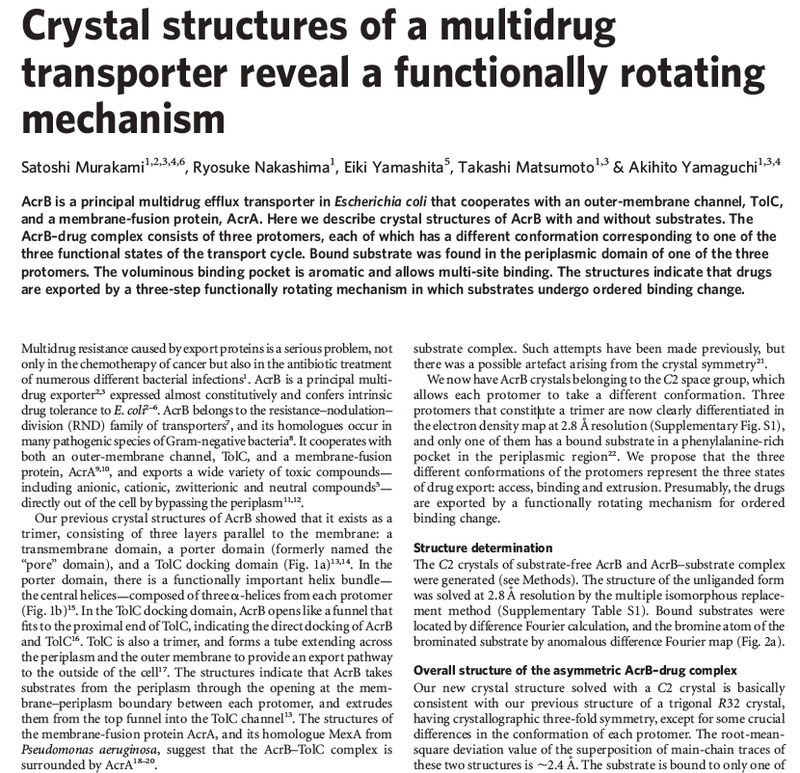
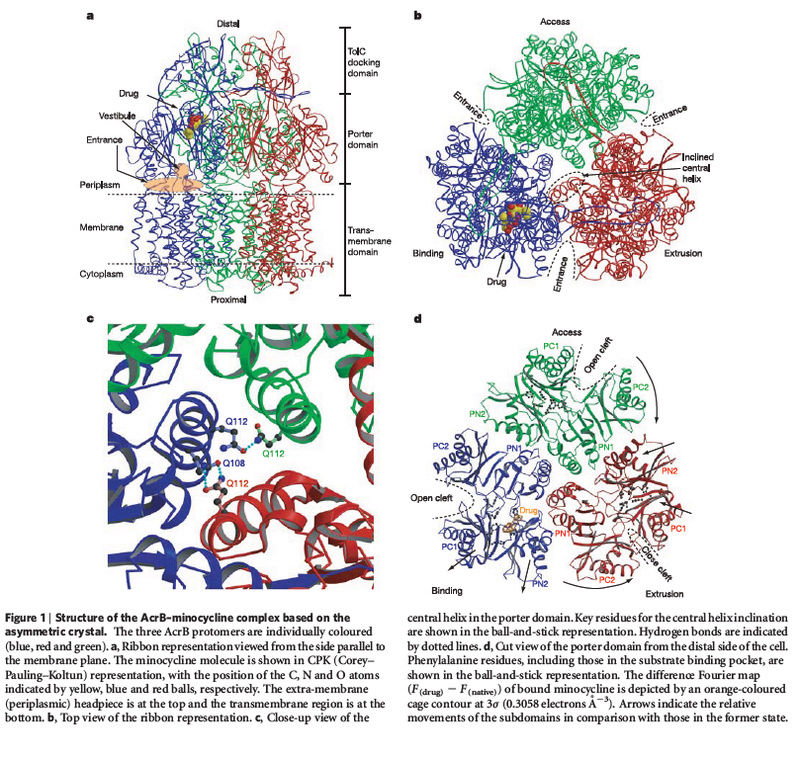
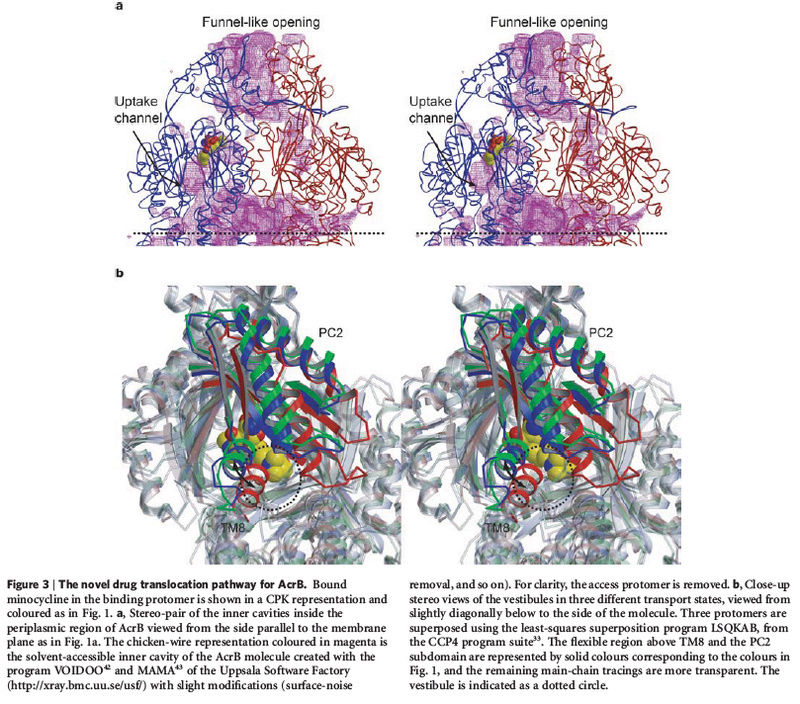
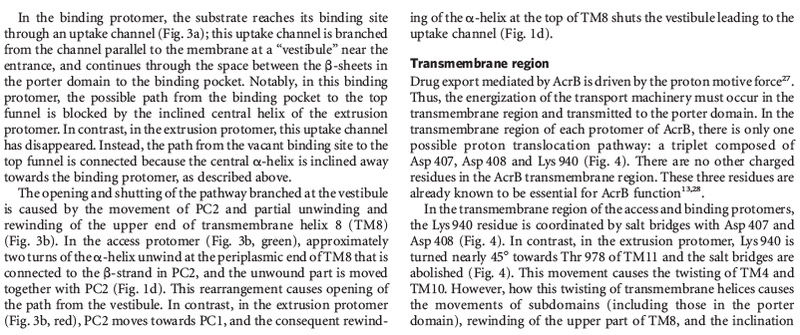
The transmembrane proteins
include the
various ion channels,
other types of channel proteins,
transporter proteins,
growth factor receptors,
and cell adhesion molecules.
There are four main classifications for transmembrane proteins,
type I,
II,
III, and
IV.
Type IV transmembrane proteins pass through the membrane several times and, therefore, they are all referred to as multiple-pass transmembrane proteins. Type I transmembrane proteins are anchored to the membrane via a sequence of hydrophobic amino acids referred to as the stop-transfer sequence and this class all have the C-terminus of the protein inside the cell and the N-terminus outside. A typical example of a type I transmembrane protein is the LDL receptor. Type II transmembrane proteins are anchored to the membrane via a signal-anchor sequence and have the C-terminus outside the cell and the N-terminus inside. An example of a type II transmembrane protein is the transferrin receptor. Type III transmembrane proteins do not have a signal sequence and the N-terminus of the protein is outside the cell. An example of a type III transmembrane protein would be any member of the cytochrome P450 family of xenobiotic metabolizing enzymes found in the liver. Type IV transmembrane proteins are typified by the G-protein coupled receptor (GPCR) superfamily of receptor proteins that span the membrane seven times. Another example of a type IV transmembrane protein is the α-subunit of a typical Na,K-ATPase (see below). Type IV transmembrane proteins are divided into type IV-A and type IV-B where the IV-A members have the N-terminus inside the cell and the C-terminus outside and the IV-B members are oriented in the opposite direction. The Na,K-ATPase α-subunit proteins are type IV-A multi-pass transmembrane proteins, whereas, all GPCRs are members of the type IV-B family.

