Metal ions in biological cells, essential for life
Sodium/Potassium Homeostasis in the Cell
It is well known that the cytoplasm of living cells, generally, contains more potassium ions than sodium ions.
Evolution of metal ions in biological systems
Major (iron, manganese, magnesium and zinc) and minor (copper, cobalt, nickel, molybdenum, tungsten) metal ions have become aligned with living organisms through the interplay of biogeochemical weathering and metabolic pathways involving the products of that weathering.
Structural Biochemistry/Enzyme/Cofactors
Metal ions are known as the common cofactors. In some enzymes, the function as a catalyst cannot be carried out if a metal ion is not available to be bound the active site. In daily nutrition, this kind of cofactor plays a role as the essential trace elements such as: iron (Fe3+), manganese (Mn2+), cobalt (Co2+), copper (Cu2+), zinc (Zn2+), selenium (Se2+), and molybdenum (Mo5+).
Metal Transporters and Metal Sensors: How Coordination Chemistry Controls Bacterial Metal Homeostasis
The transition or d-block metal ions manganese, iron, cobalt, nickel, copper, zinc, and to a more specialized degree, molybdenum, tungsten and vanadium, have been shown to be important for biological systems. These metal ions are ubiquitously found in nature, nearly exclusively as constituents of proteins. The unique properties of metal ions have been exploited by nature to perform a wide range of tasks. These include roles as structural components of biomolecules, as signaling molecules, as catalytic cofactors in reversible oxidation-reduction and hydrolytic reactions, and in structural rearrangements of organic molecules and electron transfer chemistry. Indeed, metal ions play critical roles in the cell that cannot be performed by any other entity, and are therefore essential for all of life. However, an individual metal ion is capable of performing only one or a few of these functions, but certainly not all; as a result, life requires mechanisms to effectively distinguish one metal from another. The coordination chemistry of metal ion-protein complexes is fundamental to this biological discrimination.
Metal Ion Homeostasis
Extensive regulatory and protein-coding machinery is devoted to maintaining the “homeostasis” of biologically required metal ions and underscores the essentiality of this process for cell viability. Homeostasis is defined as the maintenance of an optimal bioavailable concentration, mediated by the balancing of metal uptake and intracellular trafficking with efflux/storage processes so that the needs of the cell for that metal ion is met, i.e., the “right” metal is inserted into the “right” macromolecule at the appropriate time. Just as a scarcity of a particular metal ion induces a stress response that can lead to reprogramming of cellular metabolism to minimize the consequences of depletion of a particular metal ion, e.g., zinc in ribosome biogenesis or Cu vs. Fe in photosynthesis by Synechocystis, too much of the same metal ion can also be toxic to a cell or organism.
Metal homeostasis is governed by the formation of specific protein-metal coordination complexes used to effect uptake, efflux, intracellular trafficking within compartments, and storage (Figure 1).
