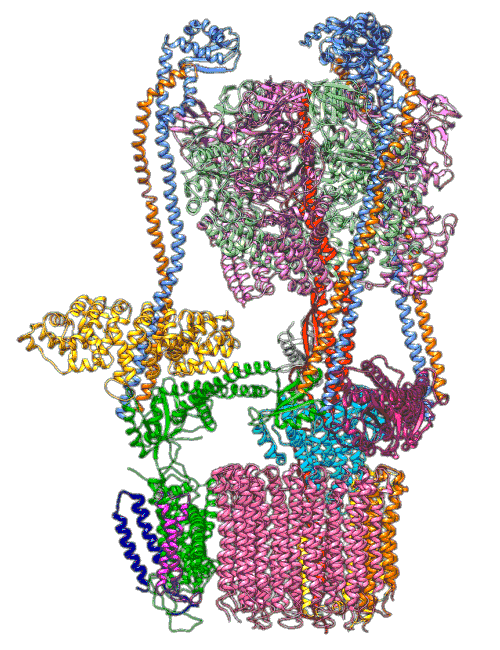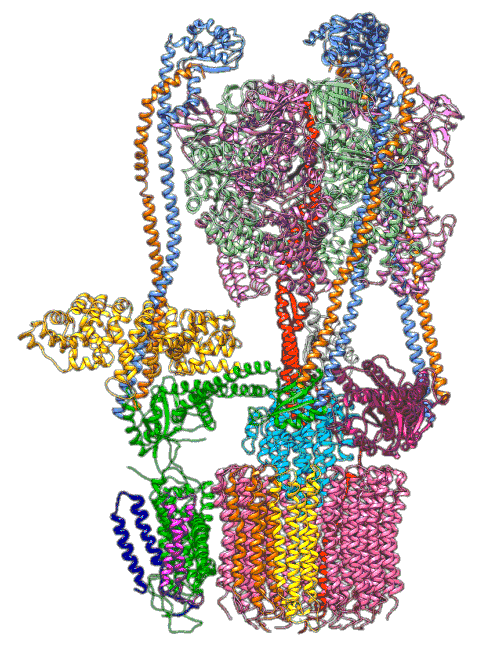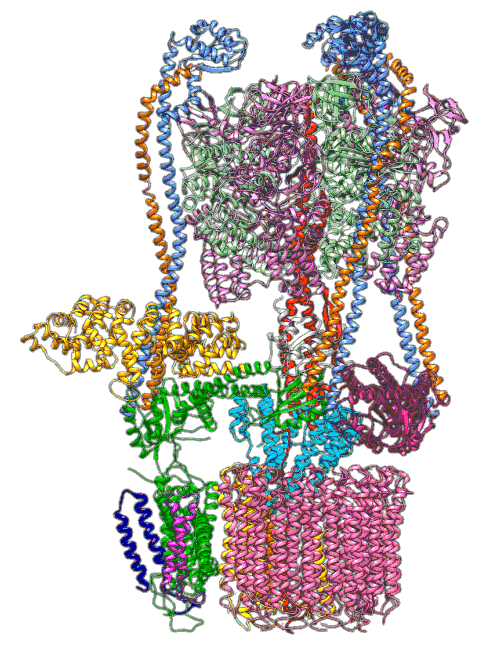
Molecular Ferris wheel delivers protons to cellular factories
https://reasonandscience.catsboard.com/t3045-molecular-ferris-wheel-delivers-protons-to-cellular-factories
All cells with nuclei, from yeast to humans, are organized like cities, with a variety of small compartments—organelles—that serve as factories where various types of work are done. Some of those factories, like the ones that break down and recycle molecules, need to continually pump in protons—hydrogen atoms with their electrons stripped off—to maintain the acidic environment they need to do their job. For this they rely on molecular Ferris wheels.