Membrane channels
- The definition of a channel (or a pore) is that of a protein structure that facilitates the translocation of molecules or ions across the membrane through the creation of a central aqueous channel in the protein. This central channel facilitates diffusion in both directions dependent upon the direction of the concentration gradient. Channel proteins do not bind or sequester the molecule or ion that is moving through the channel. Specificity of channels for ions or molecules is a function of the size and charge of the substance. The flow of molecules through a channel can be regulated by various mechanisms that result in opening or closing of the passageway. More details on the numerous types of ion channels are discussed in the sections below.
Membrane channels are of three distinct types. The α-type channels are homo- or hetero-oligomeric structures that in the latter case consist of several dissimilar proteins. This class of channel protein has between 2 and 22 transmembrane α-helical domains which explains the derivation of their class. Molecules move through α-type channels down their concentration gradients and thus, require no input of metabolic energy. Some channels of this class are highly specific with respect to the molecule translocated across the membrane while others are not. In addition, there may be differences from tissue to tissue in the channel used to transport the same molecule. As an example, there are 40 different K-specific voltage-gated channels in humans. The transport of molecules through α-type channels occurs by several different mechanisms. These mechanisms include changes in membrane potential (termed voltage-regulated or voltage-gated), phosphorylation of the channel protein, intracellular Ca, G-proteins, and organic modulators.
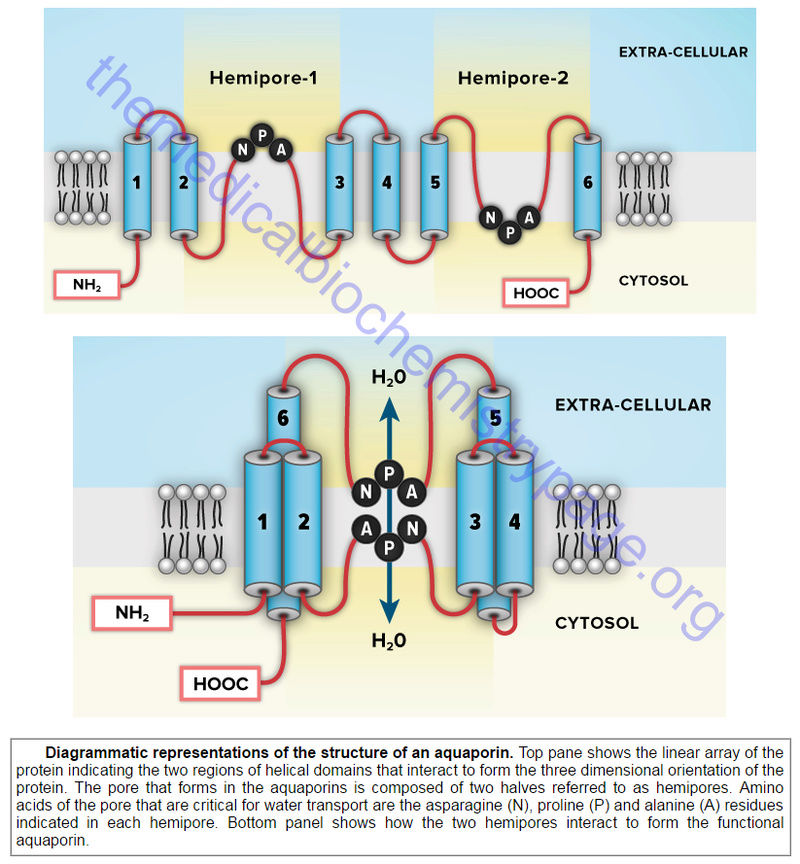
Aquaporins (AQP) are a family of α-type channels responsible for the transport of water across membranes. The β-barrel channels (also called porins) are so named because they have a transmembrane domain that consists of β-strands forming a β-barrel structure. The mitochondrial porins are voltage-gated anion channels that are involved in mitochondrial homeostasis and apoptosis.
The pore-forming toxins represent the third class of membrane channels. The defensins are a family of small cysteine-rich antibiotic proteins that are pore-forming channels found in epithelial and hematopoietic cells.