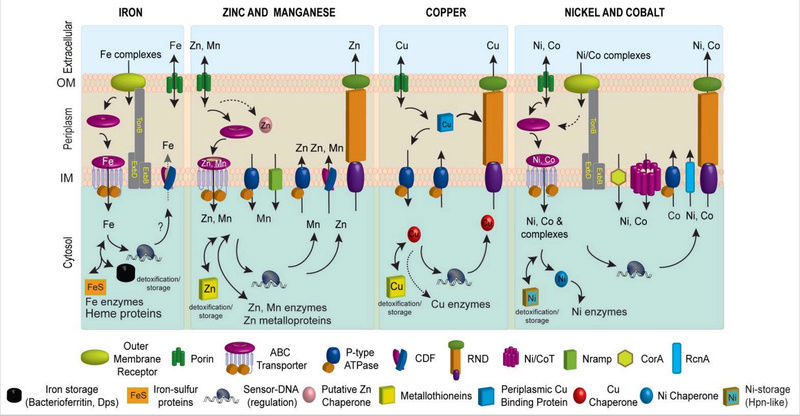
Figure 1. Schematic metal homeostasis models for iron, zinc and manganese, copper, nickel and cobalt, shown specifically in gram-negative bacteria. Homeostasis of molybdate and tungstate oxyanions are not shown, due primarily to a lack of knowledge of these systems, outside of uptake (Section 2.5) and cytosolic sensing (Section 3.8.1). This schematic is not representative for any one bacterium nor is it meant to be exhaustive, but is instead simply designed to convey the potential fates of individual metal ions in distinct cellular compartments. Not all bacteria have all components of each homeostasis system indicated. The double-headed arrows are meant to illustrate that metals can move and in and out of target protein destinations in response to proteome remodeling. Specific protein designations for individual homeostasis components are indicated in Figures Figures22 and and3.3. A putative chaperone shown for Zn(II) is YodA/ZinT,12 while actual metallochaperones for Cu(I) (Atx1221 and CopZ407), and for Ni(II) metalloenzymes urease (UreE10) or Ni-Fe hydrogenases (e.g., HypA10) are also shown. Iron metallochaperones for Fe-S cluster assembly are not shown for clarity.11 Fe(II) efflux through YiiP175 has not yet firmly established biochemically.176 The cytosolic Cu(I) quota for a non-photosynthetic bacterium may well be vanishingly small;7 as a result, transfer from a Cu(I) chaperone is indicated by the dashed double-headed arrow.
How metal ions move to and from their target destinations in the active site of a metalloenzyme or as a structural component of biomolecules also contributes to intracellular metal homeostasis (Figure 1). Metal transporters move metal ions or small molecule-metal chelates across otherwise impermeable barriers in a directional fashion, and most of these are integral membrane proteins embedded in the inner or plasma membrane (Figure 1). Specialized protein chelators designated metallochaperones traffic metals within a particular cellular compartment, e.g., the periplasm or the cytosol, and function to “hold” the metal in such a way that it can be readily transferred to an appropriate acceptor protein. This intermolecular transfer is known or is projected to occur through transiently formed, specific protein-protein complexes that mediate coordinated intermolecular metal ligand exchange. Metallochaperones have been described for copper, nickel and iron-sulfur protein biogenesis, and recent work suggests that the periplasmic Zn(II) binding protein, YodA, has characteristics consistent with a role as a zinc chaperone in E. coli (Figure 1). Finally, specialized transcriptional regulatory proteins, termed metalloregulatory or metal sensor proteins, control the expression of genes encoding these proteins that establish metal homeostasis in response to either metal deprivation or overload.
A hypothesis that emerges is that in order to effect the cellular homeostasis of a particular metal ion, each component of the homeostasis machinery must be selective for that metal ion under the prevailing conditions, to the exclusion of all others. Furthermore, individual systems must be “tuned” such that the affinity or sensitivity of each component is well-matched, either to coordinate gene expression by pairs of metal sensor proteins that coordinately shut off uptake and up-regulate efflux or detoxification systems, or to facilitate vectorial transport from metal donor to metal acceptor target protein in a metal trafficking pathway in the cell.
Introduction to Metal Transporters
All gram-negative bacteria possess an outer membrane (OM), a periplasmic space, and an inner cytoplasmic or plasma membrane through which the metal must pass before entering the cytosol; gram positive bacteria, in contrast, lack a periplasm. Trimeric β-barrel proteins called porins embedded in the outer membrane allow for non-selective passive diffusion of metal ions across the OM. In order to meet cellular metal demands, however, the cytosol must effectively concentrate metal ions. As a result, high affinity active transport systems in the OM or embedded in the plasma or inner membranes (PM or IM) function to transport and release metal ions into the cytosol. Inner membrane transport systems are driven either by the hydrolysis of ATP on the cytoplasmic side of the membrane, e.g., by ATP-binding cassette (ABC) transporters and P-type ATPases, or by coupling to an energetically favorable transfer of protons or other ions across the bilayer, e.g., by Nramp proteins and cation diffusion facilitator (CDF) proteins. The presence of additional layers of extracellular lipopolysaccharide (LPS) or complex carbohydrate matrices, as well as a biofilm, is also likely to have a considerable impact on the rates and mechanisms of metal uptake and efflux.
