The Hippo signaling pathway in organ size control, tissue regeneration and stem cell self-renewal 1
https://reasonandscience.catsboard.com/t2350-the-hippo-signaling-pathway-in-organ-size-control-tissue-regeneration-and-stem-cell-self-renewal
Precise control of organ size is crucial during animal development and regeneration. In Drosophila and mammals, studies over the past decade have uncovered a critical role for the Hippo tumour-suppressor pathway in the regulation of organ size. Dysregulation of this pathway leads to massive overgrowth of tissue. The Hippo signalling pathway is highly conserved and limits organ size by phosphorylating and inhibiting the transcription co-activators YAP and TAZ in mammals and Yki in Drosophila, key regulators of proliferation and apoptosis. The Hippo pathway also has a critical role in the self-renewal and expansion of stem cells and tissue-specific progenitor cells, and has important functions in tissue regeneration. Emerging evidence shows that the Hippo pathway is regulated by cell polarity, cell adhesion and cell junction proteins. In this review we summarize current understanding of the composition and regulation of the Hippo pathway, and discuss how cell polarity and cell adhesion proteins inform the role of this pathway in organ size control and regeneration.
Organ size regulation is a highly coordinated process involving complex mechanisms in response to physiological cues. On the organismal level, circulating factors such as hormones and insulin-like growth factors (IGF) play important roles in promoting organ size 1. In contrast, physiological perturbations, such as prolonged starvation, cause profound reduction of organ size1. Additionally, an intrinsic mechanism limits organ size, which was first demonstrated in salamander limbs by classical transplantation experiments1. The underlying mechanism of organ-autonomous size determination remained largely unknown until the past decade. Extensive research led to the identification of the Hippo tumour-suppressor pathway as a key regulator of organ size in Drosophila and mammals2. It is also known that mutations of genes that are involved in patterning, cell polarity and cell adhesion cause marked alternations of organ size3. Thus, the recent finding that the Hippo pathway is regulated by cell polarity and cell adhesion proteins is a promising basis for the potential crosstalk of the Hippo pathway and cell polarity proteins in the regulation of organ size4. Several studies have also demonstrated important roles for the Hippo pathway in stem cell/progenitor cell expansion and tissue regeneration5–13.
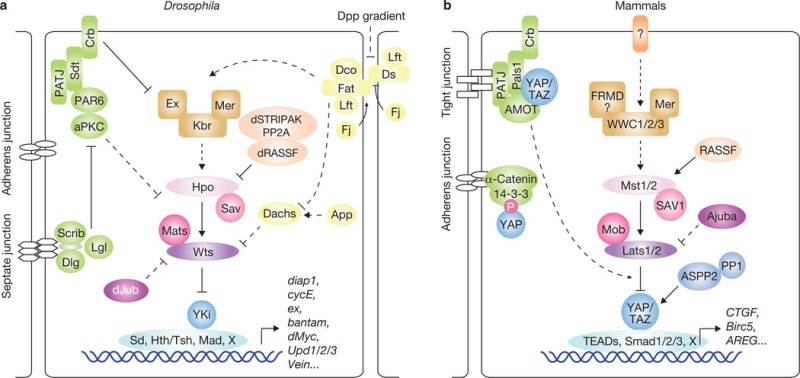
The Hippo pathway in Drosophila and mammals. Corresponding proteins in Drosophila
(a) and mammals
(b) are indicated by matching colours. Arrowed or blunted ends indicate activation or inhibition, respectively. Dashed lines indicate unknown mechanisms.
Research in the past years has uncovered many proteins that act upstream in the Drosophila Hippo pathway. Two apical cytoskeleton-binding proteins, Merlin (Mer) and Expanded (Ex)27, and their interacting protein Kibra28–30, were found to activate the Hippo pathway. The Fat protocadherin, a cell-surface molecule, was also identified as an upstream regulator of the Hippo pathway31–35. Fat activity is regulated by binding to another protocadherin, Dachsous (Ds)36, and is modulated by several proteins, such as the casein kinase Discs overgrown (Dco)37,38, the Golgiresident kinase Four-jointed (Fj)39–41 and the Fat/Ds-interacting protein Lowfat (Lft)42. Fat/Hippo pathway activity may also be influenced by Decapentaplegic (Dpp) and Wingless (Wg) morphogen gradients40,41,43, which affect the expression of Fj and Ds. It has been proposed that Fat activates the Hippo pathway by regulating the protein level and localization of the protein Ex31–33,35. Another study suggests that Fat may control the abundance of Wts through Dachs34,44. Recently, dJub, a LIM-domain-containing protein that physically interacts with Wts and Sav, was shown to negatively regulate Hippo signalling, although the detailed mechanism has not been delineated45. A number of proteins that determine cell polarity were also found to regulate the Hippo pathway. These include the Scribble (Scrib)–Discs large (Dlg)–Lethal giant larvae (Lgl) complex, atypical protein kinase C (aPKC) and Crumbs (Crb)46–49, indicating a role of cell polarity in the regulation of Hippo signalling.
