Biosynthesis of the cell membrane
The Lipids of Biological Membranes 6
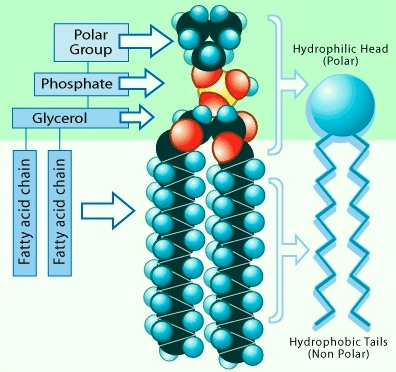
The membranes of cells and viruses, whether they are in prokaryotes, eukaryotes, or in enveloped viruses, are built on the fundamental architecture of a lipid bilayer. All biological membranes contain lipids. The lipids of biological membranes, for the most part, form lipid bilayers in the biological membrane. The structure and resulting chemical properties, along with the hydrophobic effect, of these lipids determine their ability to form bilayers in biological membranes and as purified molecules in aqueous media. Thousands of lipid species are found in biological membranes. Many of their roles in biology have yet to be fully understood. A feature in common among most lipids of biological membranes is their amphipathic structure. Much of the structure of these molecules is hydrophobic, often determined by long hydrocarbon chains of covalently bonded carbon atoms forming one portion of the molecule. The other portion of these molecules is the lipid headgroup. The lipid headgroup is usually polar and often contains charged moieties. A wide variety of chemical structures is found among lipid headgroups. With these polar headgroups at one end and the hydrophobic hydrocarbon chains at the other, the criteria are met for an amphipathic molecule. The amphipathic chemical structure of the lipids is critical to the formation of membranes.
PHOSPHOLIPIDS
A good place to begin the story of the lipids is with a look at the best known and most studied of the lipids of biological membranes, the phospholipids.
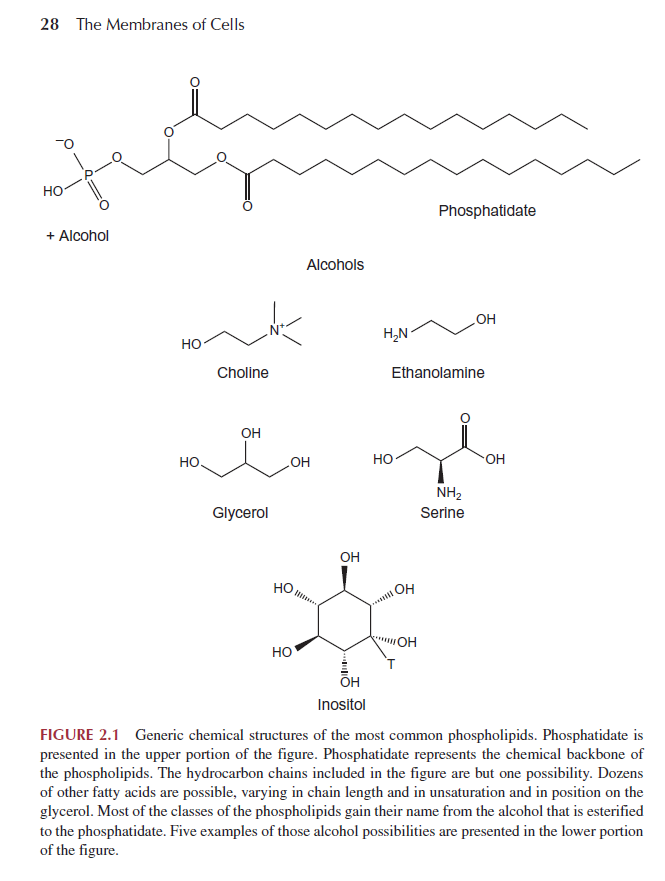
Phospholipid structures fit the general description given earlier: phospholipids consist of a polar headgroup on one end of the molecule and hydrocarbon chains on the other end. These chemical structures create an amphipathic lipid. The phospholipids derive their name in part from the phosphate group found in the polar headgroup. The general structure is shown in Fig. 2.1.
Phospholipases
Phospholipids are substrate for a class of enzymes called phospholipases. Phospholipases are enzymes that hydrolyze ( the cleavage of chemical bonds by the addition of water ) phospholipids, separating the polar and one of the hydrophobic portions of the phospholipid molecule, a fatty acid.
These enzymes are useful tools in studies of the structure and composition of biological membranes. Chemists, too, have used phospholipases to assist in the chemical synthesis of phospholipids. Phospholipases play important roles in lipid metabolism. One example is the metabolic remodeling of the phospholipid structure. A metabolic intermediate in the pathways of biosynthesis of phospholipids is diacylglycerol. In this pathway, the diacylglycerol is symmetrical and saturated in its fatty acid content. As already noted, in the mature phospholipid in cell membranes, the dominant phospholipid species have different fatty acids on the 1′ and 2′ positions of the glycerol, with the 2′ position reserved for unsaturated fatty acids. Part of this pathway involves phospholipase A2 that cleaves the ester bond at the 2′ position, allowing a subsequent reacylation of the phospholipid with an unsaturated fatty acid. This results in the common form with a saturated fatty acid at the 1′ position and an unsaturated fatty acid at the 2′ position of the glycerol. The same class of phospholipase can be stimulated
by receptors to enable the cleavage of a particular unsaturated fatty acid, for prostaglandin biosynthesis. Another class of phospholipase creates important second messengers for intracellular communication
Cell Membranes, origins through natural mechanisms, or design ?