Introduction to Metal Sensor Proteins
All cells possess a battery of regulatory proteins that mediate homeostasis of transition metal ions by regulating the expression of genes that encode metal transporters, intracellular chelators and/or other detoxification enzymes. These proteins have been coined metal sensor or metalloregulatory proteins. Each forms specific coordination complexes with metal ions that ultimately inhibits or activates operator DNA binding or directly enhances transcriptional activation. In this way, cells effectively control the expression of genes that mediate the homeostasis of metal ions.
Metal sensor proteins can be functionally divided into two groups: those that control gene expression linked to metal ion efflux and/or storage (Figure 2, top)
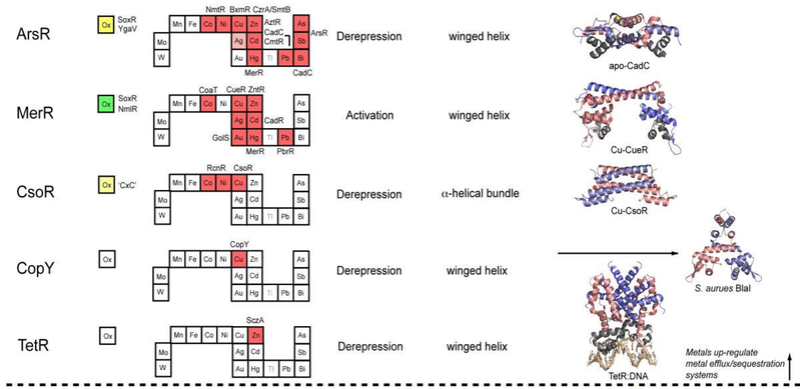
, and those that control the expression of genes required for metal ion uptake (Figure 2, bottom).
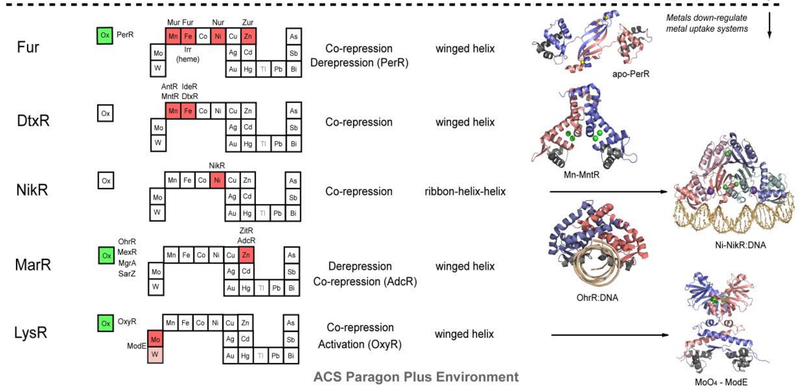
Figure 2. Structural families of metalloregulatory proteins.
For each family, boxes for metals that are known to be sensed are shaded red on the abbreviated periodic table, while green boxes on the left denote known family members that are known to sense cytosolic oxidative stress. Boxes identifying putative metal sensors and non-metal sensing oxidative stress regulators are shaded pink or yellow, respectively. The four-letter designations for individual proteins that perform the function listed in the nearby box are given (see text for details). The mechanism of regulation of gene expression is indicated as is the DNA-binding domain that mediates operator-promoter DNA binding. Ribbon representations of selected representative members are shown on the right with individual protomers shaded red and blue in each case. Structures are from top to bottom:
1) apo S. aureus pI258 CadC with structural α5-Zn(II) ions shaded yellow (1U2W pdb code);255
2) E. coli Cu(I)-sensor CueR with regulatory Cu(I) ions in red (1Q05);34
3) M. tuberculosis Cu(I)-sensor CsoR with regulatory Cu(I) ions shaded red (2HH7);244
4) S. aureus BlaI as a model for Enterococcus CopY (1SD4);320
5) TetR-Tc-Mg DNA complex structure as model for S. pneumonia SczA (3CDL);398
6) apo B. subtilis PerR with structural Zn(II) ions in yellow (2FE3);331
7) B. subtilis MntR with MnA/MnC binuclear cluster ions in green 2F5F);352
8 )Ni(II)-bound E. coli NikR-nik operator DNA complex with high affinity Ni(II) ions shown in green and regulatory K+ ions in purple (2HZV);362
9) B. subtilis OhrR-DNA complex (1Z9C);390
10) Molybdate sensor ModE with molybdate shaded green (1O71).