Functions of the Hippo pathway in organ size determination and tumour suppression have been confirmed in genetically engineered mouse models. For instance, liver-specific overexpression of YAP results in enlarged livers that return to their normal size after cessation of YAP expression12,26. However, sustained YAP overexpression leads to tumour formation26. Genomic amplification of YAP is also observed in human cancers and a mouse model of breast cancer63,64. Furthermore, elevated YAP protein levels and nuclear localization have been observed in multiple human cancers59,63,65, and the alterations of YAP may have prognostic value for certain human cancers66. Overexpression of TAZ, the paralogue of YAP, has been noted in human breast cancer samples and non-small-cell lung-cancer cell lines67,68. Ablation of the Hippo pathway components Mer and Sav and double knockout of Mst1/2 in mice also result in liver enlargement and tumour formation69–74. Remarkably, loss of one or both copies of YAP can suppress liver expansion and tumorigenesis induced by Mer deficiency69. Aberrant Mst1/2 and Lats1/2 expression andLats2, Sav1 and Mob1 mutation were also observed in human cancers or cancer cell lines2. Together, these studies highlight a significant role of the Hippo pathway in organ size regulation and tumorigenesis.
Drosophila Eye as a Model to Study Regulation of Growth Control: The Discovery of Size Control Pathways
Molecular Genetics of Axial Patterning, Growth and Disease, page 238
In the biological sense, the term growth has intricate ramifications that we have only started to comprehend. Growth is the overall increase in cell mass or size of a tissue or organism . Growth may be due to increase in cell number resulting from cell division (cell proliferation), increase in cellular mass without cell division (cell enlargement), or due to release of more extracellular matrix (cell accretion). These processes are intimately linked and it is clear that if coordinated growth has to occur in an organism, it is necessary for various biological pathways to interact and relay appropriate signals to proper cell types. Growth regulation is precisely controlled and affected by several intrinsic and extrinsic factors . The intrinsic factors mainly involve synthesis and secretion of signals or ligands, which bind to their cognate receptors to relay downstream signals. These signals consist of variety of molecules such as hormones, mitogens, apoptosis-inducing signals, patterning and axis determining signals, etc. which eventually determine organ size and tissue homeostasis . Growth of a tissue or organ is impacted not only by cell division but also by regulated cell death . In this chapter, we will focus on growth regulation in imaginal discs (epithelial sacs that are precursors of adult appendages) in D. melanogaster . The obvious advantages that Drosophila has to offer as a model organism include short life cycle, high fecundity, low cost maintenance, and lack of redundancy in genome . Furthermore, the sophisticated tools available in fly genetics provide great deal of versatility in terms of designing experiments. The plethora of knowledge thus generated through exhausting efforts of scientists has not only revealed to us the classic information about how growth occurs but has also lead to better understanding of growth-related diseases such as cancer.
Drosophila Eye as a Model to Study Regulation of Growth
The compound eyes of Drosophila arise from the eye-antennal imaginal discs, a monolayer epithelial sheet of cells that is responsible for the development of the eyes, the antennae, the ocelli, and a major part of the adult head cuticle. Each eye of the adult fruit fly on an average consists of about 800 ommatidia (Wolff and Ready 1993). Ommatidia arise from a set of 19 precursor cells that are generated by spatially and temporally coordinated cellular processes such as cell-proliferation, cell-differentiation, and cell-death in the eye imaginal discs. Eighteen of these cells contribute to the eye per se, whereas the nineteenth cell gives rise to a sensory bristle . A key feature that distinguishes eye from the rest of the organs is its ability to perceive light and relay the signal to distinct areas in the brain called the optic lobes. The eye imaginal discs arise from about 50 primordial cells that express the Drosophila PAX 6 gene eyeless (ey) during mid-to-late embryogenesis. Two such discs develop in each larva and differentiate into two compound eyes, antennae, ocelli, and the head cuticle in the adult.
Much is known about the regulation of growth and differentiation of the eyeantennal imaginal discs. Until the second larval instar of development, the cells of the eye-antennal discs proliferate without differentiation . During the second instar stage, a unique process of cell differentiation begins in the eye-antennal disc that paves the way for formation of photoreceptor neurons in the posterior region of the eye-antennal imaginal disc . The differentiation occurs in the wake of a so-called “morphogenetic furrow”—a front marked by apical constriction of epithelial cells in response to complex developmental signaling from the Hedgehog (Hh), Decapentaplegic (Dpp), Wingless (Wg), and Epidermal growth factor receptor (EGFR) pathways. Posterior to the morphogenetic furrow, the cells begin to acquire particular photoreceptor cell fates and organize into ommatidial clusters. Anterior to the furrow, the cells divide asynchronously and do not differentiate, however, in the morphogenetic furrows, cells arrest in the G1 phase of the cell cycle, synchronize, and either start to differentiate into photoreceptor cells as they leave the furrow or undergo one additional round of cell division, referred to as the second mitotic wave (SMW) before differentiating into the remaining photoreceptor, cone, pigment, and bristle cells . The cells posterior to the morphogenetic furrow enter G1 arrest caused by Dpp (decapentaplegic) signaling that is maintained by the roughex (rux) gene, which negatively regulates G1-S transition. The cells that are temporarily trapped in the G1 phase begin differentiation with specification of the R8 (photoreceptor) cell due to expression of the proneural protein Atonal (Ato) . R8 recruits other photoreceptor cells-R2, R3, R4, and R5 to form a cluster of five photoreceptor precursors. Once specified, these cells never enter cell cycle or cell division again. All other nonspecified cells re-enter cell cycle only once at the SMW . Cells in the SMW undergo G2/M phase that is mediated through local signaling from Spitz (Spi). Binding of Spi to its cognate receptor EGFR in precursor cells causes activation of downstream string (stg) that completes the G2-M transition during mitosis. Local Spi-EGFR signaling also plays an important role limiting the progression of SMW. For instance, on an average the Spi signal from one precluster can span to a length of seven cells only causing these cells to divide whereas the remaining cells remain arrested in G2 phase and fail to divide . The progression of the morphogenetic furrow is complete by the mid-third instar of larval development, and the eye-antennal disc is fully grown to about 50,000 cells. Thus, the eye-antennal disc is ideal for the study of organogenesis, morphogenesis, pattern formation, and several cell biological processes including the regulation of cell cycle, cell death, cell junctions and adhesion, transport of molecules, cell signaling, and metabolism. Recently, the eye discs have been used as an experimental system for genetic screens to discover postembryonic lethality, and for screening small molecule inhibitors in chemical and drug screens.
