https://reasonandscience.catsboard.com/t2371-how-cellular-enzymatic-and-metabolic-networks-point-to-design
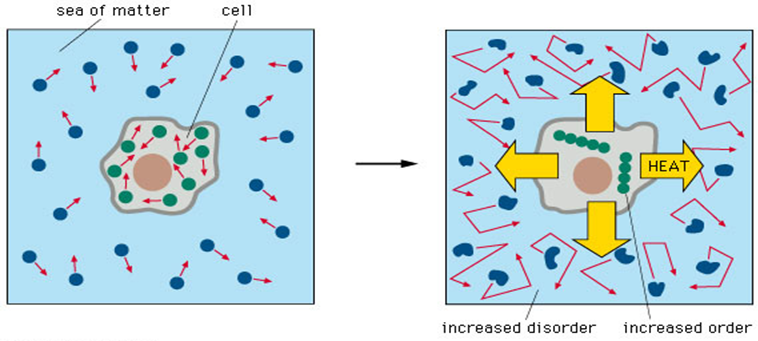
The development of the metabolic system, which, as the primordial soup thinned, must have "learned" to mobilize chemical potential and to synthesize the cellular components, poses Herculean problems.
J.Monod Chance and Necessity: An Essay on the Natural Philosophy of Modern Biology 1972
1. Cells contain high information content that directs and controls integrated metabolic pathways which if altered are inevitably damaged or destroy their function. They also require regulation and are structured in a cascade manner, similar to electronic circuit boards.
2. There is always an observable consequence if a circuit is interrupted. Since these consequences are always catastrophically bad, flexibility is minimal, and since the circuits are all interconnected, the whole network partakes in the quality that there is only one way for things to work. ( Davidson)
3. Naturalistic mechanisms or undirected causes do not suffice to explain the origin and set up of information (instructional prescribing complex information), integrated complex circuits with little tolerance of change.
4. There is no way to write the code for all the enzymes unless one knows the 3D shapes of the substrates they act upon, and one can't know this unless one sees "the big picture" of the context within which and WHY they are needed for each life-essential product (or the final end products would not be produced), it becomes very clear that believing it could "evolve" without deliberate planning, foreknowledge, etc. stretches plausibility, reason, and logic, to say the least. Therefore, intelligent design constitutes the best explanation for the origin of these systems.
The argument of an intelligent designer required to set up the Metabolic Networks for the origin of life
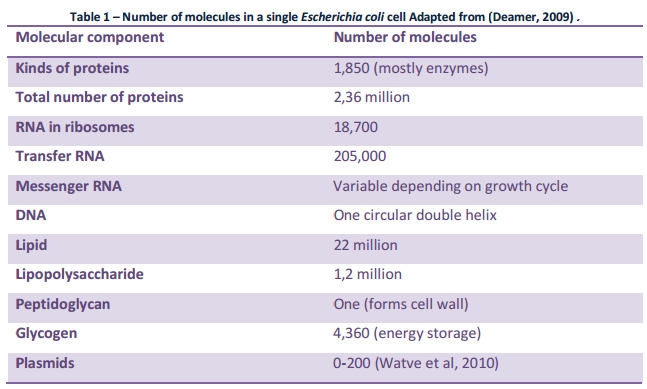
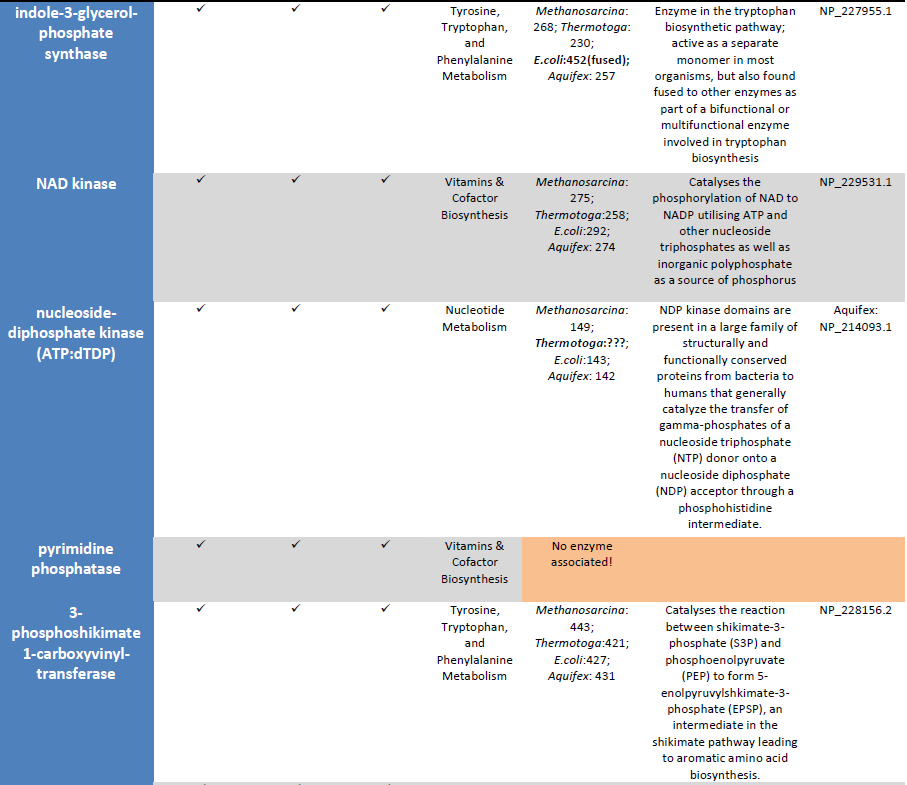
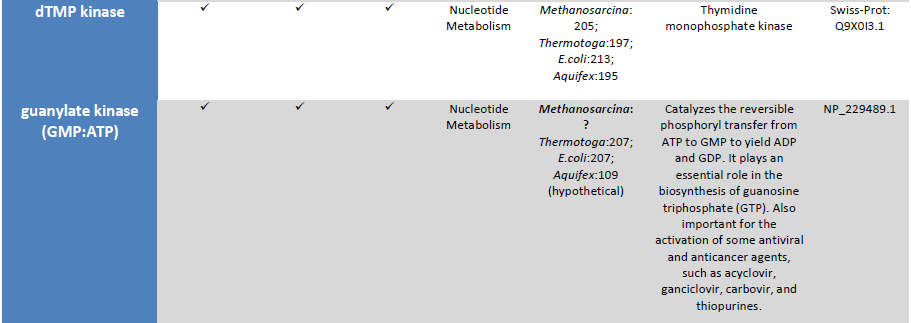
Observation: The existence of metabolic pathways is crucial for molecular and cellular function. Although bacterial genomes differ vastly in their sizes and gene repertoires, no matter how small, they must contain all the information to allow the cell to perform many essential (housekeeping) functions that give the cell the ability to maintain metabolic homeostasis, reproduce, and evolve, the three main properties of living cells. Gil et al. (2004) In fact, metabolism is one of the most conserved cellular processes. By integrating data from comparative genomics and large-scale deletion studies, the paper "Structural analyses of a hypothetical minimal metabolism" propose a minimal gene set comprising 206 protein-coding genes for a hypothetical minimal cell. The paper lists 50 enzymes/proteins required to create a metabolic network implemented by a hypothetical minimal genome for the hypothetical minimal cell. The 50 enzymes/proteins, and the metabolic network, must be fully implemented to permit a cell to keep its basic functions.