In general, metal sensor proteins that control metal uptake all bind metal ions as co-repressors, exactly analogous to the well-studied bacterial Trp repressor that controls tryptophan biosynthetic genes in a Trp-dependent manner; in other words, metal binding causes the repression of the genes that allow for metal ion uptake. Contrarily, metal sensor proteins that regulate efflux and/or intracellular storage function via a transcriptional derepression or activation mechanism. In the vast majority of cases, metal binding causes the binding affinity of the sensor protein for its specific DNA operator sequence to decrease substantially and likely dissociate; this, in turn, exposes the promoter, the DNA binding site for RNA polymerase, permitting initiation of transcription. In much that same way, lactose and lactose analogs modulate the affinity of the lactose repressor, LacI, for operator DNA (Section 3.1.3). Transcriptional activation by MerR regulators occurs via a DNA distortion or “underwinding” mechanism (Section 3.2.2), while other activators, e.g., OxyR (Section 3.8.1), appear to undergo multimerization and make direct protein-protein contacts with RNA polymerase to enhance the initiation of transcription.
Figure 2 summarizes the structural and functional properties of ten families of structurally distinguishable regulatory proteins, each of which has at least one known or projected member that senses metals directly. This summary is striking for a number of reasons. First, metal sensor proteins are related orthologs of other transcriptional regulatory proteins that control gene expression by binding small organic molecules, from lipophilic antimicrobial agents to intermediary metabolites to amino acids, rather than metal ions. In other words, there is nothing intrinsically unique about the global structures and mechanisms of action of metal sensor proteins relative to other ligand-regulated transcriptional switches, except in the evolution of “ligand” binding sites that optimize metal-specific coordination chemistry to effect specificity of the biological response (Section 3 below).
Sodium. Its Role in Metabolism 1
Na+ ( sodium ) is found in a wide range of concentrations in the natural environment. It ranges from micromolar to molar and living organisms occur throughout this range.4 In a function that is absolutely essential for survival,
both prokaryotic and eukaryotic cells exhibit homeostatic mechanisms that maintain constant but relatively low internal concentrations of Na+.
Sodium
Sodium in biology
Sodium is an essential element for all animals and some plants. In humans, sodium is an essential mineral that regulates blood volume, blood pressure, osmotic equilibrium and pH
Possible Role of Copper and Sodium Prebiotic Evolution of Peptides
Sodium chloride and copper ions can be assumed to have been ubiquitously present, as indicated by precambrian "green zones" containing malachite and azurite (basic carbonates), and in accordance with the assumption that precambrian oceans acted as accumulators of divalent ions. At the cell membrane, the interface with the external environment, generation of ion gradients depends on ATP-driven pumps, which typically transport Na1 rather than protons. H1 gradients across the cell membrane are typically generated by secondary transporters that convert the Na1 gradient into a H1 gradient via Na1/H1 antiporters. In contrast, in bacteria, the ion pumps responsible for primary energy production are themselves located in the cell membrane. In many cases, these pumps generate proton gradients, and it is the Na1 gradient that is secondary and produced by Na1/H1 antiporters. However, some bacteria are able to generate the Na1 gradient directly using the several different mechanisms.
1. Binding, Transport and Storage of Metal Ions in Biological Cells, page 32
Sodium/Potassium Homeostasis in the Cell
It is well known that the cytoplasm of living cells, generally, contains more potassium ions than sodium ions.
Evolution of metal ions in biological systems
Major (iron, manganese, magnesium and zinc) and minor (copper, cobalt, nickel, molybdenum, tungsten) metal ions have become aligned with living organisms through the interplay of biogeochemical weathering and metabolic pathways involving the products of that weathering.