The Mosaic Analysis Systems and the Drosophila Eye
Mutagenesis screens are a very well-established tool for gene discovery in flies. Over the years, the mosaic techniques have evolved to include the Flipase(FLP)-Flipase recognition target (FRT), eyGAL4 UASFlp EGUF, Flp-out clones, and Mosaic Analysis with Repressible Cell Marker MARCM . One of the first tissue-specific mosaic systems was developed in the eye-antennal discs where the mosaic clones were restricted to the eye-antennal discs by virtue of expression of the Flippase gene under the control of the Eyeless Promoter (commonly referred to as the ‘ey-FLP system’, Newsome et al. 2000). This tissue-specific system was further refined by the development of the “cell-lethal” system, where effects of loss of function of a gene could be surveyed more clearly because the wild-type twin-clones are eliminated due to the presence of cell-lethal mutations (the celllethal FLP-FRT system; Newsome et al. 2000). We focus on the genetic screens performed about 10–12 years ago (simultaneously in many labs) that led to the identification of many new genes that were shown to belong to the two major growth regulatory networks: the Hippo pathway and the Tuberous Sclerosis Complex/Target of Rapamycin—TSC-TOR pathway
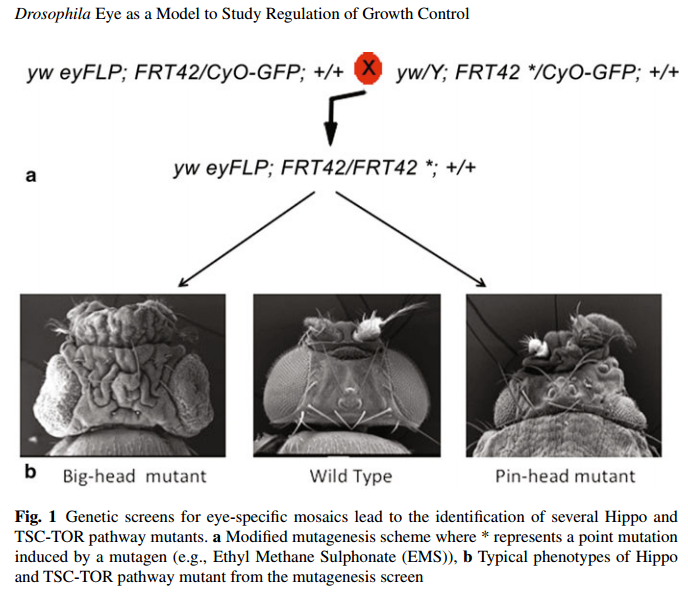
Genetic Screens for Genes That Regulate Growth: The “Big-Head” and “Pin-Head” Mutations
Barry Dickson’s group improved the traditional FLP-FRT approach developed in the Rubin Lab, to allow generation of essentially mutant eye discs by eliminating the wild-type twin clone via a cell-lethal mutation (the cell-lethal FLP-FRT system) (Fig. 1a). This so-called “cell-lethal” approach allows the mutant clones to grow to their highest potential due to elimination of competitive interactions between the mutant cells and their wild-type neighbors. Using this system, several groups carried out mutagenesis screens in flies (on the X, 2L, 2R, 3L, 3R chromosomes) and found mutations that affected patterning, growth, cell death, and differentiation. Of special interest were the genes mutations which caused a remarkable effect on growth without disrupting the patterning process . Characterization of these mutants revealed the mechanisms that regulate growth and tissue size by controlling cell number (Hippo pathway) or cell size (InR/TSC-TOR pathway) in a developing organ. Typically, loss of function mutations in positive regulators of these pathways caused development of enlarged heads that showed overgrowth—referred to as the “big head” mutations (Fig. 1b). In contrast, loss of function of negative regulators of these pathways caused reduction in head size and development of smaller organs, which may be due to cell death or reduction in cell size, and were referred to as the “pin head” mutations (Fig. 1b).