Membrane Transporters
Transporters are distinguished from channels because they catalyze (mediate) the movement of ions and molecules by physically binding to and moving the substance across the membrane. Transporters exhibit specificity for the molecule being transported as well as show defined kinetics in the transport process. Transporters can also be affected by both competitive and noncompetitive inhibitors. Transporters are also known as carriers, permeases, translocators, translocases, and porters. Mediated transporters are classified based upon the stoichiometry of the transport process.
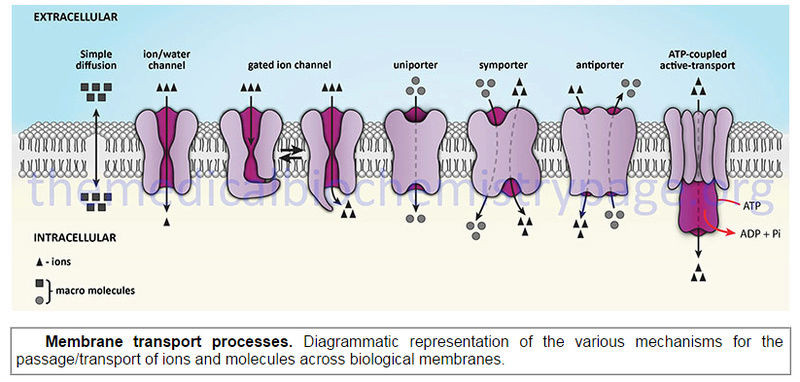
The action of transporters is divided into two classifications: passive-mediated transport (also called facilitated diffusion) and active transport. Glucose transporters are a good example of passive-mediated (facilitative diffusion) transporters. More information on the different glucose/hexose transporters can be found in the Glycolysis page. Another important class of passive-mediated transporters are the K channels (see section above). In contrast, active transporters transport specific molecules from an area of low concentration to that of high concentration. Because this process is thermodynamically unfavorable, the process must be coupled to an exergonic process, most often the hydrolysis of ATP.
The ATPase family of transporters
- There are many different classes of membrane transporters that couple the hydrolysis of ATP to the transport of specific molecules. In general these transporters are referred to as ATPases. These ATPases are so named because the ATP hydrolysis that occurs during the transport process is coupled to the autophosphorylation of the transporter. There are four primary types of ATPase transporters that function in eukaryotes. In addition to the four classes of ATPase described in this section, another important class of transporters that function via the use of ATP hydrolytic energy is the ATP-binding cassette (ABC) transporter family. The name of this family is derived from ATPases Associated with diverse cellular Activities.
E-type ATPases are cell surface transporters that hydrolyze a range of nucleoside triphosphates that includes extracellular ATP. These transporters derive their nomenclature from the fact that they are invloved in Extracellular transport. The activity of the E-type ATPases is dependent on Ca or Mg and it is insensitive to specific inhibitors of P-type, F-type, and V-type ATPases. The E-type ATPases can hydrolyze other NTPs besides ATP, and some can utilize NDPs. There are at least three classes of E-type ATPases.
F-type ATPases function in the translocation of H in the mitochondria during the process of oxidative phosphorylation. F-type transporters contain rotary motors. The nomenclature of F-type ATPases derives from phosphorylation Factor. Because these transporters transport H they are also referred to as H-transporting ATPases. Additional common nomenclature for these ATPases is F0F1-ATPase.
P-type ATPases are mostly found in the plasma membrane and are involved in the transport of H, K, Na, Ca, Cd, Cu, Mg, Co, Ag, and Zn. These transporters represent one of the largest families found in both prokaryotes and eukaryotes. The P-type ATPases are grouped into five classes designated P1–P5 with several classes further divided into subclasses designated A, B, C etc. The P-type ATPases contain a core cytoplasmic domain structure that includes a phosphorylation domain (P domain), a nucleotide-binding domain (N domain), and an actuator domain (A domain). The P-type ATPases also possess ten transmembrane helixes termed M1–M10 where helixes M1–M6 comprise the core of the membrane transport domain. The P-type ATPases are also referred to as the E1-E2 ATPases.