http://reasonandscience.heavenforum.org/t2128-membrane-structure#3798
Early evolution of polyisoprenol biosynthesis and the origin of cell walls 1
After being a matter of hot debate for years, the presence of lipid membranes in the last common ancestor of extant organisms (i.e., the cenancestor) now begins to be generally accepted. Here, we provide the first systematic phylogenomic report on the polyisoprenol biosynthesis pathways in the three domains of life. This study shows that, whereas the last steps of the polyisoprenol biosynthesis are unique to the respective domain of life of which they are characteristic, the enzymes required for basic unsaturated polyisoprenol synthesis can be traced back to the respective last common ancestor of each of the three domains of life. As a result, regardless of the topology of the tree of life that may be considered, the most parsimonious hypothesis is that these enzymes were inherited in modern lineages from the cenancestor. This observation supports the presence of an enzymatic mechanism to synthesize unsaturated polyisoprenols in the cenancestor and, since these molecules are notorious lipid carriers in glycosylation pathways involved in the synthesis of a wide diversity of prokaryotic cell walls, it provides the first indirect evidence of the existence of a hypothetical unknown cell wall synthesis mechanism in the cenancestor.
Despite being widespread in archaea, bacteria and eukaryotes, IPP and DMAPP are synthesized by respective pathways in each domain of life (Lombard & Moreira, 2011; Dellas et al., 2013; Vranová, Coman & Gruissem, 2013). A previous study on the evolution of these pathways concluded that the cenancestor had a primitive mevalonate pathway that could have synthesized IPP and DMAPP in this organism, and that each domain of life adopted its specific biosynthesis pathway later on
Or maybe each pathway was created individually by God ?
Cell walls indeed provide essential structural support and external interactions in modern organisms (Albers & Meyer, 2011), Despite the stunning diversity that exists among prokaryotic cell envelopes, the synthesis of many of their main components:
1.N-or O-glycosylated S-layer proteins
2.peptidoglycan,
3.O-antigen LPS
4.teichoic acids
5.exopolysaccharides
relies on comparable glycosylation pathways These pathways are all located in the cell membranes, are mediated by similar lipid carriers and have the same orientation across the membrane.
That raises the question: If in extant cells these pathways are located in the cell membranes, where was the location prior membranes existed ? Would it not be correct to say : It takes membranes to make membranes ?
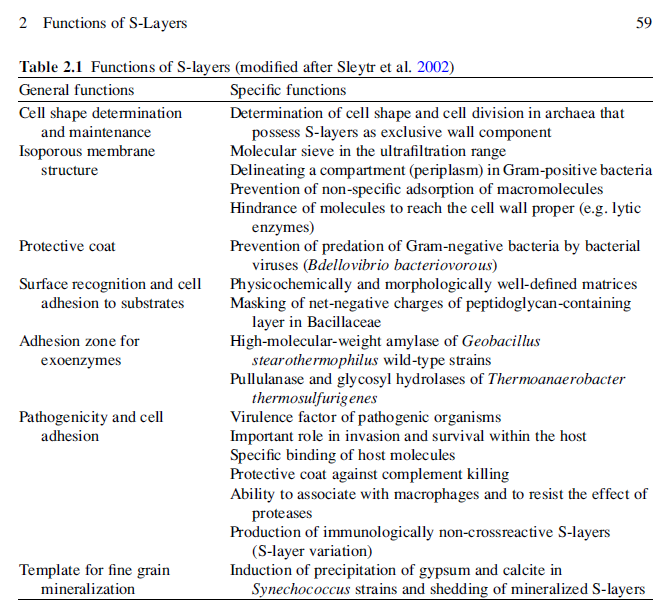
1. S-layers: principles and applications 3
Monomolecular arrays of protein or glycoprotein subunits forming surface layers (S-layers) are one of the most commonly observed prokaryotic cell envelope components. S-layers are generally the most abundantly expressed proteins, have been observed in species of nearly every taxonomical group of walled bacteria, and represent an almost universal feature of archaeal envelopes. The isoporous lattices completely covering the cell surface provide organisms with various selection advantages including functioning as protective coats, molecular sieves and ion traps, as structures involved in surface recognition and cell adhesion, and as antifouling layers. S-layers are also identified to contribute to virulence when present as a structural component of pathogens. In Archaea, most of which possess S-layers as exclusive wall component, they are involved in determining cell shape and cell division.
Introduction
With the exception of those prokaryotic organisms which have developed strategies to live under very specialized and frequently extreme environmental conditions in which monocultures are feasible, most organisms have to survive in highly competitive habitats in very complex microbiomes. Consequently, the diversity observed in the molecular architecture of bacterial and archaeal cell envelopes, particularly the structure of the outermost boundary layers, reflects evolutionary adaptations of the organism to specific environmental and ecological conditions.
Among the most commonly observed prokaryotic cell surface structures are two-dimensional arrays of proteinaceous subunits forming surface layers (termed S-layers) on prokaryotic cells (Sleytr, 1976; Sleytr et al., 1988b; TableTable1).1). Since the first ‘macromolecular monolayer’ described by Houwink and Le Poole (1952; Houwink, 1953) in the cell wall of a Spirillum sp., S-layers have now been identified in hundreds of different species of almost every taxonomic group of walled Bacteria and are an almost universal feature of Archaea .
