https://reasonandscience.catsboard.com/t2984-error-check-and-repair-during-messengerrna-translation-in-the-ribosome-by-chance-or-design
Translation is the most error-prone step of protein synthesis. 14
The control of the error rate and its effects in biological processes of information transmission is one of the key requirements to have functional living cells.
When talking about error check and repair of translation in the cell, then we have to consider several aspects. First, in order to have a functional and error-prone translation, the components involved in translation must be correctly synthesized in the cell. In translation, messenger RNA, transfer RNA, Aminoacyl-tRNA synthetase, and the Ribosome are involved. Most, if not all, are carefully error checked, either repaired when errors are detected, or discarded when misfolding through proteostasis. 13
Secondly, the process of translation is a multistep process, and error check and repair occur all along the way. They will be listed as follows:
Mistakes during DNA replication are on the order of ~10^8 and are kept to this extremely low level by a robust suite of error prevention, correction and repair mechanisms 12 Protein synthesis, offers the greatest opportunity for errors, with mistranslation events routinely occurring at a frequency of ~1 per 10,000 mRNA codons translated. The ribosome must select the correct aminoacyl-transfer RNAs (aa-tRNAs) from a large pool of near-cognate substrates fast enough to sustain an elongation rate of 10–20 amino acids per second !!.
Proofreading and editing processes are used throughout protein synthesis to ensure a faithful translation of genetic information. The maturation of tRNAs and mRNAs is monitored, as is the identity of amino acids attached to tRNAs. Accuracy is further enhanced during the selection of aminoacyl-tRNAs on the ribosome and their base-pairing with mRNA.
Ribosome biogenesis: quality control mechanisms must be in place to survey nascent ribosomes and ensure their functionality.
Chiral checkpoints during protein biosynthesis the ribosome act as “chiral checkpoints” by preferentially binding to L-amino acids or L-aminoacyl-tRNAs, thereby excluding D-amino acids 11
If misaminoacylated tRNA is successfully delivered to the ribosome, additional proofreading occurs within the A site of the ribosome based on aa-tRNA position and affinity
Ribosomal interactions with additional tRNA-specific sequences and modifications facilitate accurate selection of aa-tRNAs based on kinetic discrimination during the initial selection stage and subsequent proofreading stage.
mRNA translation regulation by nearly 100 epigenetic tRNA modifications. The finer details in this sort of regulation have been proven to differ between prokaryotic and eukaryotic organisms. ( LUCA hello ??!! 10)
tRNA structure monitoring: Export of defective or immature tRNAs is avoided by monitoring both structure and function of tRNAs in the nucleus, and only tRNAs with mature 5′ and 3′ ends are exported. 6
tRNA synthesis quality control: RTD and other mechanisms that degrade hypomodified or mutated mature yeast tRNAs serve as a surveillance system to eliminate tRNA molecules that have incorrect nucleosides or conformations
Aminoacyl-tRNA synthetase error minimization by preferential binding of the right amino acids, and selective editing and proofreading of near cognate amino acids
Aminoacyl-tRNA synthetase Pre-transfer editing: Pre-transfer editing has been described in both class I and class II aaRSs and takes place after aa-AMP synthesis but before the aminoacyl moiety is transferred to the tRNA. 9
Aminoacyl-tRNA synthetase Post-transfer editing: Post-transfer editing takes place after the transfer of the amino acid to the tRNA and involves the hydrolysis of the ester bond, in a domain separated from the active site.
Aminoacyl-tRNA synthetase Editing factors: Another important component of the translation quality control machinery is the trans-editing family, free-standing proteins that are not synthetases but are in some cases homologs to the editing domains of such enzymes. The role of these trans-editing factors is to clear the misacylated tRNA before it reaches the ribosome, acting as additional checkpoints to ensure fidelity.