Hypothesis (Prediction): The origin of biological irreducible metabolic pathways that also require regulation and which are structured like a cascade, similar to electronic circuit boards, are best explained by the creative action of an intelligent agent.
Experiment: Experimental investigations of metabolic networks indicate that they are full of nodes with enzymes/proteins, detectors, on/off switches, dimmer switches, relay switches, feedback loops etc. that require for their synthesis information-rich, language-based codes stored in DNA. Hierarchical structures have been proved to be best suited for capturing most of the features of metabolic networks (Ravasz et al, 2002). It has been found that metabolites can only be synthesized if carbon, nitrogen, phosphor, and sulfur and the basic building blocks generated from them in central metabolism are available.
This implies that regulatory networks gear metabolic activities to the availability of these basic resources. So one metabolic circuit depends on the product of other products, coming from other, central metabolic pathways, one depending on the other, like in a cascade. Further noteworthy is that Feedback loops have been found to be required to regulate metabolic flux and the activities of many or all of the enzymes in a pathway. In many cases, metabolic pathways are highly branched, in which case it is often necessary to alter fluxes through part of the network while leaving them unaltered or decreasing them in other parts of the network (Curien et al., 2009). These are interconnected in a functional way, resulting in a living cell. The biological metabolic networks are exquisitely integrated, so the significant alterations in inevitably damage or destroys the function. Changes in flux often require changes in the activities of multiple enzymes in a metabolic sequence. Synthesis of one metabolite typically requires the operation of many pathways.
Conclusion: Regardless of its initial complexity, self-maintaining chemical-based metabolic life could not have emerged in the absence of a genetic replicating mechanism ensuring the maintenance, stability, and diversification of its components. In the absence of any hereditary mechanisms, autotrophic reaction chains would have come and gone without leaving any direct descendants able to resurrect the process. Life as we know it consists of both chemistry and information. If metabolic life ever did exist on the early Earth, to convert it to life as we know it would have required the emergence of some type of information system under conditions that are favorable for the survival and maintenance of genetic informational molecules. ( Ribas de Pouplana, Ph.D.)
Progress in dissecting signaling pathways has begun to layout a circuitry that will likely mimic electronic integrated circuits in complexity and finesse, where transistors are replaced by proteins (e.g., kinases and phosphatases) and the electrons by phosphates and lipids. […] Two decades from now, having fully charted the wiring diagrams of every cellular signaling pathway, it will be possible to layout the complete ‘integrated circuit of the cell’ upon its current outline. We will then be able to apply the tools of mathematical modeling to explain how specific genetic lesions serve to reprogram this integrated circuit in each of the constituent cell types so as to manifest cancer. ( Hanahan and Weinberg, 20 0 0 , p. 59, 67)
Intelligent agents have frequently end goals in mind and use high levels of instructional complex information to meet the goal. In our experience, systems storing large amounts of specified/instructional complex information through codes and languages -- invariably originate from an intelligent source. Likewise, circuits or networks of coordinated interaction as for example of analog electronic devices can always be traced back to an intelligent causal agent. The operation of analog electronic devices maps very closely to the flow of information in chemical reactions of metabolic pathways (McAdams and Shapiro, 1995). A proposed mechanism to make metabolical networks must be capable of construct de novo, not merely modifying, a minimal set of 50 enzymes, and complex integrated metabolic circuits with the end goal to create life. A metabolic network that is not fully operational, will not permit life. We know in our experience that intelligence is able to setup circuit boards, like discrete electronic boards, and is the only known cause of irreducibly complex machines. Since evolution depends on metabolic circuits fully setup, its excluded as possible mechanism. The only two alternatives, chance/luck or physical necessity have never been observed to be able to setup circuit boards and irreducible complex systems. The origin of the basic metabolical network of the first cells is therefore best explained through the action of a intelligent agency.
The existence of metabolic pathways is crucial for molecular and cellular function. In fact, metabolism is one of the most conserved cellular processes; it is recognized that very little is known about how the chemistry of primitive enzymes arose (Perez-Jimenez et al, 2011) and which were the first enzymes appearing. Building a new living cell requires not just new genes and proteins, but at least nine different metabolic networks which are essential, and irreducible In this paper, a minimum of 14 sets are mentioned :
https://repositorio-aberto.up.pt/bitstream/10216/62065/1/000149653.pdf
These are highly complex multibranched, noded anabolic, metabolic and catabolic systems, which are functionally critical, and individually not able to turn a cell alive. Furthermore, metabolic networks support growth, the synthesis or turnover of storage compounds, or the accumulation of metabolites that have a role in coping with abiotic or biotic stress . Metabolism has been divided into discrete pathways, but we know now that it operates as a highly integrated network (Sweetlove et al., 2008). Metabolites are not synthesized in isolation from each other; rather, large sets of metabolites must be synthesized simultaneously. Hierarchical structures have been proved to be best suited for capturing most of the features of metabolic networks (Ravasz et al, 2002) Metabolites can only be synthesized if carbon, nitrogen, phosphor, and sulfur and the basic building blocks generated from them in central metabolism are available. This implies that regulatory networks gear metabolic activities to the availability of these basic resources. So one metabolic circuit depends on the product of other products, coming from other, central metabolic pathways, one depending from the other, like in a casacade. Further noteworthy is that Feedback loops are required to regulate metabolic flux, and the activities of many or all of the enzymes in a pathway. In many cases, metabolic pathways are highly branched, in which case it is often necessary to alter fluxes through part of the network while leaving them unaltered or decreasing them in other parts of the network (Curien et al., 2009). One of the most basic principles of engineering is the principle of constraints. Engineers have long understood that the more functionally integrated a system is, the more difficult it is to change any part of it without damaging or destroying the system as a whole. The biological metabolic networks are exquisitely integrated, so the significant alterations in inevitably damage or destroys the funcion. ( S.C.Meyer, Darwin's Doubt ) Changes in flux often require changes in the activities of multiple enzymes in a metabolic sequence. Synthesis of one metabolite typically requires the operation of many pathways. All of them have to be present in the first living cell, correctly interconnected and noded to provide function, internal and external communication, and the biosynthesis of various essential products and parts. These are circuits or networks of coordinated interaction, much like integrated circuits on a circuitboard. Metabolic networks work like electric circuits. The operation of analog electronic devices maps very closely to the flow of information in chemical reactions (McAdams and Shapiro, 1995). Life could not emerge without it. A proposed mechanism must be capable of constructing, not merely modifying, complex integrated metabolic circuits. The requirements for constructing the first living cells de novo cannot be done through evolution, since evolution depends on these basic initial networks, fully operational. And since there is no function for a unfinished metabolic network, then how could it ever emerge ? The only two mechanisms that remain to explain its origin if intelligent design is excluded, is chance/luck/self organisation, or physical necessity. We know of intelligence being able to construct electric circuits all the time, and even self replicating machines ( even if only experimentally , since extremely complex ). We do not know of lucky accidents with the same capacity, nor physical needs or physical/chemical laws. We can infer therefore confidently, that the origin of metabolic networks to create the first living cell was most probably designed.