V-type ATPases are located in acidic vesicles and lysosomes and have homology to the F-type ATPases and also contain rotary motors like F-type ATPases. The V nomenclature is derived from the fact that these transporters are located in Vacuoles. The V-type ATPases are involved in the processes of neurotransmitter release, protein trafficking, receptor-mediated endocytosis, and active transport of metabolites.
A fifth family of ATPase transporters is the A-type family found only in prokaryotes.
A-type ATPases are Archaeal bacterial transporters that function like the F-type class of ATPases.
Na,K-ATPases
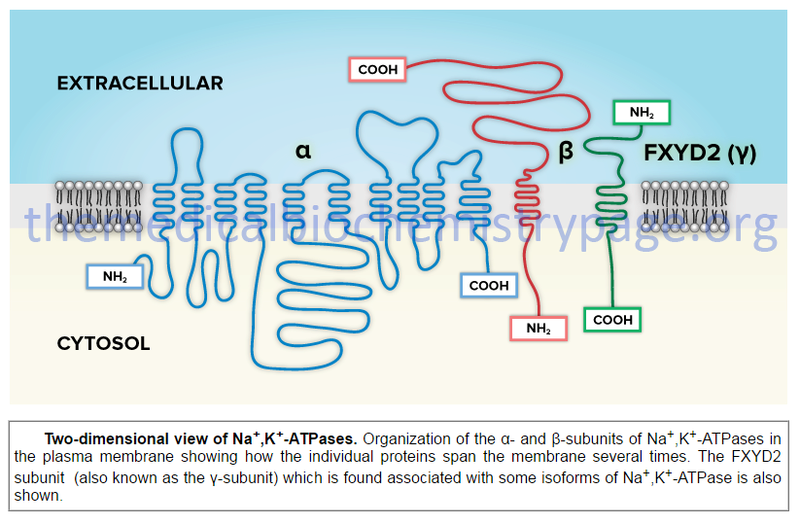
One of the most thoroughly studied classes of ATPases are the Na,K-ATPases found in plasma membranes. These transporters, sometimes called Na,K-pumps, are involved in the transport of Na out of, and K into, cells.The Na,K-ATPases belong to the P2 class and specifically to the P2C subclass of ATPases. These ATPases are composed of two subunits (α and β). The α-subunit (≈113 kD) binds ATP and both Na and K ions and contains the phosphorylation sites typical of the P-type ATPases. As discussed below, P-type ATPases are also subject to additional phosphorylation events via other kinases.
In addition to the ability to form numerous complexes through the interactions of different α- and β-subunits, the Na,K-ATPases also associate with a family of small single transmembrane-spanning proteins termed the FXYD (fix-id) proteins. Five members of this family, including FXYD1 (also known as phospholemman), FXYD2 (also known as the γ-subunit of Na,K-ATPase), FXYD3 (also known as Mat-
Two-dimensional view of Na,K-ATPases. Organization of the α- and β-subunits of Na,K-ATPases in the plasma membrane showing how the individual proteins span the membrane several times.
Three-dimensional view of Na,K-ATPases. Functional organization of the α- and β-subunits of Na,K-ATPases, along with the FXYD2 subunit, in the plasma membrane.

Biology, Robert J.Brooker , page 99:
Membrane proteins are involved in transporting ions and molecules across membranes. Other key functions of membrane proteins are ATP synthesis, photosynthesis , cell signaling ,
and cell-to-cell adhesion.
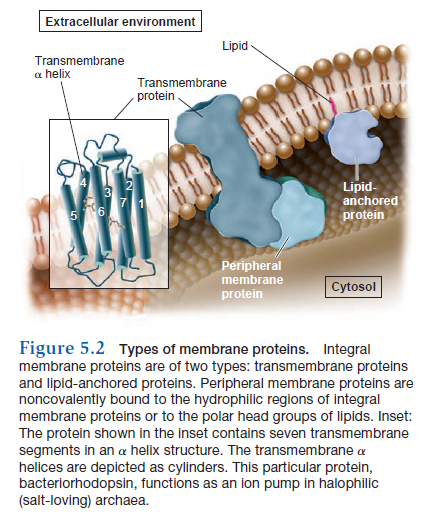
Membrane proteins have different ways of associating with a membrane (Figure 5.2). An integral membrane protein, also called an intrinsic membrane protein, cannot be released from the membrane unless the membrane is dissolved with an organic solvent or detergent—in other words, you would have to disrupt the integrity of the membrane to remove it. The most common type of integral membrane protein is a transmembrane protein, which has one or more regions that are physically inserted into the hydrophobic region of the phospholipid bilayer. These regions, the transmembrane segments, are stretches of nonpolar amino acids that span or traverse the membrane from one leaflet to the other. In most transmembrane proteins, each transmembrane segment is folded into an a helix structure. Such a segment is stable in a membrane because the nonpolar amino acids can interact favorably with the hydrophobic fatty acyl tails of the lipid molecules.