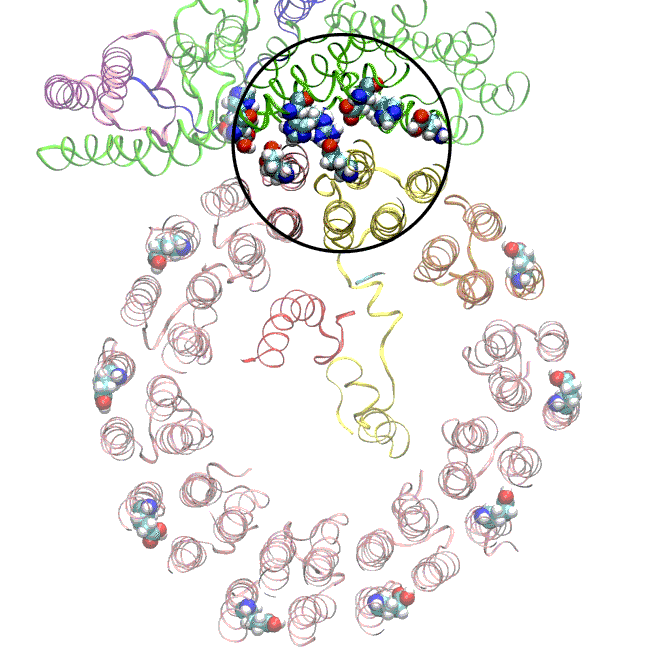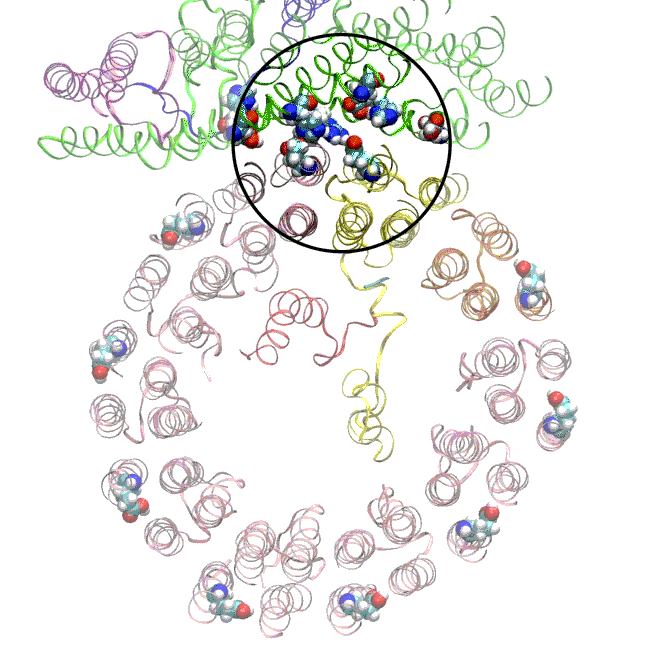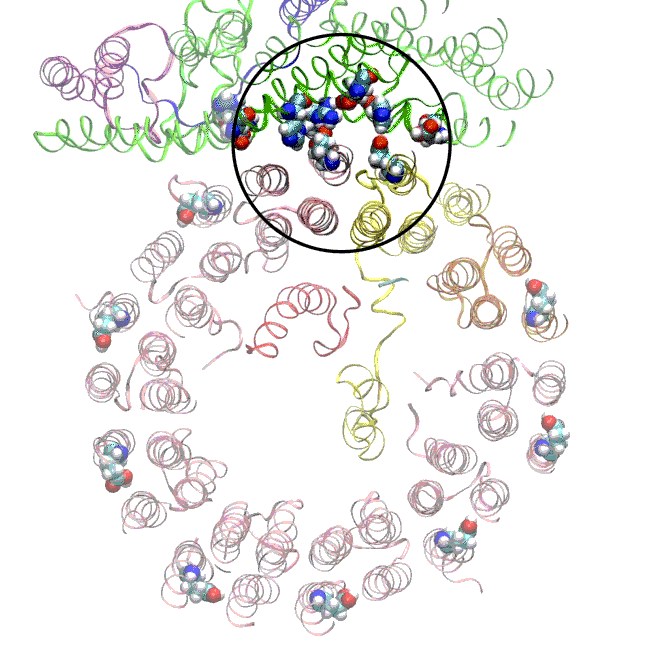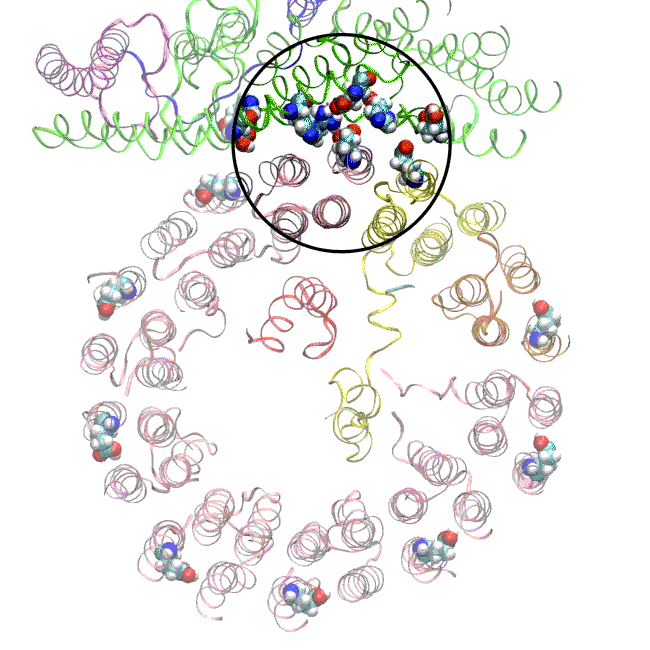
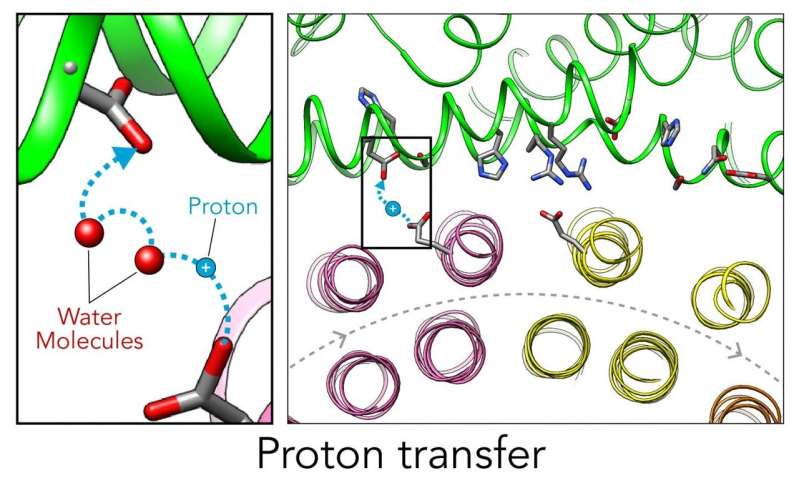
Embedded in the organelle's fatty outer membrane, these microscopic machines have rotors that spin 100 times per second, picking up protons from outside the organelle and dropping them off on the inside.
Now scientists have figured out a key step in how these Ferris wheels work in a yeast proton pump known as vacuolar ATPase (V-ATPase). There are 10 amino acid "seats" on the yeast Ferris wheel that bind protons and carry them through the membrane to the organelle's interior, as well as other amino acids that catch them when they arrive. 6
V-ATPase proton pump functions
"The V-ATPase proton pumps perform a wide range of functions, from helping transmit nerve signals to helping specialized cells secrete acid for maintaining bone. Malfunctions in these molecular machines contribute to diseases such as osteoporosis, neurodegeneration, diabetes, cancer and AIDS, so understanding them is important for human health.
Rotary vacuolar adenosine triphosphatases (V-ATPases) drive transmembrane proton transport through a Vo proton channel subcomplex. 1 Active proton transport is central to controlling the balance between cellular pH and efficiency of energy metabolism. Vacuolar adenosine triphosphatases (V-ATPases) are adenosine 5′-triphosphate (ATP) hydrolysis–driven rotary motor proton pumps that acidify the lumen of subcellular organelles in virtually every eukaryotic cell . During ATP hydrolysis–driven proton pumping in holo V-ATPase, the c-ring rotates clockwise to carry protons from the cytosolic to the luminal half-channel. Upon regulated detachment of V1-ATPase from Vo, any existing proton gradient is expected to push the c-ring in the opposite (counterclockwise) direction to allow for the reverse flow of protons.