To setup of a cell metabolic network, many different proteins/enzymes are required, correctly interconnected to provide function. Yet the individual enzymes or physical/chemical laws do not contain by themself the information of how to connect and interwine in the correct order that result in a functional metabolic circuit. Furthermore, the mechanism must be capable of construct from zero, not merely modifying, complex integrated circuits. The requirements for constructing the first living cells cannot be explained through evolution, since evolution depends on these networks fully operational. These are circuits or networks of coordinated interaction, much like integrated circuits on a circuitboard. Metabolic networks work like electric circuits. The operation of analog electronic devices maps very closely to the flow of information in chemical reactions. We know intelligence is able to setup circuit boards. There is no function for a unfinished metabolic network, which makes it extremely unlikely that new metabolic and catabolic networks would arise naturally, in non-guided manner.
Strong Irreducible Complexity of Molecular Machines and Metabolic Pathways. 5 For certain enzymes (which are themselves highly complicated molecular structures) and metabolic pathways (i.e., systems of enzymes where one enzyme passes off its product to the next, as in a production line), simplification leads not to different functions but to the complete absence of all function. Systems with this feature exhibit a strengthened form of irreducible complexity. Strong irreducible complexity, as it may be called, entails that no Darwinian account can in principle be given for the emergence of such systems. Theodosius Dobzhansky, one of the founders of the neo-Darwinian synthesis, once remarked that to talk about prebiotic natural selection is a contradiction in terms—the idea being that selection could only select for things that are already functional. Research on strong irreducible complexity finds and analyzes biological systems that cannot in principle be grist for natural selection’s mill. For this research, which is only now beginning, to be completely successful would imply the unraveling of molecular Darwinism.
The Implausibility of Metabolic Cycles on the Prebiotic Earth 3
Leslie E Orgel†
Although metabolism-first avoids the infeasibility of forming functional RNA by chance, "replication of compositional information is so inaccurate that fitter compositional genomes cannot be maintained by selection and, therefore, the system lacks evolvability (i.e., it cannot substantially depart from the asymptotic steady-state solution already built-in in the dynamical equations). We conclude that this fundamental limitation of ensemble replicators cautions against metabolism-first theories of the origin of life" [44]. Concerning the chemical cycles required, "These are chemically very difficult reactions ... One needs, therefore, to postulate highly specific catalysts for these reactions. It is likely that such catalysts could be constructed by a skilled synthetic chemist, but questionable that they could be found among naturally occurring minerals or prebiotic organic molecules. The lack of a supporting background in chemistry is even more evident in proposals that metabolic cycles can evolve to 'life-like' complexity. The most serious challenge to proponents of metabolic cycle theories—the problems presented by the lack of specificity of most non-enzymatic catalysts—has, in general, not been appreciated. If it has, it has been ignored. Theories of the origin of life based on metabolic cycles cannot be justified by the inadequacy of competing theories: they must stand on their own"
Hugh Ross , origin of life page 39
Metabolism first.
Metabolism-first proponents maintain that mineral surfaces catalyzed the formation of a diverse collection of small molecules that, with time, evolved to form an interconnected series of chemical reactions. Once in place, these interrelated chemical reactions formed the basis for the cell’s metabolic systems.21 These chemical networks eventually became encapsulated to form protocells complete with a form of protometabolism. Some metabolism-first scenarios, like the iron-sulfur world, even suggest that minerals (for example, pyrite) became encapsulated along with the protometabolic networks and thereby served as life’s first catalysts. According to the metabolism-first idea, once protometabolic systems were established, they spawned self-replicating molecules.
The Genetic Code and the Origin of Life
The metabolism theory claims that life, at least in its beginnings, was nothing more than a continuous chain of mineral surface-associated self-sustaining chemical reactions with no requirement for genetic information. A primitive type of reductive citric acid cycle is often cited as a model. There is some experimental support for the hypothesis, although the conditions for the various individual reaction steps are very different, it remains to be established if the conditions used in these laboratory experiments are geophysically plausible and are therefore relevant to the origin of life. Regardless of its initial complexity, self-maintaining chemical-based metabolic life could not have evolved in the absence of a genetic replicating mechanism insuring the maintenance, stability, and diversification of its components. In the absence of any hereditary mechanisms, autotrophic reaction chains would have come and gone without leaving any direct descendants able to resurrect the process. Life as we know it consists of both chemistry and information. Life as we know it consists of both chemistry and information. If metabolic life ever did exist on the early Earth, to convert it to life as we know it would have required the emergence of some type of information system under conditions that are favorable for the survival and maintenance of genetic informational molecules.
At a construction site, builders will make use of many materials: lumber, wires, nails, drywall, piping, and windows. Yet building materials do not determine the floor plan of the house or the arrangement of houses in a neighborhood. Similarly, electronic circuits are composed of many components, such as resistors, capacitors, and transistors. But such lower-level components do not determine their own arrangement in an integrated circuit (see Fig. 14.2).