A second type of integral membrane protein, known as a lipid-anchored protein, has a lipid molecule that is covalently attached to an amino acid side chain within the protein. The fatty acyl tails are inserted into the hydrophobic portion of the membrane and thereby keep the protein firmly attached to the membrane. Peripheral membrane proteins, also called extrinsic proteins, are another category of membrane protein. They do not interact with the hydrophobic interior of the phospholipid bilayer. Instead, they are noncovalently bound to regions of integral membrane proteins that project out from the membrane, or they are bound to the polar head groups of phospholipids. Peripheral membrane proteins are typically bound to the membrane by hydrogen and/or ionic bonds. For this reason, they usually can be removed from the membrane experimentally by varying the pH or salt concentration.
Approximately 25% of All Genes Encode Transmembrane Proteins
Membrane proteins participate in some of the most important and interesting cellular processes. These include transport, energy transduction, cell signaling, secretion, cell recognition, metabolism, and cell-to-cell contact. Research studies have revealed that cells devote a sizeable fraction of their energy and metabolic machinery to the synthesis of membrane proteins. These proteins are particularly important in human medicine—
approximately 70% of all medications exert their effects by binding to membrane proteins. These drugs bind to cyclooxygenase, a protein in the ER membrane that is necessary for the synthesis of chemicals that play a role in inflammation and pain sensation. Because membrane proteins are so important biologically and medically, researchers have analyzed the genomes of many species and asked the question, What percentage of genes encodes transmembrane proteins? To answer this question, they have developed tools to predict the likelihood that a gene encodes a transmembrane protein. For example, the occurrence of transmembrane a helices can be predicted from the amino acid sequence of a protein. All 20 amino acids can be ranked according to their tendency to enter a hydrophobic or hydrophilic environment. With these values, the amino acid sequence of a protein can be analyzed using computer software to determine the average hydrophobicity of short amino acid sequences within the protein. A stretch of 18 to 20 amino acids in an a helix is long enough to span the membrane. If such a stretch contains a high percentage of hydrophobic amino acids, it is predicted to be a transmembrane a helix. However, such computer predictions must eventually be verified by experimentation.
Using a computer approach, many research groups have attempted to calculate the percentage of genes that encode transmembrane proteins in various species. The estimated percentage of transmembrane proteins is substantial: 20–30% of all genes may encode transmembrane proteins. This trend is found throughout all domains of life, including archaea, bacteria, and eukaryotes. For example, about 30% of human genes encode transmembrane proteins. With a genome size of 20,000 to 25,000 different genes, the total number of genes that encode different transmembrane proteins is estimated at 6,000 to 7,500. The functions of many of them have yet to be determined. Identifying their functions will help researchers gain a better understanding of human biology. Likewise, medical researchers and pharmaceutical companies are interested in the identification of new transmembrane proteins that could be targets for effective new medications.
Membranes Are Semifluid
Let’s now turn our attention to the dynamic properties of membranes. Although a membrane provides a critical interface between a cell and its environment, it is not a solid, rigid structure. Rather, biomembranes exhibit properties of fluidity, which means that individual molecules remain in close association yet have the ability to readily move within the membrane. Though membranes are often described as fluid, it is more appropriate to say they are semifluid. In a fluid substance, molecules can move in three dimensions. By comparison, most phospholipids can rotate freely around their long axes and move laterally within the membrane leaflet (Figure 5.3a).