The Hippo Signaling Pathway
The Hippo signaling pathway was first discovered in flies following characterization of “big-head” mutants identified from genetic screens. Analysis of the loss of function phenotypes revealed that a fundamental function of the Hippo pathway was the regulation of organ size . Interestingly, the pathway received its name just after some growth regulatory genes (warts (wts), salvador (sav, aka shar-pie, shrp)) were characterized. Warts (wts) was named based on the bumpy “warts-like” phenotype of the mutant cells in mitotic (mosaic) clones on the body of the adult flies that were reminiscent of the warts on toads . Another group led by Xu et al. also independently found warts in the initial FLP/FRTbased screen and named it large tumor suppressor (LATS) . Two independent groups identified the gene encoding the adaptor protein Salvador (Sav aka Shar-pie, Shrp after the dog species of the same name as the mutant flies showed a characteristic phenotype of folded dark cuticle on the overgrown heads) from complementation groups isolated from the big-head genetic screens. Interestingly, both Wts and Sav regulated growth by suppressing proliferation and promoting apoptosis. Hippo (Hpo) was the name given to another complementation group from the “big-head” screens that showed a phenotype that was very similar to Wts and Sav mutants. Molecular analysis of the three genes revealed that Wts and Hpo genes encode for serine-threonine (S-T) kinases whereas Sav is a WW domain containing adaptor protein. By this time it was clear that Warts, Salvador, and Hippo all show similar loss of function phenotypes and control organ size by a common signaling pathway that promotes apoptosis and restricts cell proliferation , and the pathway got its name from the last member of this trio of genes. A complete pathway that relays a growth regulatory signal from the plasma membrane to the nucleus has emerged over the last decade. Although genetic mutagenesis screens led to the initial discovery of this pathway, several components were identified by other genetic screening strategies and biochemical approaches (e.g., yeast-two hybrid screens, TAP-TAG based protein interaction assays.Today the Hippo pathway has grown to a large network of tumor suppressor genes that function upstream and downstream of the three initial members of the Hippo pathway (also known as the core kinase cascade) that control several aspects of tissue homeostasis. Overall, the Hippo signalling pathway is a key size regulatory pathway that controls organ size in flies and vertebrates, and misregulation of Hippo signalling is implicated in several diseases including cancer.
Regulation by Core Kinase Cascade of the Hippo Pathway
The molecular analysis of the three initial members of the Hippo pathway in Drosophila revealed that Hpo codes for an S-T kinase of the mammalian Sterile- 20 family of kinases , and can physically associate with the WW-domain containing adaptor protein Sav . Wts is an S-T kinase protein of the dystrophia myotonica protein kinase (DMPK) family that associates with another adaptor protein Mob as tumor suppressor. Loss of function of these genes in genetic mosaics revealed strong overgrowth phenotype caused by increased cell proliferation and diminished sensitivity to apoptosis. Hyperactivation of the pathway by over-expression of Hpo, Sav, Wts, or Mats leads to formation of smaller organs due to increased apoptosis. Biochemical analysis showed that the Hpo kinase phosphorylates and can physically associate with Sav, Wts, and Mats to form protein complexes in vitro. However, Hpo associates with its cognate adaptor protein Sav to form the Hpo-Sav complex for efficient activation of the downstream kinase Wts . Wts itself associates with Mats to form the downstream Wts-Mats complex of the core kinase cascade of the Hippo pathway. Association of these adaptor proteins is known to stimulate the catalytic activity of the Hpo and Wts kinases. Moreover, phosphorylation of Mats by the Hpo kinase increases its affinity for the Wts kinase. Wts is activated by autophosphorylation and phosphorylation by Hpo-kinase. Activated Wts associates with Mats (thus Mats cannot simultaneously associate with Hpo and Wts), which acts as a coactivator for the kinase activity of Wts . A major output of the core kinase cascade is to inhibit the growth-promoting activity of Yorkie (Yki), the Drosophila homolog of the mammalian Yes-associated protein (YAP) oncogene that acts as a transcriptional coactivator. Yki was identified via a yeast two-hybrid screen as an interactor of Warts. Over-expression of Yki phenocopies the loss of function of hpo, sav, wts, and mats (all genes of the core kinase cascade) and causes over-growth. Loss of function of yki results in formation of smaller organs due to induction of cell death. Yki activity is regulated by controlling its subcellular localization via phosphorylation-dependent and -independent interactions with the core kinase cascade of the Hippo pathway. Yki associates with Wts, and one mechanism by which the Wts-kinase restricts Yki activity is via phosphorylation at Ser168 that creates a 14-3-3 protein-binding site . Interestingly, only phosphorylated forms ofYki can associate with 14-3-3 proteins. Yki is phosphorylated at multiple sites (e.g., Ser 111 and S250), making it less sensitive to Hpo/Wts-mediated inhibition. These phosphorylation events act in parallel to phosphoYki/14-3-3 mediated mechanisms and inhibit Yki nuclear localization and activity. It is suggested that nuclear