How messenger RNA's find their way to their destination, the Ribosome: Evidence of an intelligently designed set up
https://reasonandscience.catsboard.com/t3042-how-messenger-rna-s-find-their-way-to-their-destination-the-ribosome-evidence-of-an-intelligently-designed-set-up
Some mRNAs Are Localized to Specific Regions of the Cytosol
Once a newly made eukaryotic mRNA molecule has passed through a nuclear pore and entered the cytosol, it is typically met by ribosomes, which translate it into a polypeptide chain. Once the first round of translation “passes” the nonsense-mediated decay test, the mRNA is usually translated in earnest. If the mRNA encodes a protein that is destined to be secreted or expressed on the cell surface, a signal sequence at the protein’s N-terminus will direct it to the endoplasmic reticulum (ER). In this case, components of the cell’s protein-sorting apparatus recognize the signal sequence as soon as it emerges from the ribosome and direct the entire complex of ribosome, mRNA, and nascent protein to the membrane of the ER, where the remainder of the polypeptide chain is synthesized. In other cases, free ribosomes in the cytosol synthesize the entire protein, and signals in the completed polypeptide chain may then direct the protein to other sites in the cell. Many mRNAs are themselves directed to specific intracellular locations before their efficient translation begins, allowing the cell to position its mRNAs close to the sites where the encoded protein is needed. RNA localization has been observed in many organisms, including unicellular fungi, plants, and animals, and it is likely to be a common mechanism that cells use to concentrate high-level production of proteins at specific sites. This strategy also provides the cell with other advantages. For example, it allows the establishment of asymmetries in the cytosol of the cell, a key step in many stages of development. Localized mRNA, coupled with translational control, also allows the cell to regulate gene expression independently in different regions. This feature is particularly important in large, highly polarized cells such as neurons, where it plays a central role in synaptic function.
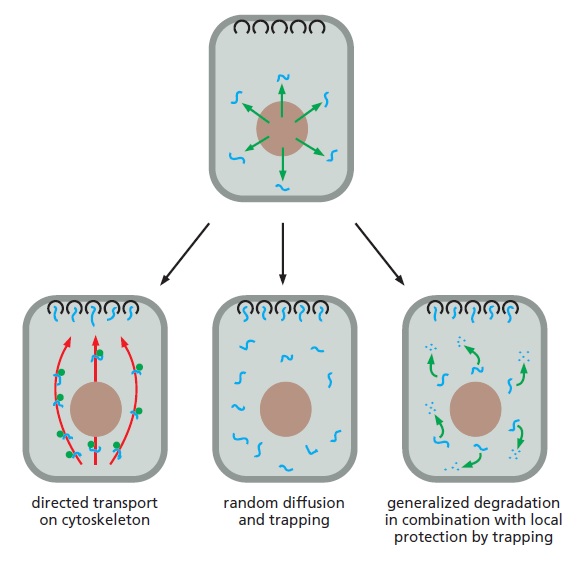

Mechanisms for the localization of mRNAs.
The mRNA to be localized leaves the nucleus through nuclear pores (top). Some localized mRNAs (left diagram) travel to their destination by associating with cytoskeletal motors, which the energy of ATP hydrolysis to move
the mRNAs unidirectionally along filaments in the cytoskeleton (red). At their destination, the mRNAs are held in place by anchor proteins (black). Other mRNAs randomly diffuse through the cytosol and are simply trapped by anchor proteins and at their sites of localization (center diagram). Some mRNAs (right diagram) are degraded in the cytosol unless they have bound, through random diffusion, a localized protein complex that anchors and protects the mRNA from degradation (black). Each mechanism requires specific signals on the mRNA, which are typically located in the 3ʹ UTR. Additional components can block the translation of the mRNA until it is properly localized.