Aminoacyl-tRNA synthetases (aaRSs), selectively hydrolyze ( chemical reaction in which a molecule of water ruptures one or more chemical bonds ) incorrectly activated non-cognate amino acids and/or misaminoacylated tRNAs. 12
In addition to misactivation of genetically encoded proteinogenic amino acids (GPAs), cells also encounter non-proteinogenic amino acids (NPAs) environmentally or as metabolic by-products, and must discriminate against these substrates to prevent aberrant use in protein synthesis.
This is a list of eleven different error check and repair mechanisms during translation. Consider, that life cannot start, unless these mechanisms are fully in place, and operational. Consider as well, that all this machinery is a pre-requirement for living cells to kick-start life. Their origin cannot be explained by evolution. The alternatives are either all these hypercomplex life essential error check and repair mechanisms emerged by a fortuitous accident, spontaneously through self-organization by unguided stochastic coincidence, natural events that turned into self-organization in an orderly manner without external direction, chemical non-biological, purely physico-dynamic kinetic processes and reactions influenced by environmental parameters, or through the direct intervention, creative force and activity of an intelligent agency, a powerful creator. Which of the two makes more sense?
1. https://www.pnas.org/content/113/12/3311
2. https://sci-hub.tw/https://www.annualreviews.org/doi/10.1146/annurev.micro.091208.073210
2. https://elifesciences.org/articles/54898
3. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2570319/
4. https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1008017
5. https://www.labome.com/method/Aminoacyl-tRNA-Synthetases.html
6. https://sci-hub.tw/https://science.sciencemag.org/content/282/5396/2082?ijkey=ace41d59cbf20b8ca66e9a068342b45398645b9b&keytype2=tf_ipsecsha
7. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3640646/
8. Quality Control Mechanisms During Translation
9. https://rnajournal.cshlp.org/content/early/2020/04/17/rna.071720.119.full.pdf
10. https://en.wikipedia.org/wiki/Translational_regulation
11. https://www.jbc.org/content/early/2019/10/07/jbc.REV119.008166.full.pdf
12. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5697424/
13. https://en.wikipedia.org/wiki/Proteostasis
14. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2764353/
Several mechanisms working together ensure that the tRNA synthetase links the correct amino acid to each tRNA. The synthetase must first select the correct amino acid, and most synthetases do so by a two-step mechanism. First, the correct amino acid has the highest affinity for the active-site pocket of its synthetase and is therefore favored over the other 19. In particular, amino acids larger than the correct one are effectively excluded from the active site. However, accurate discrimination between two similar amino acids, such as isoleucine and valine (which differ by only a methyl group), is very difficult to achieve by a one-step recognition mechanism. A second discrimination step occurs after the amino acid has been covalently linked to AMP. When tRNA binds the synthetase, it tries to force the amino acid into a second pocket in the synthetase, the precise dimensions of which exclude the correct amino acid but allow access by closely related amino acids. Once an amino acid enters this editing pocket, it is hydrolyzed from the AMP (or from the tRNA itself if the aminoacyl-tRNA bond has already formed), and is released from the enzyme. This hydrolytic editing, which is analogous to the exonucleolytic proofreading by DNA polymerases , raises the overall accuracy of tRNA charging to approximately one mistake in 40,000 couplings.
Editing significantly decreases the frequency of errors and is important for translational quality control, and many details of the various editing mechanisms and their effect on different cellular systems are now starting to emerge. 8
High Fidelity
Aminoacyl-tRNA synthetases must perform their tasks with high accuracy. Every mistake they make will result in a misplaced amino acid when new proteins are constructed. These enzymes make about one mistake in 10,000. For most amino acids, this level of accuracy is not too difficult to achieve. Most of the amino acids are quite different from one another, and, as mentioned before, many parts of the different tRNA are used for accurate recognition. But in a few cases, it is difficult to choose just the right amino acids and these enzymes must resort to special techniques.