Vacuolar-type ATPase (V-ATPase) is a highly conserved ancient enzyme with remarkably diverse functions in eukaryotic organisms. V-ATPases acidify a wide array of intracellular organelles and pump protons across the plasma membranes of numerous cell types. V-ATPases couple the energy of ATP hydrolysis to proton transport across intracellular and plasma membranes of eukaryotic cells. It is generally seen as the polar opposite of ATP synthase because ATP synthase is a proton channel that uses the energy from a proton gradient to produce ATP. V-ATPase however, is a proton pump that uses the energy from ATP hydrolysis to produce a proton gradient. 2
V-ATPases are found within the membranes of many organelles, such as endosomes, lysosomes, and secretory vesicles, where they play a variety of roles crucial for the function of these organelles. For example, the proton gradient across the yeast vacuolar membrane generated by V-ATPases drives calcium uptake into the vacuole through an H+/Ca2+ antiporter system. In synaptic transmission in neuronal cells, V-ATPase acidifies synaptic vesicles.
V-ATPases (H+ ATPases) are multisubunit, ATP-dependent proton pumps that regulate pH homeostasis in virtually all eukaryotes. They are involved in key cell biological processes including vesicle trafficking, endosomal pH sensing, membrane fusion and intracellular signaling. They also have critical systemic roles in renal acid excretion and blood pH balance, male fertility, bone remodeling, synaptic transmission, olfaction and hearing.5 While the eukaryotic V-ATPase is primarily known as a proton-pumping, rotary nano-motor that is involved in many physiological processes, it also plays an important role in: i) vesicle coat formation1 and ii) regulation of signaling and trafficking of various receptors. The V-ATPase was also identified as a sensing and signaling complex that functions in the endocytic pathway and it has a crucial role in SNARE-dependent fission/fusion process and organelle biogenesis along the exocytic pathway
No human cell can function without proton pumps, which among other things help organelles intercept viruses and other pathogens and divert them to cellular trash bins.
Origin of structure and function of V-ATPases
Claim: Proton pumping ATPases/ATPsynthases are found in all groups of present-day organisms. The structure of V- and F-type ATPases/ATP synthases is very conserved. Sequence analysis shows that the V- and F-type ATPases evolved from the same enzyme already present in the last common ancestor of all known extant life forms. The catalytic and noncatalytic subunits found in the dissociable head groups of the V/F-type ATPases are paralogous subunits, i.e., these two types of subunits evolved from a common ancestral gene. (Homologous sequences are paralogous if they were separated by a gene duplication event: if a gene in an organism is duplicated to occupy two different positions in the same genome, then the two copies are paralogous. ) The gene duplication giving rise to these two genes (i.e., encoding the catalytic and noncatalytic subunits) predates the time of the last common ancestor. https://link.springer.com/article/10.1007/BF00762534
Mapping of gene duplication events that occurred in the evolution of the proteolipid, the noncatalytic and the catalytic subunits, onto the tree of life leads to a prediction for the likely subunit structure of the encoded ATPases. A correlation between structure and function of V/F-ATPases has been established for present-day organisms. Implications resulting from this correlation for the bioenergetics operative in proto-eukaryotes and in the last common ancestor are presented. The similarities of the V/F-ATPase subunits to an ATPase-like protein that was implicated to play a role in flagellar assembly are evaluated.
Reply: Most subunits of the V-type and the F-type ATPases are not homologous.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4154653/
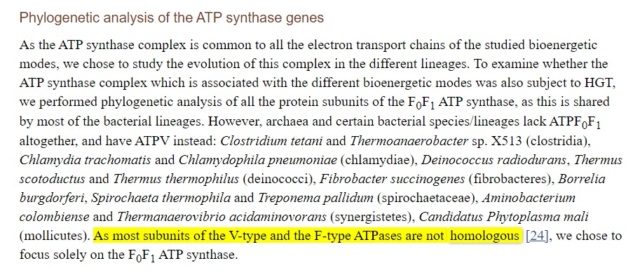
Inventing the dynamo machine: the evolution of the F‑type and V‑type ATPases 4
Sequence and structural comparisons of F- and V-type ATPases have revealed homology between their catalytic and membrane subunits, but no homology between the subunits of the central stalk that connects the catalytic and membrane components.