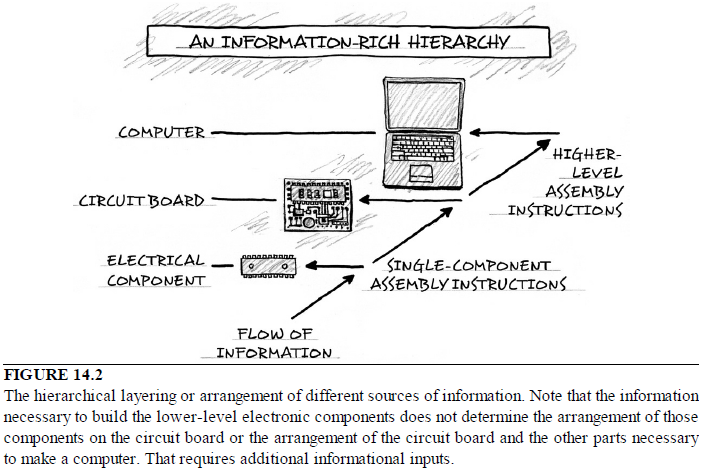
In particular, the cause must be capable of constructing, not merely modifying, complex integrated metabolic circuits. The requirements for constructing the first living cells de novo cannot be accommodated by microevolutionary or macroevolutionary theory, since evolution depends on these networks fully operational. And since there is no function for a unfinished metabolic network, then how would new metabolic and catabolic networks ever arise?
Integrated circuits in electronics are systems of individually functional components such as transistors, resistors, and capacitors that are connected together to perform an overarching function. Likewise, the functional enzymes and proteins of metabolic and anabolic networks, also form an integrated circuit, one that contributes to accomplishing the overall function of producing a working functional cell.
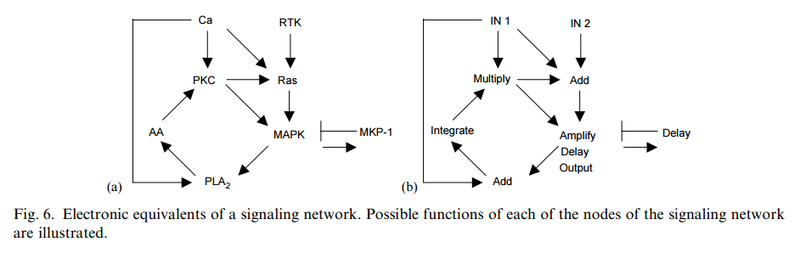
Understanding complex signaling networks through models and metaphors 1
The operation of analog electronic devices maps very closely to the flowof information in chemical reactions (McAdams and Shapiro, 1995).
The buildup of charge, for example, is analogous to the accumulation of a particular molecule. Amplifiers, like enzymes, permit a small charge (or molecular concentration) to have a large effect on another. The identity of signals in an electronic circuit is maintained by distinct, insulated wires. In signaling circuits this identity is maintained by the fact that distinct molecules convey different signals. Even at a mathematical level, the equations describing equilibration of charge and amplifier function can faithfully mimic the equations describing chemical reactions.
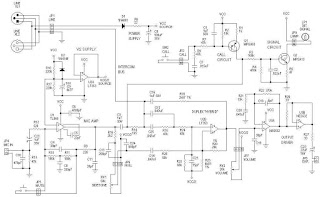
Following shows the minimal metabolic network that was required in the first supposed last universal common ancestor.
The Enzymatic and Metabolic Capabilities of Early Life
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0039912#pone.0039912-Srinivasan1
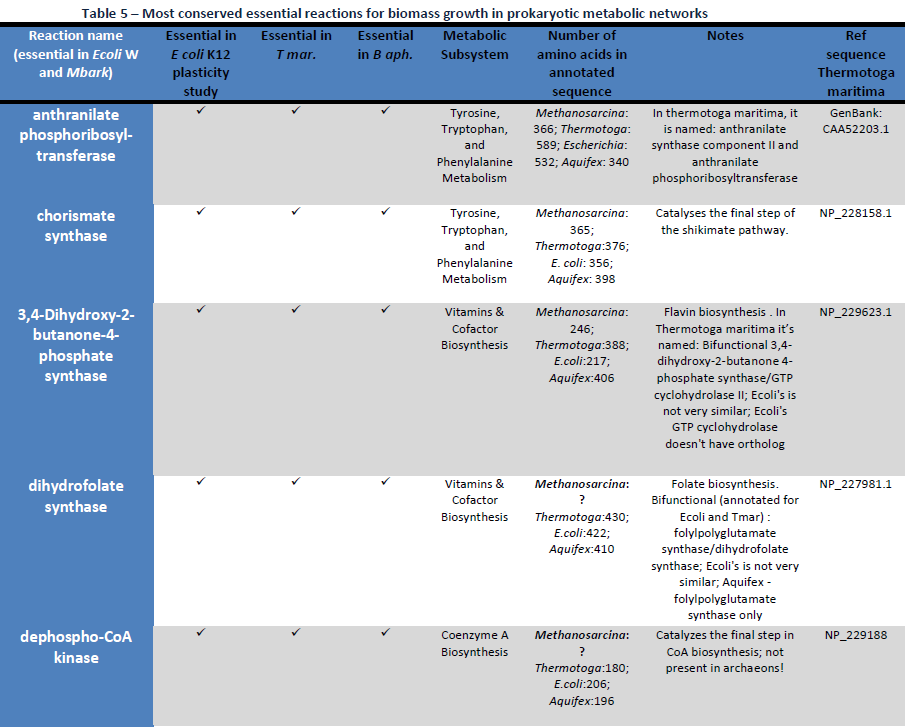
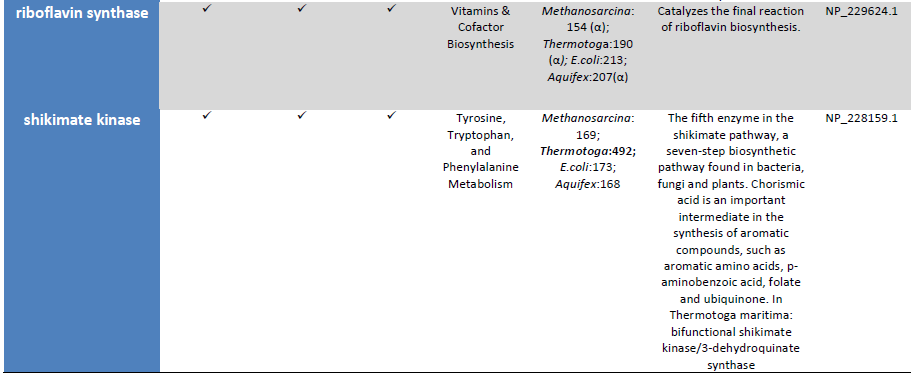
We reconstruct a representative metabolic network that may reflect the core metabolism of early life forms. Our results show that ten enzyme functions, four hydrolases, three transferases, one oxidoreductase, one lyase, and one ligase, are determined by metaconsensus to be present at least as late as the last universal common ancestor. Subnetworks within central metabolic processes related to sugar and starch metabolism, amino acid biosynthesis, phospholipid metabolism, and CoA biosynthesis, have high frequencies of these enzyme functions.