Isoleucine is a particularly difficult example. It is recognized by an isoleucine-shaped hole in the enzyme, which is too small to fit larger amino acids like methionine and phenylalanine, and too hydrophobic to bind anything with polar sidechains. But, the slightly smaller amino acid valine, different by only a single methyl group, also fits nicely into this pocket, binding instead of isoleucine in about 1 in 150 times. This is far too many errors, so corrective steps must be taken. Isoleucyl-tRNA synthetase (PDB entry 1ffy) solves this problem with a second active site, which performs an editing reaction. Isoleucine does not fit into this site, but errant valine does. The mistake is then cleaved away, leaving the tRNA ready for a properly-placed leucine amino acid. This proofreading step improves the overall error rate to about 1 in 3,000. 9
My comment: This is an amazing error proofreading technique, which adds to other repair mechanisms in the cell. Once again the question arises: How could these precise molecular machines have arisen by natural means, without intelligence involved? This seems to be one more amazing example of highly sophisticated nanomolecular machinery designed to fulfill its task with a high degree of fidelity and error minimization, which can arise only by the foresight of an incredibly intelligent creator.
I. Why wasn’t one ancestor enough when they both do the same job?
II. How did the two types of ancestral synthetases avoid competition that might have eliminated the inferior Class? 12
https://reasonandscience.catsboard.com/t2057-origin-of-translation-of-the-4-nucleic-acid-bases-and-the-20-amino-acids-and-the-universal-assignment-of-codons-to-amino-acids
Errors that occur during transcription and translation have substantial effects on gene function by producing misfolded and malfunctioning proteins. The rate of translation errors is typically an order of magnitude higher than the rate of transcription errors. 1 Errors occurring during transcription often elicit more dire consequences than those occurring during translation because individual mRNAs can be translated up to 40 times resulting in a burst of flawed proteins. Therefore, a single transcription error can result in many flawed proteins, whereas a translation error will disrupt only a single protein. Estimates of the rate of transcription errors in Escherichia coli have yielded variable estimates of transcription error rates of 10−4–10−5 per nucleotide, several orders of magnitude higher than the mutation rate . Transcript-error rates are 3 to 4 orders of magnitude higher than the corresponding genetic mutation rates. The general error rates of genomic replication (about 10−