My comment: This is absolutely fascinating. In order for messenger RNA's to get to their destination, where they can be translated, a lot of sophisticated technology is required. mRNA's need to be directed to the Ribosome. Cytoskeletal motors are like exquisitely designed molecular taxis which literally walk like humans with two "legs". While our taxis use gasoline fuel, these motor proteins ( dynein, kinesin ) use ATP. They walk and move on literal superb molecular highways, cytoskeletons, and at their destination, the mRNAs are held in place and trapped by anchor proteins and protected from degrading. Specific signals direct mRNA's to their destination. This is like an awesome GPS on a molecular level. We have here at least five different individual parts that have to work in a joint venture together to provide the required trajectory from the RNA polymerase machine which transcribes DNA to mRNA, to the Ribosome. Nobody would in its sane mind imagine that a human made GPS system could be the product of evolution. But when it comes to what goes in in biochemistry, why should someone suddenly conclude that unguided random events can produce a fully robot-like, fully programmed process like illustrated here?
1. The cell has sophisticated cargo loading, transport, and delivery systems. It is not only analogous to a man-made transport system but operates literally as such.
2. Cargo loading, transport, and delivery require specialists that operate and execute the delivery based on advanced planning, logistic software systems, GPS Positioning Systems and information management, monitoring, and control systems.
3. Once proteins are synthesized, they are tagged through the signal transduction particle ( which is life-essential), carried on molecular cargo carriers ( dynein, and kinesin motor proteins ) on molecular highways ( tubulins), and delivered at the destination where the protein cargo will be employed for operation.
4. The programming of all those steps requires with high probability of an intelligent programmer ( God).
https://www.youtube.com/watch?v=wJyUtbn0O5Y
Several mechanisms for mRNA localization have been discovered all of which require specific signals in the mRNA itself. These signals are usually concentrated in the 3ʹ untranslated region (UTR), the region of RNA that extends from the stop codon that terminates protein synthesis to the start of the poly-A tail
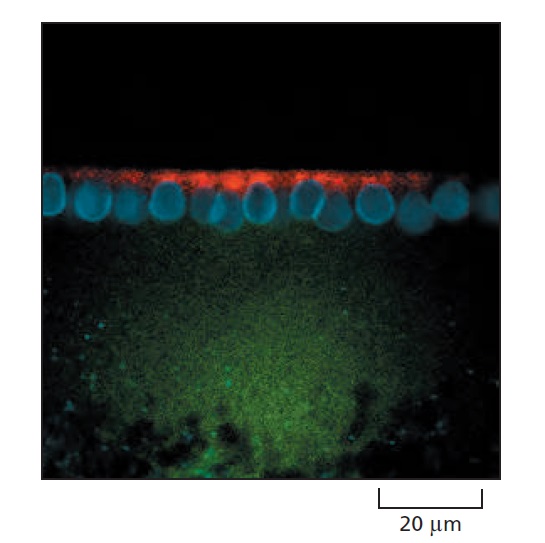
An experiment demonstrating the importance of the 3ʹ UTR in localizing mRNAs to specific regions of the cytoplasm.
For this experiment, two different fluorescently labeled RNAs were prepared by transcribing DNA in vitro in the presence of fluorescently labeled derivatives of UTP. One RNA (labeled with a red fluorochrome) contains the coding region for the Drosophila Hairy protein and includes the adjacent 3ʹ UTR. The other RNA (labeled green) contains the Hairy coding region with the 3ʹ UTR deleted. The two RNAs were mixed and injected into a Drosophila embryo at a stage of development when multiple nuclei reside in a common cytoplasm. When the fluorescent RNAs were visualized 10 minutes later, the full-length hairy RNA (red) was localized to the apical side of nuclei (blue) but the transcript missing the 3ʹ UTR (green) failed to localize. Hairy is one of many transcriptional regulators that specify positional information in the developing Drosophila embryo, and the localization of its mRNA (shown in this experiment to depend on its 3ʹ UTR) is critical for proper fly development.