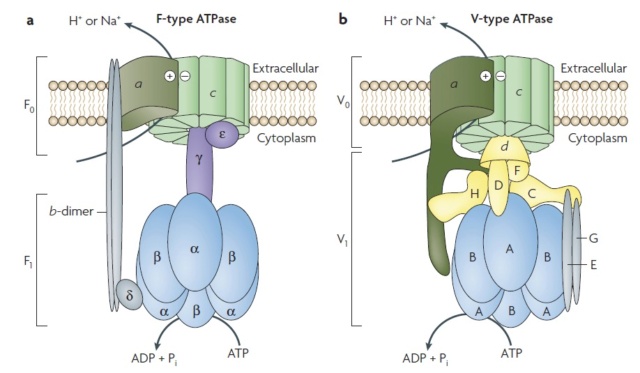
Structure of the F‑ and V‑type membrane ATPases.
Orthologous subunits are shown in the same colour and shape, and unrelated but functionally analogous subunits of the central stalk are indicated by different colours and shapes. The a-subunits, which show structural analogy but might not be homologous, are indicated by similar shapes and colours. The minimal, prokaryotic sets of subunits of the F‑ and V‑type ATPases are shown. The number of peripheral stalks in V‑type ATPases is uncertain
Structural Biochemistry/The Evolution of Membranes
F- and A/V- type ATPases are membrane-embedded proteins and were feasibly present in the LUCA (last universal common ancestor) due to their omnipresence in modern cellular life. 11
https://en.wikibooks.org/wiki/Structural_Biochemistry/The_Evolution_of_Membranes
Evolution of the F0F1 ATP Synthase Complex in Light of the Patchy Distribution of Different Bioenergetic Pathways across Prokaryotes
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4154653/ 2014 Sep 4
This highlights the patchy distribution of many pathways across different lineages, and suggests either up to 26 independent origins or 17 horizontal gene transfer events.
Comment: If one origing by unguided random events is extremely unlikely, imagine 26 independent origins !! The clearly better explanation is that an intelligent designer created these molecular turbines from the get go, fully developed, and working, and distributed them in the various life forms.
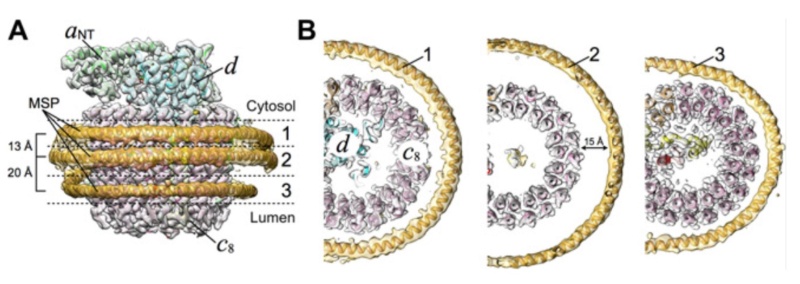
(A) Focused classification resolves three distinct belts of density that surround the membrane-exposed surface of the c-ring. The three belts were modeled as one canonical dimer and one monomer of MSP. (B) The three belts show varying distances to the c-ring. Whereas the middle belt leaves sufficient space to accommodate a layer of lipid molecules, the upper and lower belts appear to directly contact and stabilize the c-ring, presumably via hydrophobic residues of MSP that are oriented toward the c subunits.