Link to the respective Keggs database
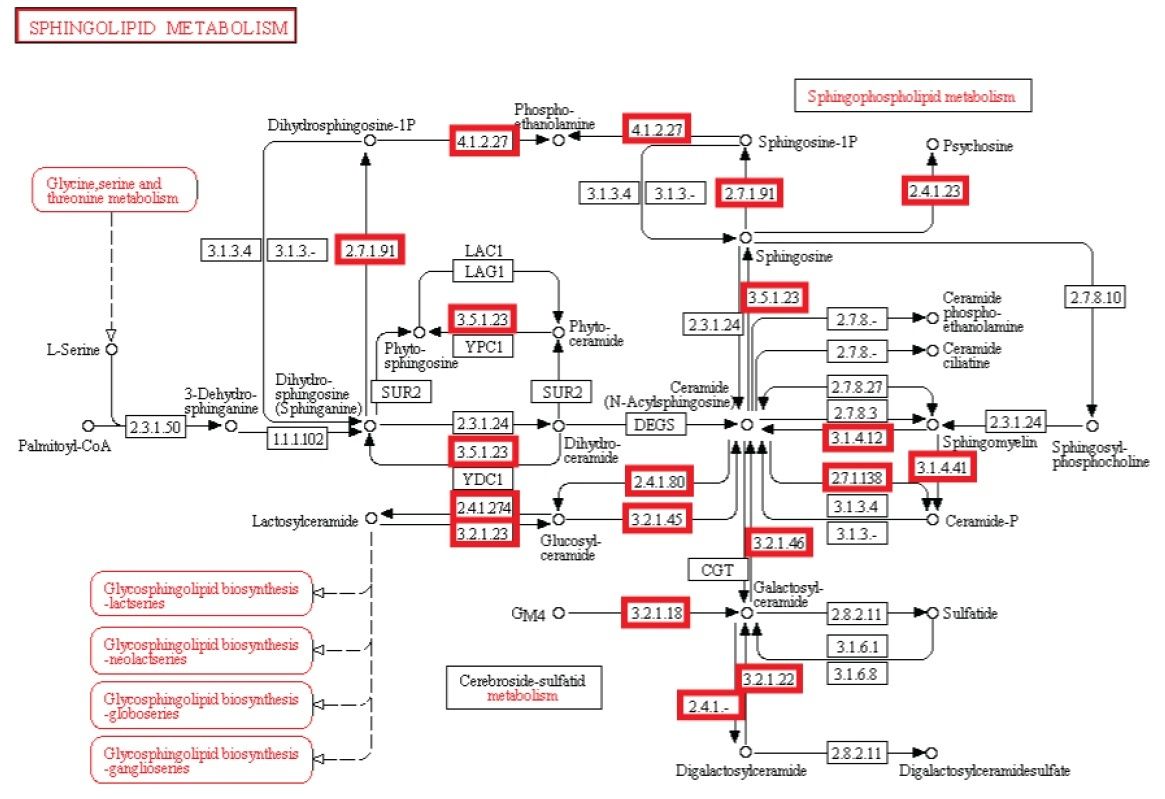
Sphingolipid metabolism
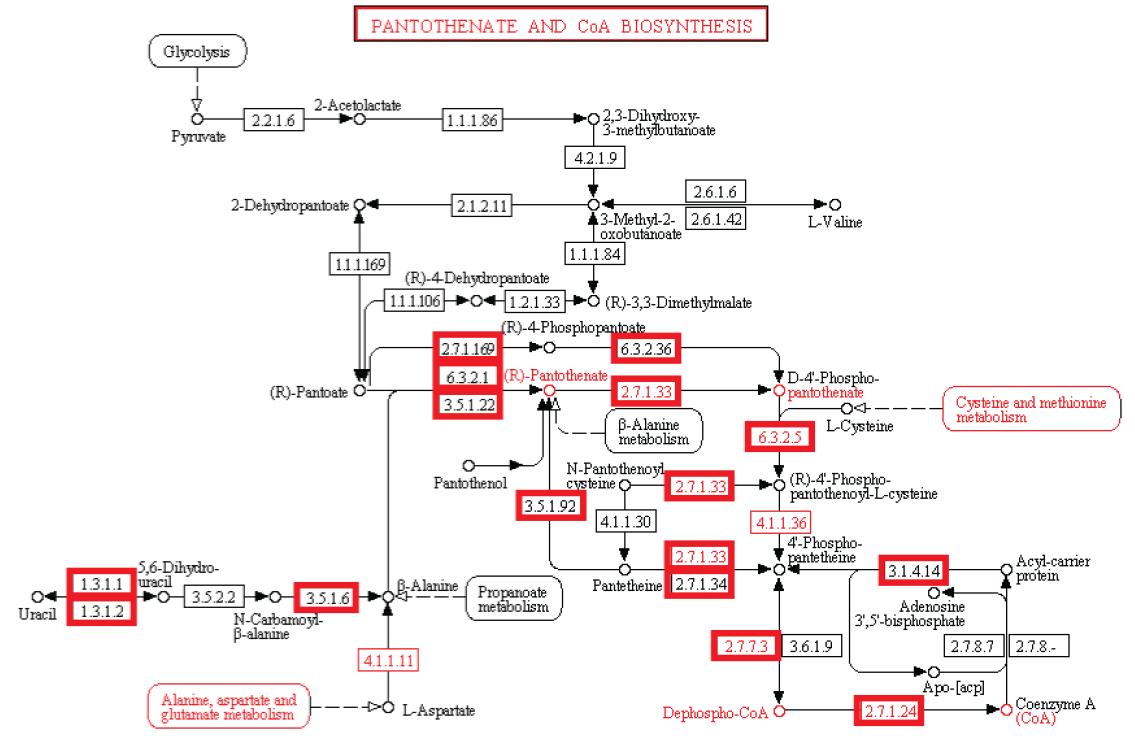
Pantothenate and CoA biosynthesis
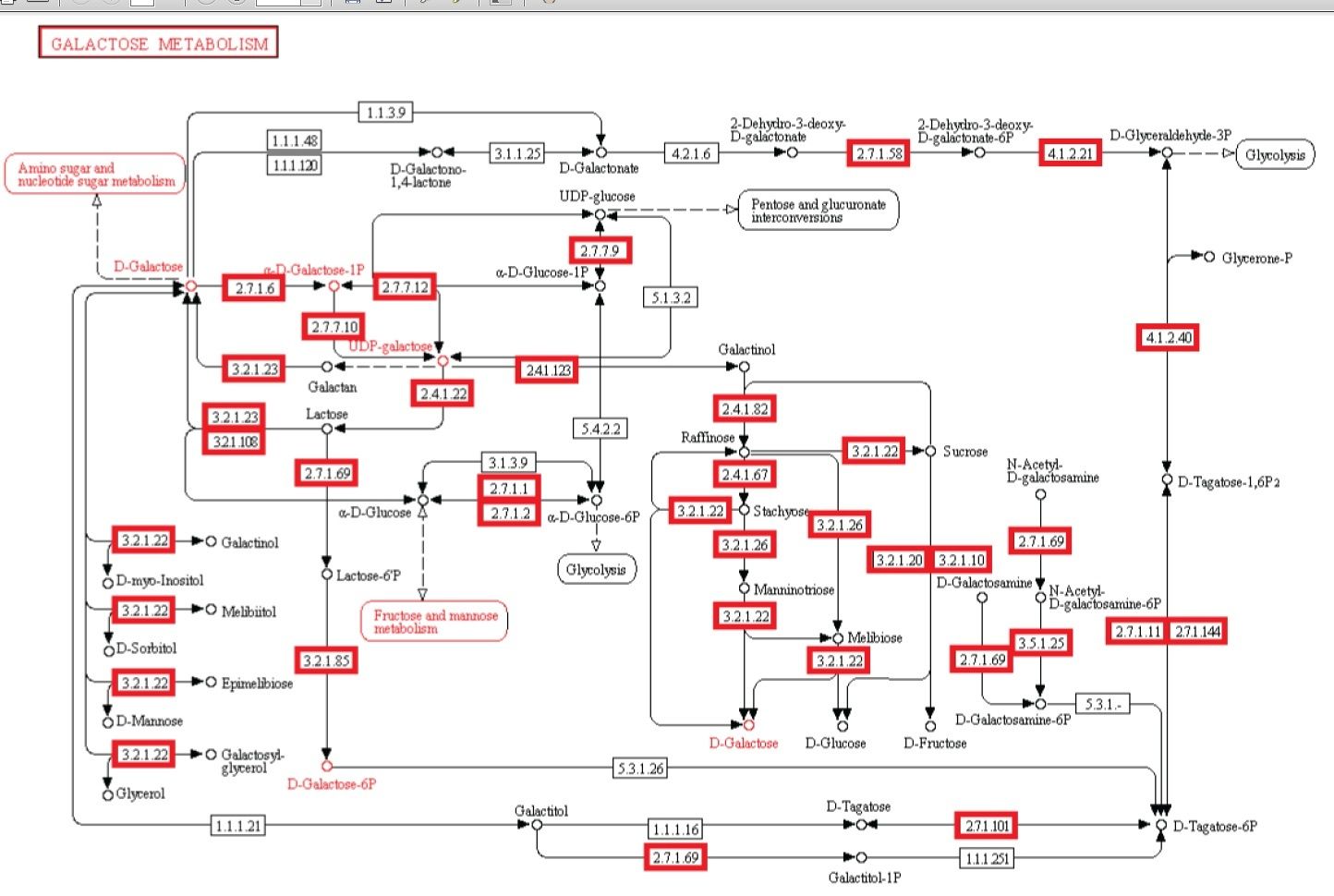
Galactose metabolism
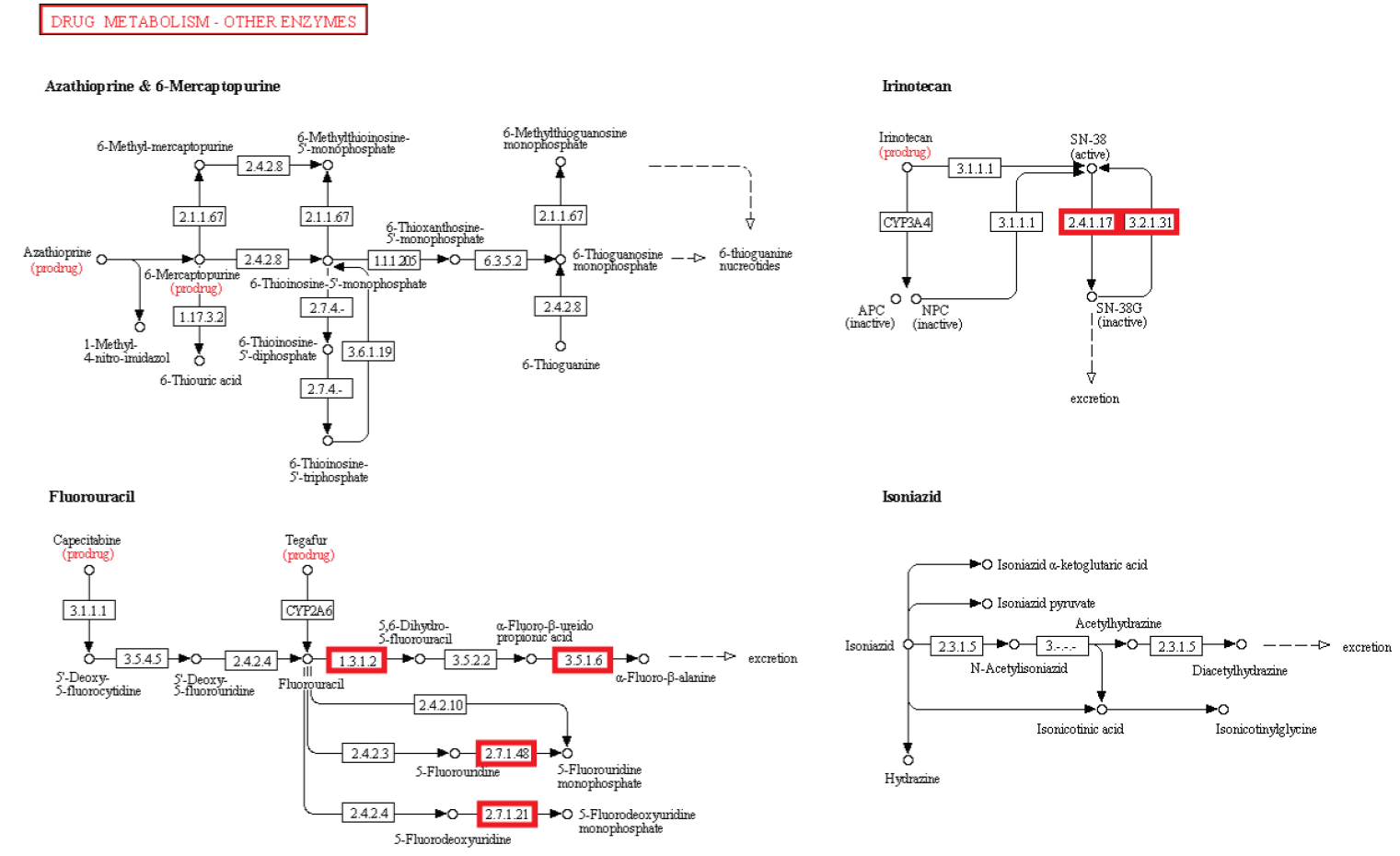
Drug metabolism - other enzymes
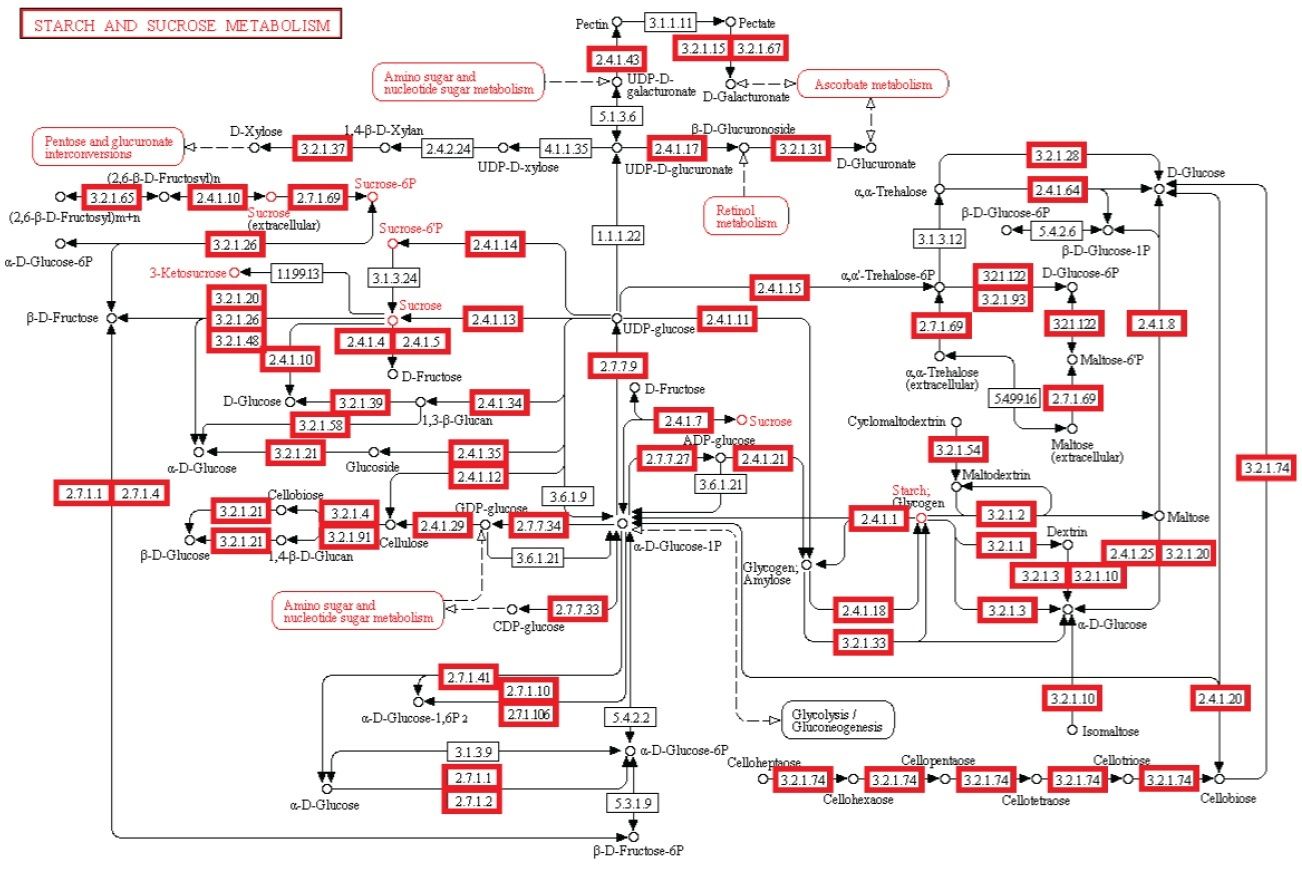
Starch and sucrose metabolism
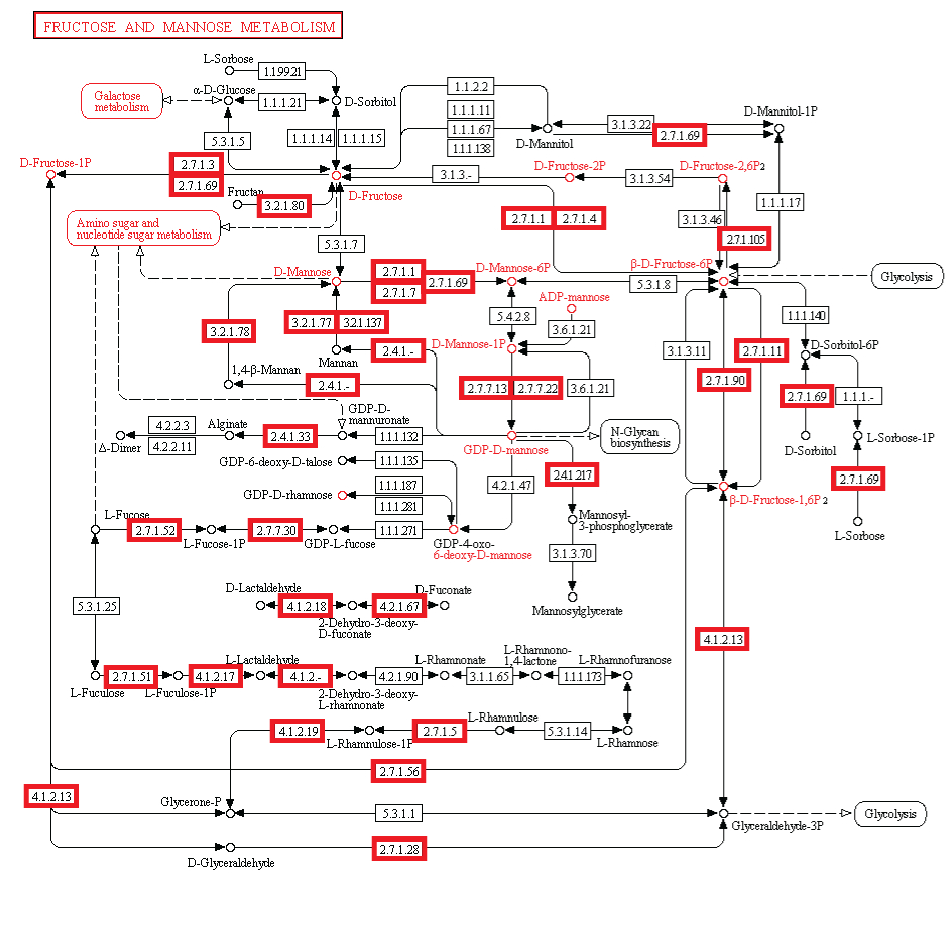
fructose and mannose metabolism