The mRNA localization is usually coupled with translational controls to ensure that the mRNA remains quiescent until it has been moved into place. The Drosophila egg exhibits an especially striking example of mRNA localization. The mRNA encoding the Bicoid transcription regulator is localized by attachment to the cytoskeleton at the anterior tip of the developing egg. When fertilization triggers the translation of this mRNA, it generates a gradient of the Bicoid protein that plays a crucial part in directing the development of the anterior part of the embryo. Many mRNAs in somatic cells are also localized in a similar way. The mRNA that encodes actin, for example, is localized to the actin-filament-rich cell cortex in mammalian fibroblasts by means of a 3ʹ UTR signal. mRNA molecules exit from the nucleus bearing numerous markings in the form of RNA modifications (the 5ʹ cap and the 3ʹ poly-A tail) and bound proteins (exon-junction complexes, for example) that signify the successful completion of the different pre-mRNA processing steps. As just described, the 3ʹ UTR of an mRNA can be thought of as a “zip code” that directs mRNAs to different places in the cell. mRNAs carry information specifying their average lifetime in the cytosol and the efficiency with which they are translated into protein. In a broad sense, the untranslated regions of eukaryotic mRNAs resemble the transcriptional control regions of genes: their nucleotide sequences contain information specifying the way the RNA is to be used, and proteins interpret this information by binding specifically to these sequences. Thus, over and above the specification of the amino acid sequences of proteins, mRNA molecules are rich with information.
The 5ʹ and 3ʹ Untranslated Regions of mRNAs Control Their Translation
Once an mRNA has been synthesized, one of the most common ways of regulating the levels of its protein product is to control the step that initiates translation. Even though the details of translation initiation differ between eukaryotes and bacteria, they each use some of the same basic regulatory strategies. In bacterial mRNAs, a conserved stretch of nucleotides, the Shine–Dalgarno sequence, is always found a few nucleotides upstream of the initiating AUG codon. In bacteria, translational control mechanisms are carried out by proteins or by RNA molecules, and they generally involve either exposing or blocking the Shine– Dalgarno sequence

Mechanisms of translational control.
Although these examples are from bacteria, many of the same principles operate in eukaryotes.
(A) Sequence-specific RNA-binding proteins repress translation of specific mRNAs by blocking access of the ribosome to the Shine–Dalgarno sequence (orange). For example, some ribosomal proteins repress translation of their own RNA. This mechanism allows the cell to maintain correctly balanced quantities of the various components needed to form ribosomes.
(B) An RNA “thermosensor” permits efficient translation initiation only at elevated temperatures at which the stem-loop structure has been melted. An example occurs in the human pathogen Listeria monocytogenes, in which the translation of its virulence genes increases at 37°C, the temperature of the host.
(C) Binding of a small molecule to a riboswitch causes a major rearrangement of the RNA forming a different set of stem-loop structures. In the bound structure, the Shine–Dalgarno sequence (orange) is sequestered and translation initiation is thereby blocked. In many bacteria, S-adenosylmethionine acts in this manner to block production of the enzymes that synthesize it.
(D) An “antisense” RNA produced elsewhere from the genome base-pairs with a specific mRNA and blocks its translation. Many bacteria regulate expression of iron-storage proteins in this way.
Eukaryotic mRNAs do not contain such a sequence. Instead, the selection of an AUG codon as a translation start site is largely determined by its proximity to the cap at the 5ʹ end of the mRNA molecule, which is the site at which the small ribosomal subunit binds to the mRNA and begins scanning for an initiating AUG codon. In eukaryotes, translational repressors can bind to the 5ʹ end of the mRNA and thereby inhibit translation initiation. Other repressors recognize nucleotide sequences in the 3ʹ UTR of specific mRNAs and decrease translation initiation by interfering with the communication between the 5ʹ cap and 3ʹ poly-A tail, a step required for efficient translation . A particularly important type of translational control in eukaryotes relies on small RNAs (termed microRNAs or miRNAs) that bind to mRNAs and reduce protein output.