Gene expression is not only controlled through altering the rate of transcription but also through varying rates of translation and mRNA decay. 3 Alterations in mRNA stability can have dramatic effects on cell physiology and as a consequence the fitness and survival of the organism. Recent evidence suggests that mRNA decay can be regulated in response to environmental cues in order to enable the organism to adapt to its changing surroundings. Bacteria have unique post-transcriptional control mechanisms to enact such adaptive responses through:
1) general mRNA decay,
2) differential mRNA degradation using small non-coding RNAs (sRNAs), and
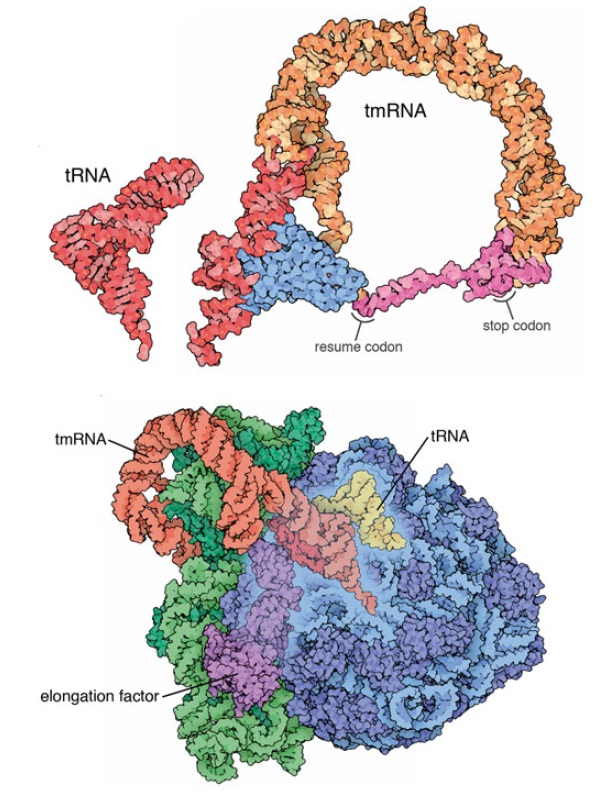
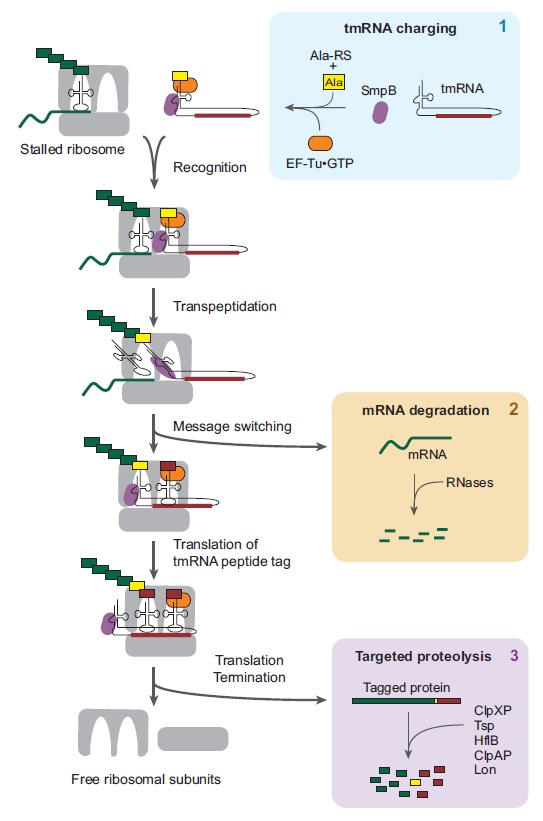
3) selective mRNA degradation using the tmRNA quality control system.
As with other biomolecules, the great variety of RNA species produced by the bacterial cell requires quality control mechanisms to ensure proper folding and function. In addition, the role of mRNA as a template for protein synthesis adds greater significance to mRNA quality control. The translation of a faulty transcript without adequate quality assurance measures might lead to the accumulation of aberrant protein products that could be detrimental to the cell. Bacterial mRNAs are not post-transcriptionally spliced, nor do they exhibit the 5’-cap structures of their eukaryotic counterparts. As such, post-transcriptional quality control processes of prokaryotic mRNA are distinct from the corresponding processes in eukaryotes. This review focuses on issues related to post-transcriptional processing, targeting, and degradation of bacterial mRNAs including those facilitated by small regulatory RNAs, with special emphasis on tmRNA and trans-translation.
Variation in the stability of transcripts has an important role in the control of protein expression within the cell, as long-lived transcripts are generally subject to more rounds of translation than those with a shorter half-life. Several factors play a role in controlling the lifespan of specific mRNAs by regulating their propensity to be degraded.
Translational decoding of the mRNA codons is constrained by factors during codon-anticodon recognition and often constitutes the rate-limiting step during protein synthesis. Besides the abundance of tRNA species, mRNA translation is regulated by nearly 100 epigenetic tRNA modifications, especially at the wobble position 4 Faithful translation of the mRNA codons into protein is essential for cellular physiology. The fidelity of the translation machinery firstly depends on the specific coupling of amino acids to their cognate tRNA species, which is catalyzed by aminoacyl-tRNA synthetases (aaRSs). Eukaryotic elongation factor 1A (eEF-1A) or prokaryotic EF-Tu delivers the aminoacyl-tRNA to the ribosome A site for elongation of nascent peptide chain after proper codon-anticodon recognition. Thus, aminoacyl-tRNA synthetase (aaRSs) are cardinal in protecting protein synthesis against misacylation ( incorrectly adding an acyl group to a compound).