Because S-layer proteins account for approximately ten percent of cellular proteins in Archaea and Bacteria, they represent interesting model systems for studying the processes involved in the synthesis, secretion, and assembly of extracellular proteins. Moreover, as the biomass of prokaryotic organisms surpasses the biomass of eukaryotic organism (Whitman et al., 1998), S-layer proteins can be considered as one of the most abundant biopolymers on our planet. S-layers also represent the simplest biological protein or glycoprotein membranes (Sleytr, 1975). They are generally composed of a single molecular species endowed with the ability to assemble on the cell surface into closed regular arrays occupying a low free-energy arrangement. Studies on the in vivo morphogenesis of S-layers demonstrated that at high growth rates, approximately 500 subunits per second must be synthesized, translocated to the cell surface, and incorporated into the existing S-layer lattice (Sleytr & Messner, 1983; Sleytr & Beveridge, 1999). It is now evident that S-layers as metabolic expensive products can provide organisms with an advantage of selection in quite diverse habitats. Although a considerable amount of knowledge has accumulated on the structure, assembly, chemistry, and genetics of S-layers, relatively little firm data are available about their specific biological functions (Sleytr et al., 2002, 2007b; Hynonen & Palva, 2013). It is now recognized that they can function as protective coats, molecular sieves, molecule and ion traps, promoters for cell adhesion, immunomodulators, surface recognition, antifouling coatings, and virulence factors in pathogenic organisms. In those Archaea that possess S-layers as the exclusive envelope component external to the cytoplasmic membrane, the lattice is involved in the determination of cell shape and as a structure aiding in the cell division process.
The S-layer glycoprotein is not just a standardized building block used to generate the two-dimensional lattice of the S-layer but rather that S-layer glycoproteins undergo a variety of post-translational modifications. 5
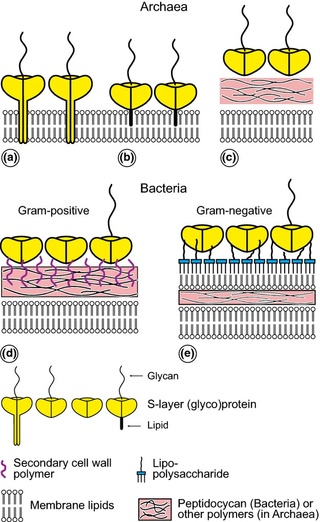
Schematic illustration of the supramolecular architecture of the major classes of prokaryotic cell envelopes containing surface (S) layers.
S-layers in archaea with glycoprotein lattices as exclusive wall component are composed either of mushroom-like subunits with pillar-like, hydrophobic trans-membrane domains (a), or lipid-modified glycoprotein subunits (b). Individual S-layers can be composed of glycoproteins possessing both types of membrane anchoring mechanisms. Few archaea possess a rigid wall layer (e.g. pseudomurein in methanogenic organisms) as intermediate layer between the plasma membrane and the S-layer (c). In Gram-positive bacteria, (d) the S-layer (glyco)proteins are bound to the rigid peptidoglycan-containing layer via secondary cell wall polymers. In Gram-negative bacteria, (e) the S-layer is closely associated with the lipopolysaccharide of the outer membrane.
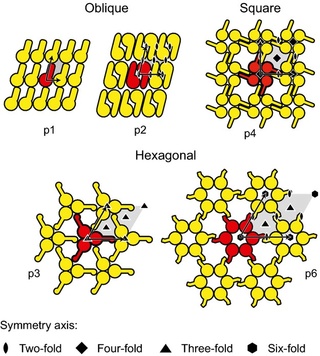
Schematic drawing of the different S-layer lattice types, their base vectors, the unit cell (shaded in gray), and the corresponding symmetry axis. The proteins at one morphological unit are shown in red.
The glycan chain and linkages of bacterial and archaeal glycoproteins are significantly different from those of eukaryotes .
The lipid carrier molecules also differ between Archaea and Bacteria. In Archaea, isoprene-based lipids such as dolichol phosphate and dolichol pyrophosphate play essential roles in the N-glycosylation process by delivering their bound glycan cargo to selected protein targets.
How was it established which target is the correct one ? how did the communication system emerge ?
S-Layer Proteins 4
Over the past 3 decades of research, it has become apparent that one of the most common surface structures on archaea and bacteria are monomolecular crystalline arrays of proteinaceous subunits termed surface layers or S-layers. S-layers represent one of the most abundant cellular proteins, it is now obvious that these metabolically expensive products must provide the organisms with an advantage of selection in very different habitats
S-layers have now been identified in hundreds of different species belonging to all major phylogenetic groups of bacteria, and they represent a feature common to almost all archaea. S-layer proteins of many archaea and those of gram-positive bacteria can possess covalently bound carbohydrate chains. The glycan chains are composed of 20 to 50 identical repeating units containing neutral hexoses, pentoses, heptoses, 6-deoxyhexoses, and amino sugars. Core oligosaccharides of S-layer glycoproteins consist of two to four sugar residues attaching the carbohydrate chains mostly via O-glycosidic linkages (galactose–tyrosine, glucose–tyrosine,N-acetylgalactosamine–serine, N-acetyl galactosamine–threonine) to the protein moiety.