The Phosphorylation of an Initiation Factor Regulates Protein Synthesis Globally
Eukaryotic cells decrease their overall rate of protein synthesis in response to a variety of situations, including deprivation of growth factors or nutrients, infection by viruses, and sudden increases in temperature. Much of this decrease is caused by the phosphorylation of the translation initiation factor eIF2 by specific protein kinases that respond to the changes in conditions. It forms a complex with GTP and mediates the binding of the methionyl initiator tRNA to the small ribosomal subunit, which then binds to the 5ʹ end of the mRNA and begins scanning along the mRNA. When an AUG codon is recognized, the eIF2 protein hydrolyzes the bound GTP to GDP, causing a conformational change in the protein and releasing it from the small ribosomal subunit. The large ribosomal subunit then joins the small one to form a complete ribosome that begins protein synthesis.
Because eIF2 binds very tightly to GDP, a guanine nucleotide exchange factor, designated eIF2B, is required to cause GDP release so that a new GTP molecule can bind and eIF2 can be reused (Figure 7–67A). The reuse of eIF2
is inhibited when it is phosphorylated—the phosphorylated eIF2 binds to eIF2B unusually tightly, inactivating eIF2B. There is more eIF2 than eIF2B in cells, and even a fraction of phosphorylated eIF2 can trap nearly all of the eIF2B. This prevents the reuse of the nonphosphorylated eIF2 and greatly slows protein synthesis (Figure B).
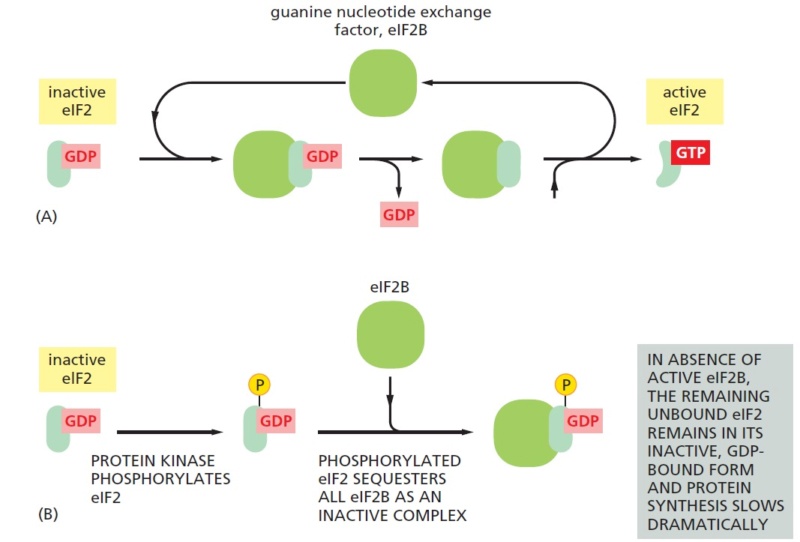
The eIF2 cycle.
(A) The recycling of used eIF2 by a guanine nucleotide exchange factor (eIF2B).
(B) eIF2 phosphorylation controls protein synthesis rates by tying up eIF2B.
Regulation of the level of active eIF2 is especially important in mammalian cells; eIF2 is part of the mechanism that allows cells to enter a nonproliferating, resting state (called G0) in which the rate of total protein synthesis is reduced to about one-fifth the rate in proliferating cells.