Structural Biochemistry/Enzyme/Cofactors
Metal ions are known as the common cofactors. In some enzymes, the function as a catalyst cannot be carried out if a metal ion is not available to be bound the active site. In daily nutrition, this kind of cofactor plays a role as the essential trace elements such as: iron (Fe3+), manganese (Mn2+), cobalt (Co2+), copper (Cu2+), zinc (Zn2+), selenium (Se2+), and molybdenum (Mo5+).
Metal Transporters and Metal Sensors: How Coordination Chemistry Controls Bacterial Metal Homeostasis
The transition or d-block metal ions manganese, iron, cobalt, nickel, copper, zinc, and to a more specialized degree, molybdenum, tungsten and vanadium, have been shown to be important for biological systems. These metal ions are ubiquitously found in nature, nearly exclusively as constituents of proteins. The unique properties of metal ions have been exploited by nature to perform a wide range of tasks. These include roles as structural components of biomolecules, as signaling molecules, as catalytic cofactors in reversible oxidation-reduction and hydrolytic reactions, and in structural rearrangements of organic molecules and electron transfer chemistry. Indeed, metal ions play critical roles in the cell that cannot be performed by any other entity, and are therefore essential for all of life. However, an individual metal ion is capable of performing only one or a few of these functions, but certainly not all; as a result, life requires mechanisms to effectively distinguish one metal from another. The coordination chemistry of metal ion-protein complexes is fundamental to this biological discrimination.
Metal Ion Homeostasis
Extensive regulatory and protein-coding machinery is devoted to maintaining the “homeostasis” of biologically required metal ions and underscores the essentiality of this process for cell viability. Homeostasis is defined as the maintenance of an optimal bioavailable concentration, mediated by the balancing of metal uptake and intracellular trafficking with efflux/storage processes so that the needs of the cell for that metal ion is met, i.e., the “right” metal is inserted into the “right” macromolecule at the appropriate time. Just as a scarcity of a particular metal ion induces a stress response that can lead to reprogramming of cellular metabolism to minimize the consequences of depletion of a particular metal ion, e.g., zinc in ribosome biogenesis or Cu vs. Fe in photosynthesis by Synechocystis, too much of the same metal ion can also be toxic to a cell or organism.
Metal homeostasis is governed by the formation of specific protein-metal coordination complexes used to effect uptake, efflux, intracellular trafficking within compartments, and storage (Figure 1).

Figure 1. Schematic metal homeostasis models for iron, zinc and manganese, copper, nickel and cobalt, shown specifically in gram-negative bacteria. Homeostasis of molybdate and tungstate oxyanions are not shown, due primarily to a lack of knowledge of these systems, outside of uptake (Section 2.5) and cytosolic sensing (Section 3.8.1). This schematic is not representative for any one bacterium nor is it meant to be exhaustive, but is instead simply designed to convey the potential fates of individual metal ions in distinct cellular compartments. Not all bacteria have all components of each homeostasis system indicated. The double-headed arrows are meant to illustrate that metals can move and in and out of target protein destinations in response to proteome remodeling. Specific protein designations for individual homeostasis components are indicated in Figures Figures22 and and3.3. A putative chaperone shown for Zn(II) is YodA/ZinT,12 while actual metallochaperones for Cu(I) (Atx1221 and CopZ407), and for Ni(II) metalloenzymes urease (UreE10) or Ni-Fe hydrogenases (e.g., HypA10) are also shown. Iron metallochaperones for Fe-S cluster assembly are not shown for clarity.11 Fe(II) efflux through YiiP175 has not yet firmly established biochemically.176 The cytosolic Cu(I) quota for a non-photosynthetic bacterium may well be vanishingly small;7 as a result, transfer from a Cu(I) chaperone is indicated by the dashed double-headed arrow.