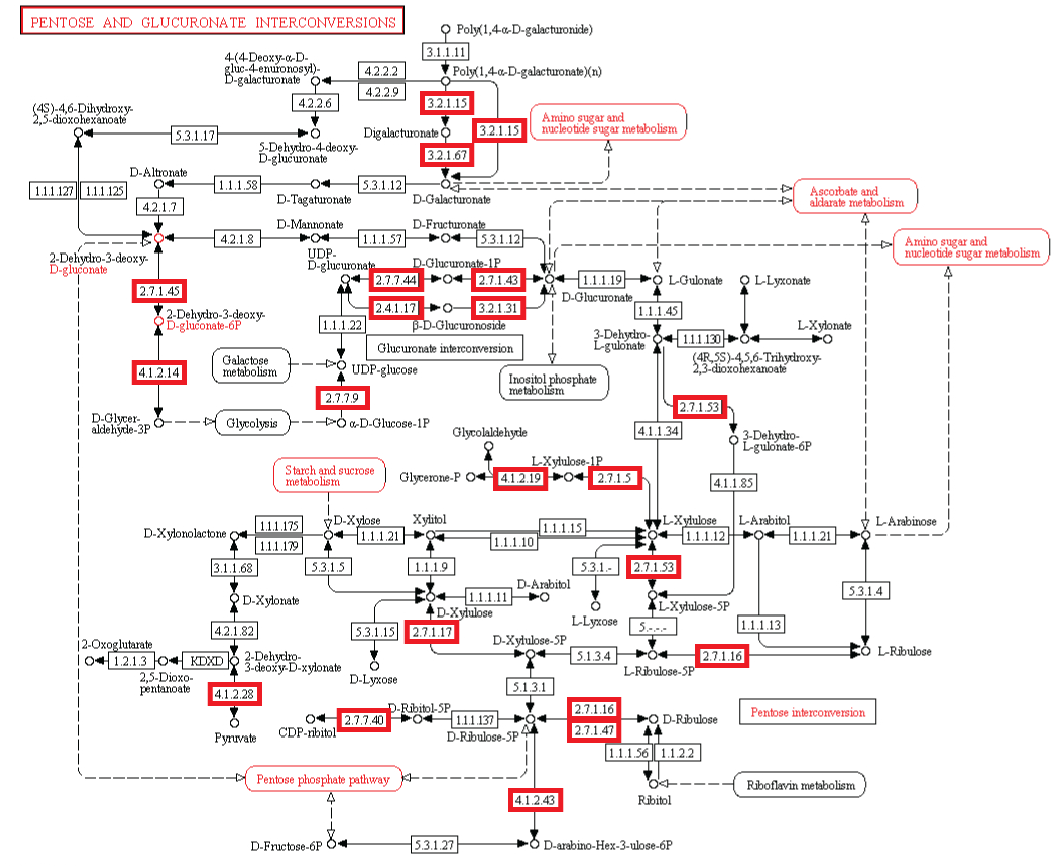
pentose and glucuronate interconversions
Lipopolysaccaride biosynthesis
cyanoamino acid metabolism
#1 - http://religiopoliticaltalk.com/wp-content/uploads/2016/07/01-Sphingolipid.jpg
#2 - http://religiopoliticaltalk.com/wp-content/uploads/2016/07/02-Pantothenate.jpg
#3 - http://religiopoliticaltalk.com/wp-content/uploads/2016/07/03-Galactose.jpg
#4 - http://religiopoliticaltalk.com/wp-content/uploads/2016/07/04-Drug.jpg
#5 - http://religiopoliticaltalk.com/wp-content/uploads/2016/07/05-Starch.jpg
#6 - http://religiopoliticaltalk.com/wp-content/uploads/2016/07/06-Fructose.jpg
#7 - http://religiopoliticaltalk.com/wp-content/uploads/2016/07/07-Fructose.jpg
#8 - http://religiopoliticaltalk.com/wp-content/uploads/2016/07/08-Lipopolysaccharide.jpg
#9 - http://religiopoliticaltalk.com/wp-content/uploads/2016/07/010-Cyanoamino.jpg

1. http://www.nada.kth.se/kurser/kth/2D1436/2005/lasmaterial/f8c/Bhalla.pdf
2. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0039912#pone.0039912-Srinivasan1
3. http://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0060018
4. http://www.plantphysiol.org/content/152/2/428.full?sid=c0bd645e-881c-4bed-9f5b-0e4d595f140d
5. http://www.ideacenter.org/contentmgr/showdetails.php/id/1181
Last edited by Otangelo on Sun Sep 04, 2022 8:50 pm; edited 48 times in total