export is required for shuttling Yki to the nucleus in response to Hpo signaling, and binding of 14-3-3 proteins is thought to impede nuclear import and/or promote nuclear export thereby facilitating nucleocytoplasmic shuttling of target proteins. Nuclear transport of Yki depends on its binding with cognate transcription factors as Yki does not have an intrinsic nuclear localization signal (NLS). Currently, it is unclear if binding of 14-3-3 proteins to Yki prevents its binding with cognate transcription factors, or masks the NLSs or promotes export from the nucleus. Nevertheless, coactivator Yki/YAP is the critical downstream regulatory target of the Hpo kinase cascade, and regulation of its subcellular localization is the primary mechanism by which the Hpo pathway influences target gene expression. Yki (like Sav) is a WW-domain-containing protein and interacts with the PPxY (where P = Proline; x = any amino acid; Y = Tyrosine) motifs in Wts. Besides Wts, the WW-domains of Yki interact with the PPxY motifs present in other components of Hippo signaling pathway like Expanded (Ex), Hpo, WW-domain-binding protein 2, and Myopic to regulate Hippo signaling via phosphorylation-independent mechanisms. Another protein that acts via its WW-domains is Kibra which associates with the PPxY motifs in Ex . The identification of multiple proteins that act through the interaction between WW-domains and PPxY motifs in the Hippo pathway suggests that these motif-specific interactions are important for regulation of Hippo signaling
1) http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3987945/
https://reasonandscience.catsboard.com/t2350-the-hippo-signaling-pathway-in-organ-size-control-tissue-regeneration-and-stem-cell-self-renewal
Precise control of organ size is crucial during animal development and regeneration. In Drosophila and mammals, studies over the past decade have uncovered a critical role for the Hippo tumour-suppressor pathway in the regulation of organ size. Dysregulation of this pathway leads to massive overgrowth of tissue. The Hippo signalling pathway is highly conserved and limits organ size by phosphorylating and inhibiting the transcription co-activators YAP and TAZ in mammals and Yki in Drosophila, key regulators of proliferation and apoptosis. The Hippo pathway also has a critical role in the self-renewal and expansion of stem cells and tissue-specific progenitor cells, and has important functions in tissue regeneration. Emerging evidence shows that the Hippo pathway is regulated by cell polarity, cell adhesion and cell junction proteins. In this review we summarize current understanding of the composition and regulation of the Hippo pathway, and discuss how cell polarity and cell adhesion proteins inform the role of this pathway in organ size control and regeneration.
Organ size regulation is a highly coordinated process involving complex mechanisms in response to physiological cues. On the organismal level, circulating factors such as hormones and insulin-like growth factors (IGF) play important roles in promoting organ size 1. In contrast, physiological perturbations, such as prolonged starvation, cause profound reduction of organ size1. Additionally, an intrinsic mechanism limits organ size, which was first demonstrated in salamander limbs by classical transplantation experiments1. The underlying mechanism of organ-autonomous size determination remained largely unknown until the past decade. Extensive research led to the identification of the Hippo tumour-suppressor pathway as a key regulator of organ size in Drosophila and mammals2. It is also known that mutations of genes that are involved in patterning, cell polarity and cell adhesion cause marked alternations of organ size3. Thus, the recent finding that the Hippo pathway is regulated by cell polarity and cell adhesion proteins is a promising basis for the potential crosstalk of the Hippo pathway and cell polarity proteins in the regulation of organ size4. Several studies have also demonstrated important roles for the Hippo pathway in stem cell/progenitor cell expansion and tissue regeneration5–13.
The Hippo pathway in Drosophila
In Drosophila, the first core components of the Hippo pathway to be identified, using genetic mosaic screens, were the tumour-suppressor genes warts (wts)14,15, hippo (hpo)16–20 and salvador (sav)21,22. These genes belong to the hyperplastic group of Drosophila tumour-suppressors. Mutation of these genes results in robust tissue overgrowth without alteration of cell fate determination or cell polarity. Biochemical studies revealed that Hpo directly interacts with Sav to phosphorylate and activate the complex formed by Wts and another core Hippo pathway protein, Mats16,23 (Fig. 1a). The kinase activity of Hpo is antagonized by a PP2A phosphatase complex, dSTRIPAK24. The Hippo pathway is known to limit organ size partly by transcriptional regulation of cyclin E and diap1 (refs 16,17,20,21,23), suggesting the existence of a transcriptional regulator as a downstream effector of the pathway. By performing a yeast two-hybrid screen using Wts as bait, the transcription co-activator Yorkie (Yki) was identified as a potent effector of the Hippo pathway25. Subsequent biochemical studies showed that Wts directly phosphorylates and inhibits Yki26.
The Hippo pathway in Drosophila and mammals. Corresponding proteins in Drosophila
(a) and mammals
(b) are indicated by matching colours. Arrowed or blunted ends indicate activation or inhibition, respectively. Dashed lines indicate unknown mechanisms.