Their respective mechanisms can be described in three major steps:
(1) synthesis of an oligomer on a lipid carrier in the cytoplasmic side of the cell membranes;
(2) ‘‘flipping’’ of the oligomer-linked lipid carrier to the opposite side of the membrane; and
(3) oligomer transfer from the lipid carrier to the acceptor molecule (e.g., protein or external polymer)
The proteins involved in these glycosylation pathways belong to a small number of protein superfamilies, some of which could be traced back to the cenancestor (Lombard, 2016). the prokaryotic cell wall synthesis mechanisms have at least one element in common: they all use polyisoprenol phosphate lipid carriers (Hartley & Imperiali, 2012).
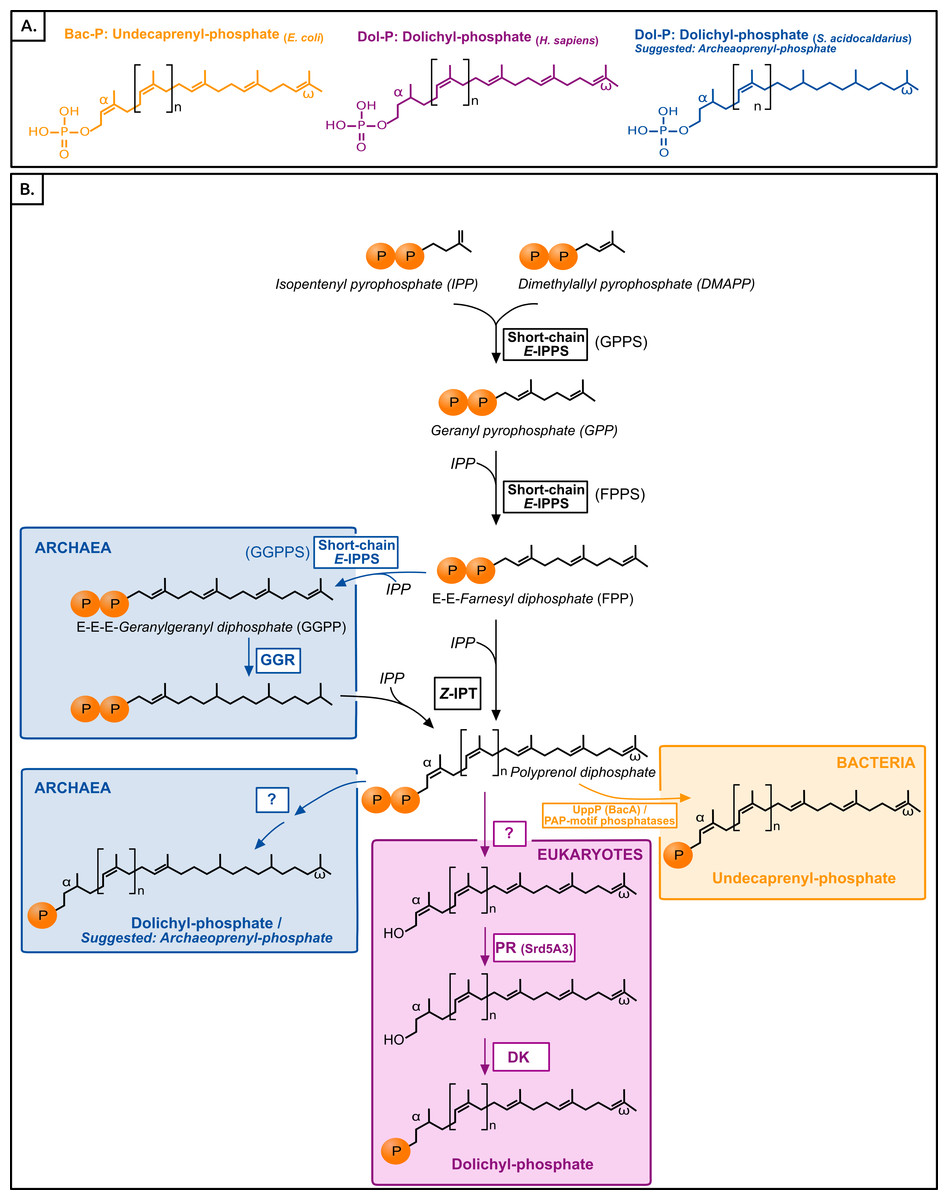
Figure 1:
(A) Typical polyisoprenol lipid carriers in the three domains of life.
(B) Polyisoprenol-phosphate biosynthesis pathways in the three domains of life.
Steps outside boxes are common to all pathways, whereas steps in boxes are characteristic of only one domain of life. Unknown steps are labeled with a question mark. GPPS, geranyl diphosphate synthase; FPPS, farnesyl diphosphate synthase; GGPPS, geranylgeranyl diphosphate synthase; GGR, geranylgeranylreductase; IPT, isopentenyl transferase; UppP, undecaprenyl pyrophosphate phosphatase; PR, polyprenol reductase; DK, dolichol kinase.