1. https://advances.sciencemag.org/content/6/41/eabb9605
2. https://en.wikipedia.org/wiki/V-ATPase
3. https://link.springer.com/article/10.1007/BF00762534
4. https://sci-hub.st/https://www.nature.com/articles/nrmicro1767
5. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4595830/
6. https://phys.org/news/2020-10-molecular-ferris-wheel-protons-cellular.html
7. https://sci-hub.st/https://www.sciencedirect.com/science/article/pii/S1097276518301047
https://reasonandscience.catsboard.com/t3045-molecular-ferris-wheel-delivers-protons-to-cellular-factories
All cells with nuclei, from yeast to humans, are organized like cities, with a variety of small compartments—organelles—that serve as factories where various types of work are done. Some of those factories, like the ones that break down and recycle molecules, need to continually pump in protons—hydrogen atoms with their electrons stripped off—to maintain the acidic environment they need to do their job. For this they rely on molecular Ferris wheels.
Embedded in the organelle's fatty outer membrane, these microscopic machines have rotors that spin 100 times per second, picking up protons from outside the organelle and dropping them off on the inside.
Now scientists have figured out a key step in how these Ferris wheels work in a yeast proton pump known as vacuolar ATPase (V-ATPase). There are 10 amino acid "seats" on the yeast Ferris wheel that bind protons and carry them through the membrane to the organelle's interior, as well as other amino acids that catch them when they arrive. 6
V-ATPase proton pump functions
"The V-ATPase proton pumps perform a wide range of functions, from helping transmit nerve signals to helping specialized cells secrete acid for maintaining bone. Malfunctions in these molecular machines contribute to diseases such as osteoporosis, neurodegeneration, diabetes, cancer and AIDS, so understanding them is important for human health.
Rotary vacuolar adenosine triphosphatases (V-ATPases) drive transmembrane proton transport through a Vo proton channel subcomplex. 1 Active proton transport is central to controlling the balance between cellular pH and efficiency of energy metabolism. Vacuolar adenosine triphosphatases (V-ATPases) are adenosine 5′-triphosphate (ATP) hydrolysis–driven rotary motor proton pumps that acidify the lumen of subcellular organelles in virtually every eukaryotic cell . During ATP hydrolysis–driven proton pumping in holo V-ATPase, the c-ring rotates clockwise to carry protons from the cytosolic to the luminal half-channel. Upon regulated detachment of V1-ATPase from Vo, any existing proton gradient is expected to push the c-ring in the opposite (counterclockwise) direction to allow for the reverse flow of protons.
Vacuolar-type ATPase (V-ATPase) is a highly conserved ancient enzyme with remarkably diverse functions in eukaryotic organisms. V-ATPases acidify a wide array of intracellular organelles and pump protons across the plasma membranes of numerous cell types. V-ATPases couple the energy of ATP hydrolysis to proton transport across intracellular and plasma membranes of eukaryotic cells. It is generally seen as the polar opposite of ATP synthase because ATP synthase is a proton channel that uses the energy from a proton gradient to produce ATP. V-ATPase however, is a proton pump that uses the energy from ATP hydrolysis to produce a proton gradient. 2
V-ATPases are found within the membranes of many organelles, such as endosomes, lysosomes, and secretory vesicles, where they play a variety of roles crucial for the function of these organelles. For example, the proton gradient across the yeast vacuolar membrane generated by V-ATPases drives calcium uptake into the vacuole through an H+/Ca2+ antiporter system. In synaptic transmission in neuronal cells, V-ATPase acidifies synaptic vesicles.
V-ATPases (H+ ATPases) are multisubunit, ATP-dependent proton pumps that regulate pH homeostasis in virtually all eukaryotes. They are involved in key cell biological processes including vesicle trafficking, endosomal pH sensing, membrane fusion and intracellular signaling. They also have critical systemic roles in renal acid excretion and blood pH balance, male fertility, bone remodeling, synaptic transmission, olfaction and hearing.5 While the eukaryotic V-ATPase is primarily known as a proton-pumping, rotary nano-motor that is involved in many physiological processes, it also plays an important role in: i) vesicle coat formation1 and ii) regulation of signaling and trafficking of various receptors. The V-ATPase was also identified as a sensing and signaling complex that functions in the endocytic pathway and it has a crucial role in SNARE-dependent fission/fusion process and organelle biogenesis along the exocytic pathway
No human cell can function without proton pumps, which among other things help organelles intercept viruses and other pathogens and divert them to cellular trash bins.