Research in the past years has uncovered many proteins that act upstream in the Drosophila Hippo pathway. Two apical cytoskeleton-binding proteins, Merlin (Mer) and Expanded (Ex)27, and their interacting protein Kibra28–30, were found to activate the Hippo pathway. The Fat protocadherin, a cell-surface molecule, was also identified as an upstream regulator of the Hippo pathway31–35. Fat activity is regulated by binding to another protocadherin, Dachsous (Ds)36, and is modulated by several proteins, such as the casein kinase Discs overgrown (Dco)37,38, the Golgiresident kinase Four-jointed (Fj)39–41 and the Fat/Ds-interacting protein Lowfat (Lft)42. Fat/Hippo pathway activity may also be influenced by Decapentaplegic (Dpp) and Wingless (Wg) morphogen gradients40,41,43, which affect the expression of Fj and Ds. It has been proposed that Fat activates the Hippo pathway by regulating the protein level and localization of the protein Ex31–33,35. Another study suggests that Fat may control the abundance of Wts through Dachs34,44. Recently, dJub, a LIM-domain-containing protein that physically interacts with Wts and Sav, was shown to negatively regulate Hippo signalling, although the detailed mechanism has not been delineated45. A number of proteins that determine cell polarity were also found to regulate the Hippo pathway. These include the Scribble (Scrib)–Discs large (Dlg)–Lethal giant larvae (Lgl) complex, atypical protein kinase C (aPKC) and Crumbs (Crb)46–49, indicating a role of cell polarity in the regulation of Hippo signalling.
The Hippo pathway in mammals
The core components and downstream effectors of the Drosophila Hippo pathway are highly conserved in mammals: Mst1/2 (homologues of Hpo), Sav1 (Sav homologue), Lats1/2 (Wts homologues), MOBKL1A and MOBKL1B (collectively referred to as Mob1; homologues of Mats), and YAP and its paralogue TAZ (also called WWTR1; homologues of Yki) (Fig. 1b). Expression of human YAP, Lats1, Mst2 and Mob1 can rescue the phenotypes of their corresponding Drosophila mutants in vivo16,23,25,50. The core components Mst1/2 are pro-apoptotic kinases that are activated by caspase cleavage under apoptotic stress51. Sav1 interacts with Mst1/2 through the SARAH domains present in both Sav1 and Mst1/2 (ref.52). Although Sav1 has been shown to activate Mst1/2, the underlying mechanism is unclear, but might involve regulation of Mst1 nuclear translocation53. Mst1/2 is also activated by binding to Ras association domain family (RASSF) proteins54, possibly owing to alteration of Mst1/2 subcellular localization55. InDrosophila, however, dRASSF inhibits Hpo possibly through competition with Sav for Hpo binding56 and through recruitment of the dSTRIPAK–PP2A complex24. Activation of Mst1/2 leads to phosphorylation and activation of their direct substrates, Lats1/2 (ref. 57). Mob1, which forms a complex with Lats1/2, is also phosphorylated by Mst1/2, resulting in an enhanced Lats1/2–Mob1 interaction58. Activated Lats1/2 in turn phosphorylate and inhibit YAP/TAZ transcription co-activators26,59–62.Functions of the Hippo pathway in organ size determination and tumour suppression have been confirmed in genetically engineered mouse models. For instance, liver-specific overexpression of YAP results in enlarged livers that return to their normal size after cessation of YAP expression12,26. However, sustained YAP overexpression leads to tumour formation26. Genomic amplification of YAP is also observed in human cancers and a mouse model of breast cancer63,64. Furthermore, elevated YAP protein levels and nuclear localization have been observed in multiple human cancers59,63,65, and the alterations of YAP may have prognostic value for certain human cancers66. Overexpression of TAZ, the paralogue of YAP, has been noted in human breast cancer samples and non-small-cell lung-cancer cell lines67,68. Ablation of the Hippo pathway components Mer and Sav and double knockout of Mst1/2 in mice also result in liver enlargement and tumour formation69–74. Remarkably, loss of one or both copies of YAP can suppress liver expansion and tumorigenesis induced by Mer deficiency69. Aberrant Mst1/2 and Lats1/2 expression andLats2, Sav1 and Mob1 mutation were also observed in human cancers or cancer cell lines2. Together, these studies highlight a significant role of the Hippo pathway in organ size regulation and tumorigenesis.
Drosophila Eye as a Model to Study Regulation of Growth Control: The Discovery of Size Control Pathways
Molecular Genetics of Axial Patterning, Growth and Disease, page 238
In the biological sense, the term growth has intricate ramifications that we have only started to comprehend. Growth is the overall increase in cell mass or size of a tissue or organism . Growth may be due to increase in cell number resulting from cell division (cell proliferation), increase in cellular mass without cell division (cell enlargement), or due to release of more extracellular matrix (cell accretion). These processes are intimately linked and it is clear that if coordinated growth has to occur in an organism, it is necessary for various biological pathways to interact and relay appropriate signals to proper cell types. Growth regulation is precisely controlled and affected by several intrinsic and extrinsic factors . The intrinsic factors mainly involve synthesis and secretion of signals or ligands, which bind to their cognate receptors to relay downstream signals. These signals consist of variety of molecules such as hormones, mitogens, apoptosis-inducing signals, patterning and axis determining signals, etc. which eventually determine organ size and tissue homeostasis . Growth of a tissue or organ is impacted not only by cell division but also by regulated cell death . In this chapter, we will focus on growth regulation in imaginal discs (epithelial sacs that are precursors of adult appendages) in D. melanogaster . The obvious advantages that Drosophila has to offer as a model organism include short life cycle, high fecundity, low cost maintenance, and lack of redundancy in genome . Furthermore, the sophisticated tools available in fly genetics provide great deal of versatility in terms of designing experiments. The plethora of knowledge thus generated through exhausting efforts of scientists has not only revealed to us the classic information about how growth occurs but has also lead to better understanding of growth-related diseases such as cancer.