Initiation at AUG Codons Upstream of the Translation Start Can Regulate Eukaryotic Translation Initiation
We saw in Chapter 6 that eukaryotic translation typically begins at the first AUG downstream of the 5ʹ end of the mRNA, which is the first AUG encountered by a scanning small ribosomal subunit. But the nucleotides immediately surrounding the AUG also influence the efficiency of translation initiation. If the recognition site is poor enough, scanning ribosomal subunits will sometimes ignore the first AUG codon in the mRNA and skip to the second or third AUG codon instead. This phenomenon, known as “leaky scanning,” is a strategy frequently used to produce two or more closely related proteins, differing only in their N-termini, from the same mRNA. A particularly important use of this mechanism is the production of the same protein with and without a signal sequence attached at its N-terminus. This allows the protein to be directed to two different locations in the cell (for example, to both mitochondria and the cytosol). Cells can regulate the relative abundance of the protein isoforms produced by leaky scanning; for example, a cell-type-specific increase in the abundance of the initiation factor eIF4F favors the use of the AUG closest to the 5ʹ end of the mRNA. Another type of control found in eukaryotes uses one or more short open reading frames—short stretches of DNA that begin with a start codon (ATG) and end with a stop codon, with no stop codons in between—that lie between the 5ʹ end of the mRNA and the beginning of the gene. Often, the amino acid sequences coded by these upstream open reading frames (uORFs) are not important; rather, the uORFs serve a purely regulatory function. An uORF present on an mRNA molecule will generally decrease translation of the downstream gene by trapping a scanning ribosome initiation complex and causing the ribosome to translate the uORF and dissociate from the mRNA before it reaches the bona fide protein-coding sequence. When the activity of a general translation factor (such as the eIF2 discussed above) is reduced, one might expect that the translation of all mRNAs would be reduced equally. Contrary to this expectation, however, the phosphorylation of eIF2 can have selective effects, even enhancing the translation of specific mRNAs that contain uORFs. This can enable cells, for example, to adapt to starvation for specific nutrients by shutting down the synthesis of all proteins except those that are required for synthesis of the missing nutrients. The details of this mechanism have been worked out for a specific yeast mRNA that encodes a protein called Gcn4, a transcription regulator that activates many genes that encode proteins that are important for amino acid synthesis. The Gcn4 mRNA contains several short uORFs, and when amino acids are abundant, ribosomes translate the uORFs and generally dissociate before they reach the Gcn4 coding region. A global decrease in eIF2 activity brought about by amino acid starvation makes it more likely that a scanning small ribosomal subunit will move across the uORFs (without translating them) before it acquires a molecule of eIF2. Such a ribosomal subunit is then free to initiate translation on the actual Gcn4 sequences. The increased level of this transcription regulator increases production of the amino acid biosynthetic enzymes.
https://reasonandscience.catsboard.com/t3042-how-messenger-rna-s-find-their-way-to-their-destination-the-ribosome-evidence-of-an-intelligently-designed-set-up
Some mRNAs Are Localized to Specific Regions of the Cytosol
Once a newly made eukaryotic mRNA molecule has passed through a nuclear pore and entered the cytosol, it is typically met by ribosomes, which translate it into a polypeptide chain. Once the first round of translation “passes” the nonsense-mediated decay test, the mRNA is usually translated in earnest. If the mRNA encodes a protein that is destined to be secreted or expressed on the cell surface, a signal sequence at the protein’s N-terminus will direct it to the endoplasmic reticulum (ER). In this case, components of the cell’s protein-sorting apparatus recognize the signal sequence as soon as it emerges from the ribosome and direct the entire complex of ribosome, mRNA, and nascent protein to the membrane of the ER, where the remainder of the polypeptide chain is synthesized. In other cases, free ribosomes in the cytosol synthesize the entire protein, and signals in the completed polypeptide chain may then direct the protein to other sites in the cell. Many mRNAs are themselves directed to specific intracellular locations before their efficient translation begins, allowing the cell to position its mRNAs close to the sites where the encoded protein is needed. RNA localization has been observed in many organisms, including unicellular fungi, plants, and animals, and it is likely to be a common mechanism that cells use to concentrate high-level production of proteins at specific sites. This strategy also provides the cell with other advantages. For example, it allows the establishment of asymmetries in the cytosol of the cell, a key step in many stages of development. Localized mRNA, coupled with translational control, also allows the cell to regulate gene expression independently in different regions. This feature is particularly important in large, highly polarized cells such as neurons, where it plays a central role in synaptic function.


Mechanisms for the localization of mRNAs.
The mRNA to be localized leaves the nucleus through nuclear pores (top). Some localized mRNAs (left diagram) travel to their destination by associating with cytoskeletal motors, which the energy of ATP hydrolysis to move
the mRNAs unidirectionally along filaments in the cytoskeleton (red). At their destination, the mRNAs are held in place by anchor proteins (black). Other mRNAs randomly diffuse through the cytosol and are simply trapped by anchor proteins and at their sites of localization (center diagram). Some mRNAs (right diagram) are degraded in the cytosol unless they have bound, through random diffusion, a localized protein complex that anchors and protects the mRNA from degradation (black). Each mechanism requires specific signals on the mRNA, which are typically located in the 3ʹ UTR. Additional components can block the translation of the mRNA until it is properly localized.