The early evolution of lipid membranes and the three domains of life 2
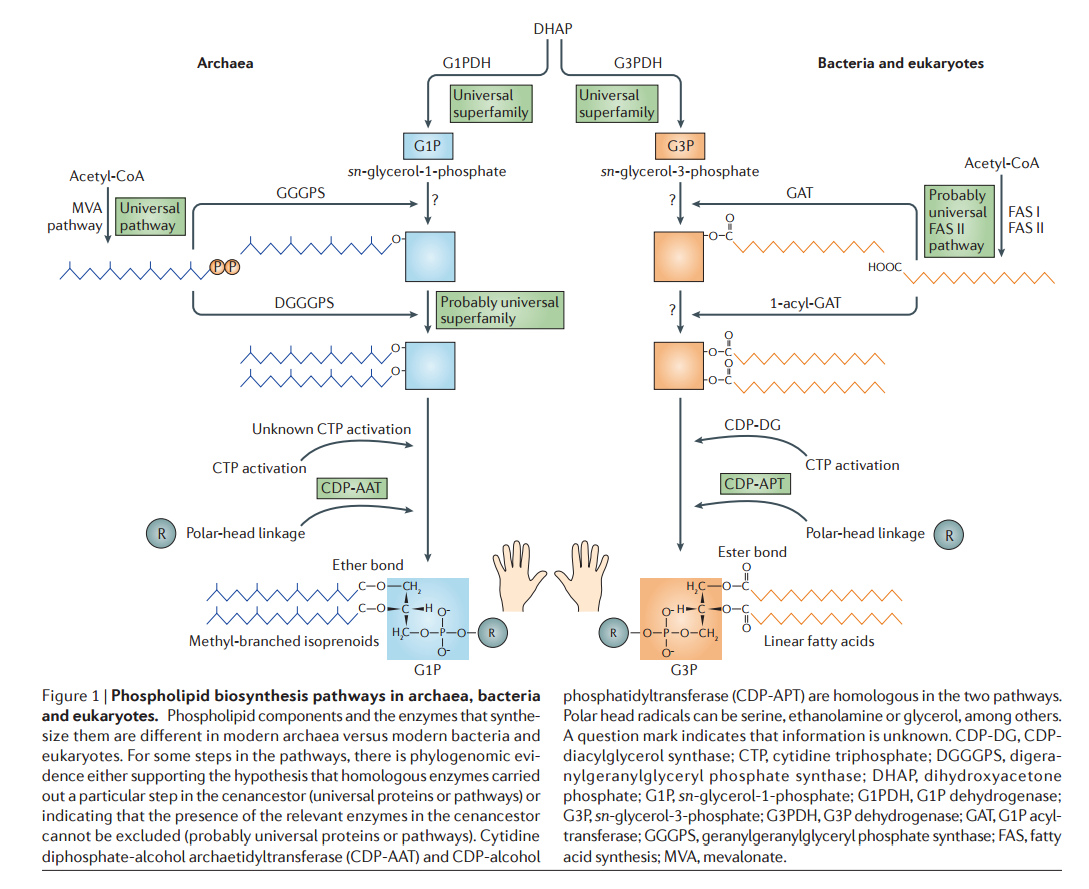
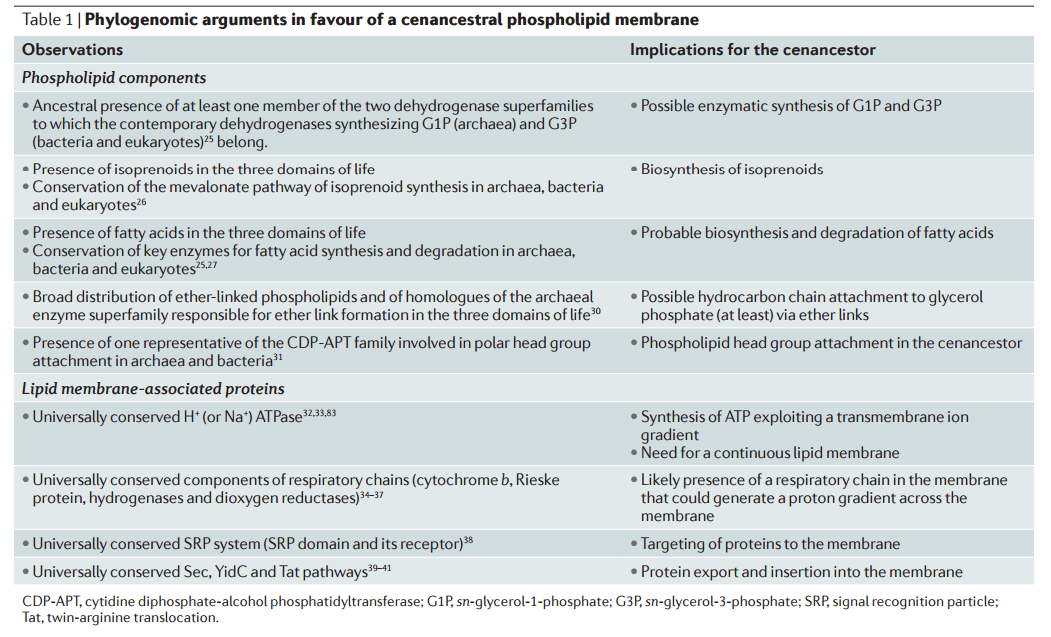
Various hypotheses have addressed the fundamental differences between archaeal and bacterial–eukaryotic phospholipids and, more specifically, the apparently unrelated nature of the pathways that synthesize the two opposed glycerol phosphate stereoisomers Although this hypothesis accounts directly for the differences between archaeal and bacterial membrane phospholipids, it is at odds with the increasing evidence pointing to a complex cenancestor that contained several hundred genes. Such complexity would have required Darwinian evolution (based on natural selection and other evolutionary forces) to operate on individualized entities, a situation that would have been achieved by membrane compartmentation.