Drosophila Eye as a Model to Study Regulation of Growth
The compound eyes of Drosophila arise from the eye-antennal imaginal discs, a monolayer epithelial sheet of cells that is responsible for the development of the eyes, the antennae, the ocelli, and a major part of the adult head cuticle. Each eye of the adult fruit fly on an average consists of about 800 ommatidia (Wolff and Ready 1993). Ommatidia arise from a set of 19 precursor cells that are generated by spatially and temporally coordinated cellular processes such as cell-proliferation, cell-differentiation, and cell-death in the eye imaginal discs. Eighteen of these cells contribute to the eye per se, whereas the nineteenth cell gives rise to a sensory bristle . A key feature that distinguishes eye from the rest of the organs is its ability to perceive light and relay the signal to distinct areas in the brain called the optic lobes. The eye imaginal discs arise from about 50 primordial cells that express the Drosophila PAX 6 gene eyeless (ey) during mid-to-late embryogenesis. Two such discs develop in each larva and differentiate into two compound eyes, antennae, ocelli, and the head cuticle in the adult.
Much is known about the regulation of growth and differentiation of the eyeantennal imaginal discs. Until the second larval instar of development, the cells of the eye-antennal discs proliferate without differentiation . During the second instar stage, a unique process of cell differentiation begins in the eye-antennal disc that paves the way for formation of photoreceptor neurons in the posterior region of the eye-antennal imaginal disc . The differentiation occurs in the wake of a so-called “morphogenetic furrow”—a front marked by apical constriction of epithelial cells in response to complex developmental signaling from the Hedgehog (Hh), Decapentaplegic (Dpp), Wingless (Wg), and Epidermal growth factor receptor (EGFR) pathways. Posterior to the morphogenetic furrow, the cells begin to acquire particular photoreceptor cell fates and organize into ommatidial clusters. Anterior to the furrow, the cells divide asynchronously and do not differentiate, however, in the morphogenetic furrows, cells arrest in the G1 phase of the cell cycle, synchronize, and either start to differentiate into photoreceptor cells as they leave the furrow or undergo one additional round of cell division, referred to as the second mitotic wave (SMW) before differentiating into the remaining photoreceptor, cone, pigment, and bristle cells . The cells posterior to the morphogenetic furrow enter G1 arrest caused by Dpp (decapentaplegic) signaling that is maintained by the roughex (rux) gene, which negatively regulates G1-S transition. The cells that are temporarily trapped in the G1 phase begin differentiation with specification of the R8 (photoreceptor) cell due to expression of the proneural protein Atonal (Ato) . R8 recruits other photoreceptor cells-R2, R3, R4, and R5 to form a cluster of five photoreceptor precursors. Once specified, these cells never enter cell cycle or cell division again. All other nonspecified cells re-enter cell cycle only once at the SMW . Cells in the SMW undergo G2/M phase that is mediated through local signaling from Spitz (Spi). Binding of Spi to its cognate receptor EGFR in precursor cells causes activation of downstream string (stg) that completes the G2-M transition during mitosis. Local Spi-EGFR signaling also plays an important role limiting the progression of SMW. For instance, on an average the Spi signal from one precluster can span to a length of seven cells only causing these cells to divide whereas the remaining cells remain arrested in G2 phase and fail to divide . The progression of the morphogenetic furrow is complete by the mid-third instar of larval development, and the eye-antennal disc is fully grown to about 50,000 cells. Thus, the eye-antennal disc is ideal for the study of organogenesis, morphogenesis, pattern formation, and several cell biological processes including the regulation of cell cycle, cell death, cell junctions and adhesion, transport of molecules, cell signaling, and metabolism. Recently, the eye discs have been used as an experimental system for genetic screens to discover postembryonic lethality, and for screening small molecule inhibitors in chemical and drug screens.
The Mosaic Analysis Systems and the Drosophila Eye
Mutagenesis screens are a very well-established tool for gene discovery in flies. Over the years, the mosaic techniques have evolved to include the Flipase(FLP)-Flipase recognition target (FRT), eyGAL4 UASFlp EGUF, Flp-out clones, and Mosaic Analysis with Repressible Cell Marker MARCM . One of the first tissue-specific mosaic systems was developed in the eye-antennal discs where the mosaic clones were restricted to the eye-antennal discs by virtue of expression of the Flippase gene under the control of the Eyeless Promoter (commonly referred to as the ‘ey-FLP system’, Newsome et al. 2000). This tissue-specific system was further refined by the development of the “cell-lethal” system, where effects of loss of function of a gene could be surveyed more clearly because the wild-type twin-clones are eliminated due to the presence of cell-lethal mutations (the celllethal FLP-FRT system; Newsome et al. 2000). We focus on the genetic screens performed about 10–12 years ago (simultaneously in many labs) that led to the identification of many new genes that were shown to belong to the two major growth regulatory networks: the Hippo pathway and the Tuberous Sclerosis Complex/Target of Rapamycin—TSC-TOR pathway
Genetic Screens for Genes That Regulate Growth: The “Big-Head” and “Pin-Head” Mutations
Barry Dickson’s group improved the traditional FLP-FRT approach developed in the Rubin Lab, to allow generation of essentially mutant eye discs by eliminating the wild-type twin clone via a cell-lethal mutation (the cell-lethal FLP-FRT system) (Fig. 1a). This so-called “cell-lethal” approach allows the mutant clones to grow to their highest potential due to elimination of competitive interactions between the mutant cells and their wild-type neighbors. Using this system, several groups carried out mutagenesis screens in flies (on the X, 2L, 2R, 3L, 3R chromosomes) and found mutations that affected patterning, growth, cell death, and differentiation. Of special interest were the genes mutations which caused a remarkable effect on growth without disrupting the patterning process . Characterization of these mutants revealed the mechanisms that regulate growth and tissue size by controlling cell number (Hippo pathway) or cell size (InR/TSC-TOR pathway) in a developing organ. Typically, loss of function mutations in positive regulators of these pathways caused development of enlarged heads that showed overgrowth—referred to as the “big head” mutations (Fig. 1b). In contrast, loss of function of negative regulators of these pathways caused reduction in head size and development of smaller organs, which may be due to cell death or reduction in cell size, and were referred to as the “pin head” mutations (Fig. 1b).