tRNA synthesis quality control
Hypomodified tRNAs (modified to an abnormally small degree) can be degraded by the RNA degradosome, a multicomponent RNA degradation complex. Thus, the degradosome serves as a previously unrecognized bacterial tRNA quality control system that mediates clearance of hypomodified tRNAs.can be degraded by the RNA degradosome, a multicomponent RNA degradation complex. Thus, the degradosome serves as a bacterial tRNA quality control system that mediates clearance of hypomodified tRNAs. 1 Within the anticodon, the first or “wobble” position is particularly subject to modification, and these modifications are often critical for efficient translation; consequently, mutations that prevent such modification can result in loss of viability. tRNAs’ highly stable structures are thought to contribute to their intracellular stability; in general, tRNAs exhibit a high melting temperature in vitro and a long half-life within the cell. RTD and other mechanisms that degrade hypomodified or mutated mature yeast tRNAs serve as a surveillance system to eliminate tRNA molecules that have incorrect nucleosides or conformations
In order to ensure fidelity, some of the synthetases perform editing functions to reduce the occurrence of errors during protein synthesis. 5
Quality control mechanisms during ribosome maturation
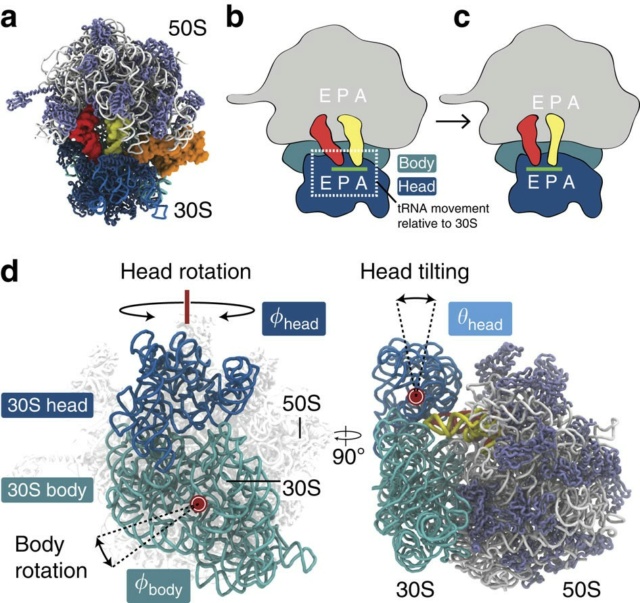
Protein synthesis on ribosomes is carefully quality controlled to ensure the faithful transmission of genetic information from mRNA to protein. Many of these mechanisms rely on communication between distant sites on the ribosomes, and thus on the integrity of the ribosome structure. Furthermore, haploinsufficiency of ribosomal proteins, which increases the chances of forming incompletely assembled ribosomes, can predispose to cancer. Finally, release of inactive ribosomes into the translating pool will lead to their degradation together with the degradation of the bound mRNA. Together, these findings suggest that quality control mechanisms must be in place to survey nascent ribosomes and ensure their functionality. This review gives an account of these mechanisms as currently known. 7
Proof-reading
Yeast employs structural proofreading of ribosome functional centres during nuclear steps of assembly
Accumulating evidence indicates a tight coupling between nuclear export, cytoplasmic maturation, and final proofreading of the ribosome.
We propose that conformational proofreading exerted via Rps20 constitutes a checkpoint permitting assembly factor release and progression of pre-40S maturation only after completion of all earlier maturation steps.
To minimize production of dysfunctional ribosomes, yeast employs structural proofreading of ribosome functional centres during nuclear steps of assembly
Arx1 binding may serve to proofread the proper accommodation of these ribosomal proteins into the tunnel exit
time window for the quality-control machinery to functionally proofread preribosomes.
Last edited by Otangelo on Tue Mar 16, 2021 7:20 pm; edited 4 times in total