How metal ions move to and from their target destinations in the active site of a metalloenzyme or as a structural component of biomolecules also contributes to intracellular metal homeostasis (Figure 1). Metal transporters move metal ions or small molecule-metal chelates across otherwise impermeable barriers in a directional fashion, and most of these are integral membrane proteins embedded in the inner or plasma membrane (Figure 1). Specialized protein chelators designated metallochaperones traffic metals within a particular cellular compartment, e.g., the periplasm or the cytosol, and function to “hold” the metal in such a way that it can be readily transferred to an appropriate acceptor protein. This intermolecular transfer is known or is projected to occur through transiently formed, specific protein-protein complexes that mediate coordinated intermolecular metal ligand exchange. Metallochaperones have been described for copper, nickel and iron-sulfur protein biogenesis, and recent work suggests that the periplasmic Zn(II) binding protein, YodA, has characteristics consistent with a role as a zinc chaperone in E. coli (Figure 1). Finally, specialized transcriptional regulatory proteins, termed metalloregulatory or metal sensor proteins, control the expression of genes encoding these proteins that establish metal homeostasis in response to either metal deprivation or overload.
A hypothesis that emerges is that in order to effect the cellular homeostasis of a particular metal ion, each component of the homeostasis machinery must be selective for that metal ion under the prevailing conditions, to the exclusion of all others. Furthermore, individual systems must be “tuned” such that the affinity or sensitivity of each component is well-matched, either to coordinate gene expression by pairs of metal sensor proteins that coordinately shut off uptake and up-regulate efflux or detoxification systems, or to facilitate vectorial transport from metal donor to metal acceptor target protein in a metal trafficking pathway in the cell.
Introduction to Metal Transporters
All gram-negative bacteria possess an outer membrane (OM), a periplasmic space, and an inner cytoplasmic or plasma membrane through which the metal must pass before entering the cytosol; gram positive bacteria, in contrast, lack a periplasm. Trimeric β-barrel proteins called porins embedded in the outer membrane allow for non-selective passive diffusion of metal ions across the OM. In order to meet cellular metal demands, however, the cytosol must effectively concentrate metal ions. As a result, high affinity active transport systems in the OM or embedded in the plasma or inner membranes (PM or IM) function to transport and release metal ions into the cytosol. Inner membrane transport systems are driven either by the hydrolysis of ATP on the cytoplasmic side of the membrane, e.g., by ATP-binding cassette (ABC) transporters and P-type ATPases, or by coupling to an energetically favorable transfer of protons or other ions across the bilayer, e.g., by Nramp proteins and cation diffusion facilitator (CDF) proteins. The presence of additional layers of extracellular lipopolysaccharide (LPS) or complex carbohydrate matrices, as well as a biofilm, is also likely to have a considerable impact on the rates and mechanisms of metal uptake and efflux.
Introduction to Metal Sensor Proteins
All cells possess a battery of regulatory proteins that mediate homeostasis of transition metal ions by regulating the expression of genes that encode metal transporters, intracellular chelators and/or other detoxification enzymes. These proteins have been coined metal sensor or metalloregulatory proteins. Each forms specific coordination complexes with metal ions that ultimately inhibits or activates operator DNA binding or directly enhances transcriptional activation. In this way, cells effectively control the expression of genes that mediate the homeostasis of metal ions.
Metal sensor proteins can be functionally divided into two groups: those that control gene expression linked to metal ion efflux and/or storage (Figure 2, top)

, and those that control the expression of genes required for metal ion uptake (Figure 2, bottom).

Figure 2. Structural families of metalloregulatory proteins.