The cell membrane had to emerge prior life began, and be fully setup , with all membrane proteins which are essential for the proper working of the cell, fully functioning as well. So the author of the paper tries to insert evolution as the mechanism where it does not belong. And if evolution had to operate on individual entities, cell membranes would have had to emerge separately 3 times individually in a convergent manner. If once is extremely difficult, imagine 3 times... Once more, the evidence brings us very close to the inference, that each animal was created individually by God.
The are fundamental differences between archaeal and bacterial–eukaryotic phospholipids and, more specifically, the apparently unrelated nature of the pathways that synthesize the two opposed glycerol phosphate stereoisomers 13 The asymmetry of the glycerol phosphate stereoisomers — G1P in archaea and G3P in bacteria and eukaryotes —that are synthesized by non-homologous glycerol phosphate dehydrogenases is the only inviolate difference.
If its inviolable, it means the membranes had to emerge separately.
1. https://peerj.com/articles/2626.pdf
2. http://fire.biol.wwu.edu/cmoyer/zztemp_fire/biol345_F13/papers/Lombard_membranes_3domains_natrevmicro12.pdf
3. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4232325/
4. http://jb.asm.org/content/182/4/859
5. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4245038/
6. The Membranes of Cells page 32
The Lipids of Biological Membranes 6
The membranes of cells and viruses, whether they are in prokaryotes, eukaryotes, or in enveloped viruses, are built on the fundamental architecture of a lipid bilayer. All biological membranes contain lipids. The lipids of biological membranes, for the most part, form lipid bilayers in the biological membrane. The structure and resulting chemical properties, along with the hydrophobic effect, of these lipids determine their ability to form bilayers in biological membranes and as purified molecules in aqueous media. Thousands of lipid species are found in biological membranes. Many of their roles in biology have yet to be fully understood. A feature in common among most lipids of biological membranes is their amphipathic structure. Much of the structure of these molecules is hydrophobic, often determined by long hydrocarbon chains of covalently bonded carbon atoms forming one portion of the molecule. The other portion of these molecules is the lipid headgroup. The lipid headgroup is usually polar and often contains charged moieties. A wide variety of chemical structures is found among lipid headgroups. With these polar headgroups at one end and the hydrophobic hydrocarbon chains at the other, the criteria are met for an amphipathic molecule. The amphipathic chemical structure of the lipids is critical to the formation of membranes.
PHOSPHOLIPIDS
A good place to begin the story of the lipids is with a look at the best known and most studied of the lipids of biological membranes, the phospholipids.

Phospholipid structures fit the general description given earlier: phospholipids consist of a polar headgroup on one end of the molecule and hydrocarbon chains on the other end. These chemical structures create an amphipathic lipid. The phospholipids derive their name in part from the phosphate group found in the polar headgroup. The general structure is shown in Fig. 2.1.
Phospholipases
Phospholipids are substrate for a class of enzymes called phospholipases. Phospholipases are enzymes that hydrolyze ( the cleavage of chemical bonds by the addition of water ) phospholipids, separating the polar and one of the hydrophobic portions of the phospholipid molecule, a fatty acid.

These enzymes are useful tools in studies of the structure and composition of biological membranes. Chemists, too, have used phospholipases to assist in the chemical synthesis of phospholipids. Phospholipases play important roles in lipid metabolism. One example is the metabolic remodeling of the phospholipid structure. A metabolic intermediate in the pathways of biosynthesis of phospholipids is diacylglycerol. In this pathway, the diacylglycerol is symmetrical and saturated in its fatty acid content. As already noted, in the mature phospholipid in cell membranes, the dominant phospholipid species have different fatty acids on the 1′ and 2′ positions of the glycerol, with the 2′ position reserved for unsaturated fatty acids. Part of this pathway involves phospholipase A2 that cleaves the ester bond at the 2′ position, allowing a subsequent reacylation of the phospholipid with an unsaturated fatty acid. This results in the common form with a saturated fatty acid at the 1′ position and an unsaturated fatty acid at the 2′ position of the glycerol. The same class of phospholipase can be stimulated
by receptors to enable the cleavage of a particular unsaturated fatty acid, for prostaglandin biosynthesis. Another class of phospholipase creates important second messengers for intracellular communication
Cell Membranes, origins through natural mechanisms, or design ?
http://reasonandscience.heavenforum.org/t2128-membrane-structure#3798
Early evolution of polyisoprenol biosynthesis and the origin of cell walls 1
After being a matter of hot debate for years, the presence of lipid membranes in the last common ancestor of extant organisms (i.e., the cenancestor) now begins to be generally accepted. Here, we provide the first systematic phylogenomic report on the polyisoprenol biosynthesis pathways in the three domains of life. This study shows that, whereas the last steps of the polyisoprenol biosynthesis are unique to the respective domain of life of which they are characteristic, the enzymes required for basic unsaturated polyisoprenol synthesis can be traced back to the respective last common ancestor of each of the three domains of life. As a result, regardless of the topology of the tree of life that may be considered, the most parsimonious hypothesis is that these enzymes were inherited in modern lineages from the cenancestor. This observation supports the presence of an enzymatic mechanism to synthesize unsaturated polyisoprenols in the cenancestor and, since these molecules are notorious lipid carriers in glycosylation pathways involved in the synthesis of a wide diversity of prokaryotic cell walls, it provides the first indirect evidence of the existence of a hypothetical unknown cell wall synthesis mechanism in the cenancestor.