My comment: This is absolutely fascinating. In order for messenger RNA's to get to their destination, where they can be translated, a lot of sophisticated technology is required. mRNA's need to be directed to the Ribosome. Cytoskeletal motors are like exquisitely designed molecular taxis which literally walk like humans with two "legs". While our taxis use gasoline fuel, these motor proteins ( dynein, kinesin ) use ATP. They walk and move on literal superb molecular highways, cytoskeletons, and at their destination, the mRNAs are held in place and trapped by anchor proteins and protected from degrading. Specific signals direct mRNA's to their destination. This is like an awesome GPS on a molecular level. We have here at least five different individual parts that have to work in a joint venture together to provide the required trajectory from the RNA polymerase machine which transcribes DNA to mRNA, to the Ribosome. Nobody would in its sane mind imagine that a human made GPS system could be the product of evolution. But when it comes to what goes in in biochemistry, why should someone suddenly conclude that unguided random events can produce a fully robot-like, fully programmed process like illustrated here?
1. The cell has sophisticated cargo loading, transport, and delivery systems. It is not only analogous to a man-made transport system but operates literally as such.
2. Cargo loading, transport, and delivery require specialists that operate and execute the delivery based on advanced planning, logistic software systems, GPS Positioning Systems and information management, monitoring, and control systems.
3. Once proteins are synthesized, they are tagged through the signal transduction particle ( which is life-essential), carried on molecular cargo carriers ( dynein, and kinesin motor proteins ) on molecular highways ( tubulins), and delivered at the destination where the protein cargo will be employed for operation.
4. The programming of all those steps requires with high probability of an intelligent programmer ( God).
https://www.youtube.com/watch?v=wJyUtbn0O5Y
Several mechanisms for mRNA localization have been discovered all of which require specific signals in the mRNA itself. These signals are usually concentrated in the 3ʹ untranslated region (UTR), the region of RNA that extends from the stop codon that terminates protein synthesis to the start of the poly-A tail

An experiment demonstrating the importance of the 3ʹ UTR in localizing mRNAs to specific regions of the cytoplasm.
For this experiment, two different fluorescently labeled RNAs were prepared by transcribing DNA in vitro in the presence of fluorescently labeled derivatives of UTP. One RNA (labeled with a red fluorochrome) contains the coding region for the Drosophila Hairy protein and includes the adjacent 3ʹ UTR. The other RNA (labeled green) contains the Hairy coding region with the 3ʹ UTR deleted. The two RNAs were mixed and injected into a Drosophila embryo at a stage of development when multiple nuclei reside in a common cytoplasm. When the fluorescent RNAs were visualized 10 minutes later, the full-length hairy RNA (red) was localized to the apical side of nuclei (blue) but the transcript missing the 3ʹ UTR (green) failed to localize. Hairy is one of many transcriptional regulators that specify positional information in the developing Drosophila embryo, and the localization of its mRNA (shown in this experiment to depend on its 3ʹ UTR) is critical for proper fly development.