For each family, boxes for metals that are known to be sensed are shaded red on the abbreviated periodic table, while green boxes on the left denote known family members that are known to sense cytosolic oxidative stress. Boxes identifying putative metal sensors and non-metal sensing oxidative stress regulators are shaded pink or yellow, respectively. The four-letter designations for individual proteins that perform the function listed in the nearby box are given (see text for details). The mechanism of regulation of gene expression is indicated as is the DNA-binding domain that mediates operator-promoter DNA binding. Ribbon representations of selected representative members are shown on the right with individual protomers shaded red and blue in each case. Structures are from top to bottom:
1) apo S. aureus pI258 CadC with structural α5-Zn(II) ions shaded yellow (1U2W pdb code);255
2) E. coli Cu(I)-sensor CueR with regulatory Cu(I) ions in red (1Q05);34
3) M. tuberculosis Cu(I)-sensor CsoR with regulatory Cu(I) ions shaded red (2HH7);244
4) S. aureus BlaI as a model for Enterococcus CopY (1SD4);320
5) TetR-Tc-Mg DNA complex structure as model for S. pneumonia SczA (3CDL);398
6) apo B. subtilis PerR with structural Zn(II) ions in yellow (2FE3);331
7) B. subtilis MntR with MnA/MnC binuclear cluster ions in green 2F5F);352
8 )Ni(II)-bound E. coli NikR-nik operator DNA complex with high affinity Ni(II) ions shown in green and regulatory K+ ions in purple (2HZV);362
9) B. subtilis OhrR-DNA complex (1Z9C);390
10) Molybdate sensor ModE with molybdate shaded green (1O71).
In general, metal sensor proteins that control metal uptake all bind metal ions as co-repressors, exactly analogous to the well-studied bacterial Trp repressor that controls tryptophan biosynthetic genes in a Trp-dependent manner; in other words, metal binding causes the repression of the genes that allow for metal ion uptake. Contrarily, metal sensor proteins that regulate efflux and/or intracellular storage function via a transcriptional derepression or activation mechanism. In the vast majority of cases, metal binding causes the binding affinity of the sensor protein for its specific DNA operator sequence to decrease substantially and likely dissociate; this, in turn, exposes the promoter, the DNA binding site for RNA polymerase, permitting initiation of transcription. In much that same way, lactose and lactose analogs modulate the affinity of the lactose repressor, LacI, for operator DNA (Section 3.1.3). Transcriptional activation by MerR regulators occurs via a DNA distortion or “underwinding” mechanism (Section 3.2.2), while other activators, e.g., OxyR (Section 3.8.1), appear to undergo multimerization and make direct protein-protein contacts with RNA polymerase to enhance the initiation of transcription.
Figure 2 summarizes the structural and functional properties of ten families of structurally distinguishable regulatory proteins, each of which has at least one known or projected member that senses metals directly. This summary is striking for a number of reasons. First, metal sensor proteins are related orthologs of other transcriptional regulatory proteins that control gene expression by binding small organic molecules, from lipophilic antimicrobial agents to intermediary metabolites to amino acids, rather than metal ions. In other words, there is nothing intrinsically unique about the global structures and mechanisms of action of metal sensor proteins relative to other ligand-regulated transcriptional switches, except in the evolution of “ligand” binding sites that optimize metal-specific coordination chemistry to effect specificity of the biological response (Section 3 below).
Sodium. Its Role in Metabolism 1
Na+ ( sodium ) is found in a wide range of concentrations in the natural environment. It ranges from micromolar to molar and living organisms occur throughout this range.4 In a function that is absolutely essential for survival,
both prokaryotic and eukaryotic cells exhibit homeostatic mechanisms that maintain constant but relatively low internal concentrations of Na+.
Sodium
Sodium in biology
Sodium is an essential element for all animals and some plants. In humans, sodium is an essential mineral that regulates blood volume, blood pressure, osmotic equilibrium and pH
Possible Role of Copper and Sodium Prebiotic Evolution of Peptides
Sodium chloride and copper ions can be assumed to have been ubiquitously present, as indicated by precambrian "green zones" containing malachite and azurite (basic carbonates), and in accordance with the assumption that precambrian oceans acted as accumulators of divalent ions. At the cell membrane, the interface with the external environment, generation of ion gradients depends on ATP-driven pumps, which typically transport Na1 rather than protons. H1 gradients across the cell membrane are typically generated by secondary transporters that convert the Na1 gradient into a H1 gradient via Na1/H1 antiporters. In contrast, in bacteria, the ion pumps responsible for primary energy production are themselves located in the cell membrane. In many cases, these pumps generate proton gradients, and it is the Na1 gradient that is secondary and produced by Na1/H1 antiporters. However, some bacteria are able to generate the Na1 gradient directly using the several different mechanisms.
1. Binding, Transport and Storage of Metal Ions in Biological Cells, page 32