Despite being widespread in archaea, bacteria and eukaryotes, IPP and DMAPP are synthesized by respective pathways in each domain of life (Lombard & Moreira, 2011; Dellas et al., 2013; Vranová, Coman & Gruissem, 2013). A previous study on the evolution of these pathways concluded that the cenancestor had a primitive mevalonate pathway that could have synthesized IPP and DMAPP in this organism, and that each domain of life adopted its specific biosynthesis pathway later on
Or maybe each pathway was created individually by God ?
Cell walls indeed provide essential structural support and external interactions in modern organisms (Albers & Meyer, 2011), Despite the stunning diversity that exists among prokaryotic cell envelopes, the synthesis of many of their main components:
1.N-or O-glycosylated S-layer proteins
2.peptidoglycan,
3.O-antigen LPS
4.teichoic acids
5.exopolysaccharides
relies on comparable glycosylation pathways These pathways are all located in the cell membranes, are mediated by similar lipid carriers and have the same orientation across the membrane.
That raises the question: If in extant cells these pathways are located in the cell membranes, where was the location prior membranes existed ? Would it not be correct to say : It takes membranes to make membranes ?
1. S-layers: principles and applications 3
Monomolecular arrays of protein or glycoprotein subunits forming surface layers (S-layers) are one of the most commonly observed prokaryotic cell envelope components. S-layers are generally the most abundantly expressed proteins, have been observed in species of nearly every taxonomical group of walled bacteria, and represent an almost universal feature of archaeal envelopes. The isoporous lattices completely covering the cell surface provide organisms with various selection advantages including functioning as protective coats, molecular sieves and ion traps, as structures involved in surface recognition and cell adhesion, and as antifouling layers. S-layers are also identified to contribute to virulence when present as a structural component of pathogens. In Archaea, most of which possess S-layers as exclusive wall component, they are involved in determining cell shape and cell division.
Introduction
With the exception of those prokaryotic organisms which have developed strategies to live under very specialized and frequently extreme environmental conditions in which monocultures are feasible, most organisms have to survive in highly competitive habitats in very complex microbiomes. Consequently, the diversity observed in the molecular architecture of bacterial and archaeal cell envelopes, particularly the structure of the outermost boundary layers, reflects evolutionary adaptations of the organism to specific environmental and ecological conditions.
Among the most commonly observed prokaryotic cell surface structures are two-dimensional arrays of proteinaceous subunits forming surface layers (termed S-layers) on prokaryotic cells (Sleytr, 1976; Sleytr et al., 1988b; TableTable1).1). Since the first ‘macromolecular monolayer’ described by Houwink and Le Poole (1952; Houwink, 1953) in the cell wall of a Spirillum sp., S-layers have now been identified in hundreds of different species of almost every taxonomic group of walled Bacteria and are an almost universal feature of Archaea .
Because S-layer proteins account for approximately ten percent of cellular proteins in Archaea and Bacteria, they represent interesting model systems for studying the processes involved in the synthesis, secretion, and assembly of extracellular proteins. Moreover, as the biomass of prokaryotic organisms surpasses the biomass of eukaryotic organism (Whitman et al., 1998), S-layer proteins can be considered as one of the most abundant biopolymers on our planet. S-layers also represent the simplest biological protein or glycoprotein membranes (Sleytr, 1975). They are generally composed of a single molecular species endowed with the ability to assemble on the cell surface into closed regular arrays occupying a low free-energy arrangement. Studies on the in vivo morphogenesis of S-layers demonstrated that at high growth rates, approximately 500 subunits per second must be synthesized, translocated to the cell surface, and incorporated into the existing S-layer lattice (Sleytr & Messner, 1983; Sleytr & Beveridge, 1999). It is now evident that S-layers as metabolic expensive products can provide organisms with an advantage of selection in quite diverse habitats. Although a considerable amount of knowledge has accumulated on the structure, assembly, chemistry, and genetics of S-layers, relatively little firm data are available about their specific biological functions (Sleytr et al., 2002, 2007b; Hynonen & Palva, 2013). It is now recognized that they can function as protective coats, molecular sieves, molecule and ion traps, promoters for cell adhesion, immunomodulators, surface recognition, antifouling coatings, and virulence factors in pathogenic organisms. In those Archaea that possess S-layers as the exclusive envelope component external to the cytoplasmic membrane, the lattice is involved in the determination of cell shape and as a structure aiding in the cell division process.
The S-layer glycoprotein is not just a standardized building block used to generate the two-dimensional lattice of the S-layer but rather that S-layer glycoproteins undergo a variety of post-translational modifications. 5

Schematic illustration of the supramolecular architecture of the major classes of prokaryotic cell envelopes containing surface (S) layers.