The mRNA localization is usually coupled with translational controls to ensure that the mRNA remains quiescent until it has been moved into place. The Drosophila egg exhibits an especially striking example of mRNA localization. The mRNA encoding the Bicoid transcription regulator is localized by attachment to the cytoskeleton at the anterior tip of the developing egg. When fertilization triggers the translation of this mRNA, it generates a gradient of the Bicoid protein that plays a crucial part in directing the development of the anterior part of the embryo. Many mRNAs in somatic cells are also localized in a similar way. The mRNA that encodes actin, for example, is localized to the actin-filament-rich cell cortex in mammalian fibroblasts by means of a 3ʹ UTR signal. mRNA molecules exit from the nucleus bearing numerous markings in the form of RNA modifications (the 5ʹ cap and the 3ʹ poly-A tail) and bound proteins (exon-junction complexes, for example) that signify the successful completion of the different pre-mRNA processing steps. As just described, the 3ʹ UTR of an mRNA can be thought of as a “zip code” that directs mRNAs to different places in the cell. mRNAs carry information specifying their average lifetime in the cytosol and the efficiency with which they are translated into protein. In a broad sense, the untranslated regions of eukaryotic mRNAs resemble the transcriptional control regions of genes: their nucleotide sequences contain information specifying the way the RNA is to be used, and proteins interpret this information by binding specifically to these sequences. Thus, over and above the specification of the amino acid sequences of proteins, mRNA molecules are rich with information.
The 5ʹ and 3ʹ Untranslated Regions of mRNAs Control Their Translation
Once an mRNA has been synthesized, one of the most common ways of regulating the levels of its protein product is to control the step that initiates translation. Even though the details of translation initiation differ between eukaryotes and bacteria, they each use some of the same basic regulatory strategies. In bacterial mRNAs, a conserved stretch of nucleotides, the Shine–Dalgarno sequence, is always found a few nucleotides upstream of the initiating AUG codon. In bacteria, translational control mechanisms are carried out by proteins or by RNA molecules, and they generally involve either exposing or blocking the Shine– Dalgarno sequence

Mechanisms of translational control.
Although these examples are from bacteria, many of the same principles operate in eukaryotes.
(A) Sequence-specific RNA-binding proteins repress translation of specific mRNAs by blocking access of the ribosome to the Shine–Dalgarno sequence (orange). For example, some ribosomal proteins repress translation of their own RNA. This mechanism allows the cell to maintain correctly balanced quantities of the various components needed to form ribosomes.
(B) An RNA “thermosensor” permits efficient translation initiation only at elevated temperatures at which the stem-loop structure has been melted. An example occurs in the human pathogen Listeria monocytogenes, in which the translation of its virulence genes increases at 37°C, the temperature of the host.
(C) Binding of a small molecule to a riboswitch causes a major rearrangement of the RNA forming a different set of stem-loop structures. In the bound structure, the Shine–Dalgarno sequence (orange) is sequestered and translation initiation is thereby blocked. In many bacteria, S-adenosylmethionine acts in this manner to block production of the enzymes that synthesize it.
(D) An “antisense” RNA produced elsewhere from the genome base-pairs with a specific mRNA and blocks its translation. Many bacteria regulate expression of iron-storage proteins in this way.
Eukaryotic mRNAs do not contain such a sequence. Instead, the selection of an AUG codon as a translation start site is largely determined by its proximity to the cap at the 5ʹ end of the mRNA molecule, which is the site at which the small ribosomal subunit binds to the mRNA and begins scanning for an initiating AUG codon. In eukaryotes, translational repressors can bind to the 5ʹ end of the mRNA and thereby inhibit translation initiation. Other repressors recognize nucleotide sequences in the 3ʹ UTR of specific mRNAs and decrease translation initiation by interfering with the communication between the 5ʹ cap and 3ʹ poly-A tail, a step required for efficient translation . A particularly important type of translational control in eukaryotes relies on small RNAs (termed microRNAs or miRNAs) that bind to mRNAs and reduce protein output.
The Phosphorylation of an Initiation Factor Regulates Protein Synthesis Globally
Eukaryotic cells decrease their overall rate of protein synthesis in response to a variety of situations, including deprivation of growth factors or nutrients, infection by viruses, and sudden increases in temperature. Much of this decrease is caused by the phosphorylation of the translation initiation factor eIF2 by specific protein kinases that respond to the changes in conditions. It forms a complex with GTP and mediates the binding of the methionyl initiator tRNA to the small ribosomal subunit, which then binds to the 5ʹ end of the mRNA and begins scanning along the mRNA. When an AUG codon is recognized, the eIF2 protein hydrolyzes the bound GTP to GDP, causing a conformational change in the protein and releasing it from the small ribosomal subunit. The large ribosomal subunit then joins the small one to form a complete ribosome that begins protein synthesis.