Origin of structure and function of V-ATPases
Claim: Proton pumping ATPases/ATPsynthases are found in all groups of present-day organisms. The structure of V- and F-type ATPases/ATP synthases is very conserved. Sequence analysis shows that the V- and F-type ATPases evolved from the same enzyme already present in the last common ancestor of all known extant life forms. The catalytic and noncatalytic subunits found in the dissociable head groups of the V/F-type ATPases are paralogous subunits, i.e., these two types of subunits evolved from a common ancestral gene. (Homologous sequences are paralogous if they were separated by a gene duplication event: if a gene in an organism is duplicated to occupy two different positions in the same genome, then the two copies are paralogous. ) The gene duplication giving rise to these two genes (i.e., encoding the catalytic and noncatalytic subunits) predates the time of the last common ancestor. https://link.springer.com/article/10.1007/BF00762534
Mapping of gene duplication events that occurred in the evolution of the proteolipid, the noncatalytic and the catalytic subunits, onto the tree of life leads to a prediction for the likely subunit structure of the encoded ATPases. A correlation between structure and function of V/F-ATPases has been established for present-day organisms. Implications resulting from this correlation for the bioenergetics operative in proto-eukaryotes and in the last common ancestor are presented. The similarities of the V/F-ATPase subunits to an ATPase-like protein that was implicated to play a role in flagellar assembly are evaluated.
Reply: Most subunits of the V-type and the F-type ATPases are not homologous.

https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4154653/
Inventing the dynamo machine: the evolution of the F‑type and V‑type ATPases 4
Sequence and structural comparisons of F- and V-type ATPases have revealed homology between their catalytic and membrane subunits, but no homology between the subunits of the central stalk that connects the catalytic and membrane components.

Structure of the F‑ and V‑type membrane ATPases.
Orthologous subunits are shown in the same colour and shape, and unrelated but functionally analogous subunits of the central stalk are indicated by different colours and shapes. The a-subunits, which show structural analogy but might not be homologous, are indicated by similar shapes and colours. The minimal, prokaryotic sets of subunits of the F‑ and V‑type ATPases are shown. The number of peripheral stalks in V‑type ATPases is uncertain
Structural Biochemistry/The Evolution of Membranes
F- and A/V- type ATPases are membrane-embedded proteins and were feasibly present in the LUCA (last universal common ancestor) due to their omnipresence in modern cellular life. 11
https://en.wikibooks.org/wiki/Structural_Biochemistry/The_Evolution_of_Membranes
Evolution of the F0F1 ATP Synthase Complex in Light of the Patchy Distribution of Different Bioenergetic Pathways across Prokaryotes
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4154653/ 2014 Sep 4
This highlights the patchy distribution of many pathways across different lineages, and suggests either up to 26 independent origins or 17 horizontal gene transfer events.
Comment: If one origing by unguided random events is extremely unlikely, imagine 26 independent origins !! The clearly better explanation is that an intelligent designer created these molecular turbines from the get go, fully developed, and working, and distributed them in the various life forms.

(A) Focused classification resolves three distinct belts of density that surround the membrane-exposed surface of the c-ring. The three belts were modeled as one canonical dimer and one monomer of MSP. (B) The three belts show varying distances to the c-ring. Whereas the middle belt leaves sufficient space to accommodate a layer of lipid molecules, the upper and lower belts appear to directly contact and stabilize the c-ring, presumably via hydrophobic residues of MSP that are oriented toward the c subunits.
1. https://advances.sciencemag.org/content/6/41/eabb9605
2. https://en.wikipedia.org/wiki/V-ATPase
3. https://link.springer.com/article/10.1007/BF00762534
4. https://sci-hub.st/https://www.nature.com/articles/nrmicro1767
5. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4595830/
6. https://phys.org/news/2020-10-molecular-ferris-wheel-protons-cellular.html
7. https://sci-hub.st/https://www.sciencedirect.com/science/article/pii/S1097276518301047
Last edited by Admin on Sat Oct 10, 2020 9:12 am; edited 7 times in total