S-layers in archaea with glycoprotein lattices as exclusive wall component are composed either of mushroom-like subunits with pillar-like, hydrophobic trans-membrane domains (a), or lipid-modified glycoprotein subunits (b). Individual S-layers can be composed of glycoproteins possessing both types of membrane anchoring mechanisms. Few archaea possess a rigid wall layer (e.g. pseudomurein in methanogenic organisms) as intermediate layer between the plasma membrane and the S-layer (c). In Gram-positive bacteria, (d) the S-layer (glyco)proteins are bound to the rigid peptidoglycan-containing layer via secondary cell wall polymers. In Gram-negative bacteria, (e) the S-layer is closely associated with the lipopolysaccharide of the outer membrane.

Schematic drawing of the different S-layer lattice types, their base vectors, the unit cell (shaded in gray), and the corresponding symmetry axis. The proteins at one morphological unit are shown in red.
The glycan chain and linkages of bacterial and archaeal glycoproteins are significantly different from those of eukaryotes .
The lipid carrier molecules also differ between Archaea and Bacteria. In Archaea, isoprene-based lipids such as dolichol phosphate and dolichol pyrophosphate play essential roles in the N-glycosylation process by delivering their bound glycan cargo to selected protein targets.
How was it established which target is the correct one ? how did the communication system emerge ?

S-Layer Proteins 4
Over the past 3 decades of research, it has become apparent that one of the most common surface structures on archaea and bacteria are monomolecular crystalline arrays of proteinaceous subunits termed surface layers or S-layers. S-layers represent one of the most abundant cellular proteins, it is now obvious that these metabolically expensive products must provide the organisms with an advantage of selection in very different habitats
S-layers have now been identified in hundreds of different species belonging to all major phylogenetic groups of bacteria, and they represent a feature common to almost all archaea. S-layer proteins of many archaea and those of gram-positive bacteria can possess covalently bound carbohydrate chains. The glycan chains are composed of 20 to 50 identical repeating units containing neutral hexoses, pentoses, heptoses, 6-deoxyhexoses, and amino sugars. Core oligosaccharides of S-layer glycoproteins consist of two to four sugar residues attaching the carbohydrate chains mostly via O-glycosidic linkages (galactose–tyrosine, glucose–tyrosine,N-acetylgalactosamine–serine, N-acetyl galactosamine–threonine) to the protein moiety.
Their respective mechanisms can be described in three major steps:
(1) synthesis of an oligomer on a lipid carrier in the cytoplasmic side of the cell membranes;
(2) ‘‘flipping’’ of the oligomer-linked lipid carrier to the opposite side of the membrane; and
(3) oligomer transfer from the lipid carrier to the acceptor molecule (e.g., protein or external polymer)
The proteins involved in these glycosylation pathways belong to a small number of protein superfamilies, some of which could be traced back to the cenancestor (Lombard, 2016). the prokaryotic cell wall synthesis mechanisms have at least one element in common: they all use polyisoprenol phosphate lipid carriers (Hartley & Imperiali, 2012).

Figure 1:
(A) Typical polyisoprenol lipid carriers in the three domains of life.
(B) Polyisoprenol-phosphate biosynthesis pathways in the three domains of life.
Steps outside boxes are common to all pathways, whereas steps in boxes are characteristic of only one domain of life. Unknown steps are labeled with a question mark. GPPS, geranyl diphosphate synthase; FPPS, farnesyl diphosphate synthase; GGPPS, geranylgeranyl diphosphate synthase; GGR, geranylgeranylreductase; IPT, isopentenyl transferase; UppP, undecaprenyl pyrophosphate phosphatase; PR, polyprenol reductase; DK, dolichol kinase.
The early evolution of lipid membranes and the three domains of life 2
Various hypotheses have addressed the fundamental differences between archaeal and bacterial–eukaryotic phospholipids and, more specifically, the apparently unrelated nature of the pathways that synthesize the two opposed glycerol phosphate stereoisomers Although this hypothesis accounts directly for the differences between archaeal and bacterial membrane phospholipids, it is at odds with the increasing evidence pointing to a complex cenancestor that contained several hundred genes. Such complexity would have required Darwinian evolution (based on natural selection and other evolutionary forces) to operate on individualized entities, a situation that would have been achieved by membrane compartmentation.
The cell membrane had to emerge prior life began, and be fully setup , with all membrane proteins which are essential for the proper working of the cell, fully functioning as well. So the author of the paper tries to insert evolution as the mechanism where it does not belong. And if evolution had to operate on individual entities, cell membranes would have had to emerge separately 3 times individually in a convergent manner. If once is extremely difficult, imagine 3 times... Once more, the evidence brings us very close to the inference, that each animal was created individually by God.
The are fundamental differences between archaeal and bacterial–eukaryotic phospholipids and, more specifically, the apparently unrelated nature of the pathways that synthesize the two opposed glycerol phosphate stereoisomers 13 The asymmetry of the glycerol phosphate stereoisomers — G1P in archaea and G3P in bacteria and eukaryotes —that are synthesized by non-homologous glycerol phosphate dehydrogenases is the only inviolate difference.
If its inviolable, it means the membranes had to emerge separately.


1. https://peerj.com/articles/2626.pdf
2. http://fire.biol.wwu.edu/cmoyer/zztemp_fire/biol345_F13/papers/Lombard_membranes_3domains_natrevmicro12.pdf
3. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4232325/
4. http://jb.asm.org/content/182/4/859
5. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4245038/
6. The Membranes of Cells page 32