Because eIF2 binds very tightly to GDP, a guanine nucleotide exchange factor, designated eIF2B, is required to cause GDP release so that a new GTP molecule can bind and eIF2 can be reused (Figure 7–67A). The reuse of eIF2
is inhibited when it is phosphorylated—the phosphorylated eIF2 binds to eIF2B unusually tightly, inactivating eIF2B. There is more eIF2 than eIF2B in cells, and even a fraction of phosphorylated eIF2 can trap nearly all of the eIF2B. This prevents the reuse of the nonphosphorylated eIF2 and greatly slows protein synthesis (Figure B).

The eIF2 cycle.
(A) The recycling of used eIF2 by a guanine nucleotide exchange factor (eIF2B).
(B) eIF2 phosphorylation controls protein synthesis rates by tying up eIF2B.
Regulation of the level of active eIF2 is especially important in mammalian cells; eIF2 is part of the mechanism that allows cells to enter a nonproliferating, resting state (called G0) in which the rate of total protein synthesis is reduced to about one-fifth the rate in proliferating cells.
Initiation at AUG Codons Upstream of the Translation Start Can Regulate Eukaryotic Translation Initiation
We saw in Chapter 6 that eukaryotic translation typically begins at the first AUG downstream of the 5ʹ end of the mRNA, which is the first AUG encountered by a scanning small ribosomal subunit. But the nucleotides immediately surrounding the AUG also influence the efficiency of translation initiation. If the recognition site is poor enough, scanning ribosomal subunits will sometimes ignore the first AUG codon in the mRNA and skip to the second or third AUG codon instead. This phenomenon, known as “leaky scanning,” is a strategy frequently used to produce two or more closely related proteins, differing only in their N-termini, from the same mRNA. A particularly important use of this mechanism is the production of the same protein with and without a signal sequence attached at its N-terminus. This allows the protein to be directed to two different locations in the cell (for example, to both mitochondria and the cytosol). Cells can regulate the relative abundance of the protein isoforms produced by leaky scanning; for example, a cell-type-specific increase in the abundance of the initiation factor eIF4F favors the use of the AUG closest to the 5ʹ end of the mRNA. Another type of control found in eukaryotes uses one or more short open reading frames—short stretches of DNA that begin with a start codon (ATG) and end with a stop codon, with no stop codons in between—that lie between the 5ʹ end of the mRNA and the beginning of the gene. Often, the amino acid sequences coded by these upstream open reading frames (uORFs) are not important; rather, the uORFs serve a purely regulatory function. An uORF present on an mRNA molecule will generally decrease translation of the downstream gene by trapping a scanning ribosome initiation complex and causing the ribosome to translate the uORF and dissociate from the mRNA before it reaches the bona fide protein-coding sequence. When the activity of a general translation factor (such as the eIF2 discussed above) is reduced, one might expect that the translation of all mRNAs would be reduced equally. Contrary to this expectation, however, the phosphorylation of eIF2 can have selective effects, even enhancing the translation of specific mRNAs that contain uORFs. This can enable cells, for example, to adapt to starvation for specific nutrients by shutting down the synthesis of all proteins except those that are required for synthesis of the missing nutrients. The details of this mechanism have been worked out for a specific yeast mRNA that encodes a protein called Gcn4, a transcription regulator that activates many genes that encode proteins that are important for amino acid synthesis. The Gcn4 mRNA contains several short uORFs, and when amino acids are abundant, ribosomes translate the uORFs and generally dissociate before they reach the Gcn4 coding region. A global decrease in eIF2 activity brought about by amino acid starvation makes it more likely that a scanning small ribosomal subunit will move across the uORFs (without translating them) before it acquires a molecule of eIF2. Such a ribosomal subunit is then free to initiate translation on the actual Gcn4 sequences. The increased level of this transcription regulator increases production of the amino acid biosynthetic enzymes.