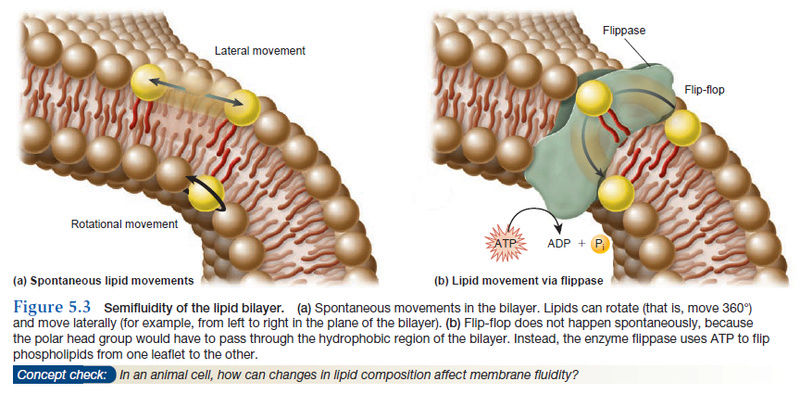
This type of motion is considered two-dimensional, which means it occurs within the plane of the membrane. Because rotational and lateral movements keep the fatty acyl tails within the hydrophobic interior, such movements are energetically favorable. At 37°C, a typical lipid molecule exchanges places with its neighbors about 107 times per second, and it can move several micrometers per second. At this rate, a lipid could traverse the length of a bacterial cell (approximately 1 μm) in only 1 second and the length of a typical animal cell in 10–20 seconds. In contrast to rotational and lateral movements, the “flipflop” of lipids from one leaflet to the opposite leaflet does not occur spontaneously. Energetically, such movements are unfavorable because the polar head of a phospholipid would have to be transported through the hydrophobic interior of the membrane. How are lipids moved from one leaflet to the other? The transport of lipids between leaflets requires the action of the enzyme flippase, which provides energy from the hydrolysis of ATP (Figure 5.3b).
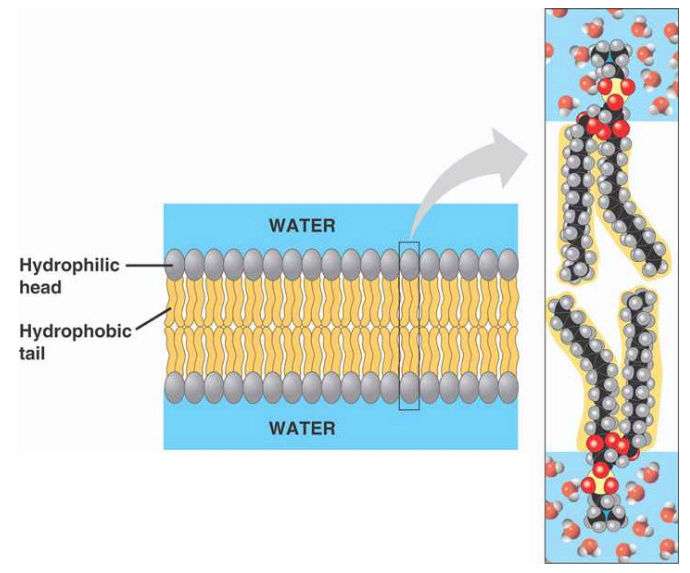
Cell membranes are bi-layer
There have been several molecular dynamic simulations carried out to investigate the biophysicochemistry of spontaneous bilayer assembly. There, lipids start in random orientations. The ordered bilayers we know spontaneously assemble in under 100ns. What's actually happening at a molecular dynamics level is the self-association of the hydrophobic lipid tail groups driven entropically by water. In other words the polar (hydrophilic) head-groups "prefer" interacting with the water (called the interfacial region) and the the hydrophobic tail groups "prefer" not interacting with the water. With those two preferences in play, the lipid bilayer forms.
Flipping lipids: why an’ what’s the reason for? 1
1.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2784161/#R2
2.http://biology.stackexchange.com/questions/9261/why-do-cell-membranes-have-a-lipid-bilayer-instead-of-a-monolayer
3. http://themedicalbiochemistrypage.org/membranes.php
Last edited by Admin on Mon Dec 04, 2017 1:48 am; edited 8 times in total