The Hippo Signaling Pathway
The Hippo signaling pathway was first discovered in flies following characterization of “big-head” mutants identified from genetic screens. Analysis of the loss of function phenotypes revealed that a fundamental function of the Hippo pathway was the regulation of organ size . Interestingly, the pathway received its name just after some growth regulatory genes (warts (wts), salvador (sav, aka shar-pie, shrp)) were characterized. Warts (wts) was named based on the bumpy “warts-like” phenotype of the mutant cells in mitotic (mosaic) clones on the body of the adult flies that were reminiscent of the warts on toads . Another group led by Xu et al. also independently found warts in the initial FLP/FRTbased screen and named it large tumor suppressor (LATS) . Two independent groups identified the gene encoding the adaptor protein Salvador (Sav aka Shar-pie, Shrp after the dog species of the same name as the mutant flies showed a characteristic phenotype of folded dark cuticle on the overgrown heads) from complementation groups isolated from the big-head genetic screens. Interestingly, both Wts and Sav regulated growth by suppressing proliferation and promoting apoptosis. Hippo (Hpo) was the name given to another complementation group from the “big-head” screens that showed a phenotype that was very similar to Wts and Sav mutants. Molecular analysis of the three genes revealed that Wts and Hpo genes encode for serine-threonine (S-T) kinases whereas Sav is a WW domain containing adaptor protein. By this time it was clear that Warts, Salvador, and Hippo all show similar loss of function phenotypes and control organ size by a common signaling pathway that promotes apoptosis and restricts cell proliferation , and the pathway got its name from the last member of this trio of genes. A complete pathway that relays a growth regulatory signal from the plasma membrane to the nucleus has emerged over the last decade. Although genetic mutagenesis screens led to the initial discovery of this pathway, several components were identified by other genetic screening strategies and biochemical approaches (e.g., yeast-two hybrid screens, TAP-TAG based protein interaction assays.Today the Hippo pathway has grown to a large network of tumor suppressor genes that function upstream and downstream of the three initial members of the Hippo pathway (also known as the core kinase cascade) that control several aspects of tissue homeostasis. Overall, the Hippo signalling pathway is a key size regulatory pathway that controls organ size in flies and vertebrates, and misregulation of Hippo signalling is implicated in several diseases including cancer.
Regulation by Core Kinase Cascade of the Hippo Pathway
The molecular analysis of the three initial members of the Hippo pathway in Drosophila revealed that Hpo codes for an S-T kinase of the mammalian Sterile- 20 family of kinases , and can physically associate with the WW-domain containing adaptor protein Sav . Wts is an S-T kinase protein of the dystrophia myotonica protein kinase (DMPK) family that associates with another adaptor protein Mob as tumor suppressor. Loss of function of these genes in genetic mosaics revealed strong overgrowth phenotype caused by increased cell proliferation and diminished sensitivity to apoptosis. Hyperactivation of the pathway by over-expression of Hpo, Sav, Wts, or Mats leads to formation of smaller organs due to increased apoptosis. Biochemical analysis showed that the Hpo kinase phosphorylates and can physically associate with Sav, Wts, and Mats to form protein complexes in vitro. However, Hpo associates with its cognate adaptor protein Sav to form the Hpo-Sav complex for efficient activation of the downstream kinase Wts . Wts itself associates with Mats to form the downstream Wts-Mats complex of the core kinase cascade of the Hippo pathway. Association of these adaptor proteins is known to stimulate the catalytic activity of the Hpo and Wts kinases. Moreover, phosphorylation of Mats by the Hpo kinase increases its affinity for the Wts kinase. Wts is activated by autophosphorylation and phosphorylation by Hpo-kinase. Activated Wts associates with Mats (thus Mats cannot simultaneously associate with Hpo and Wts), which acts as a coactivator for the kinase activity of Wts . A major output of the core kinase cascade is to inhibit the growth-promoting activity of Yorkie (Yki), the Drosophila homolog of the mammalian Yes-associated protein (YAP) oncogene that acts as a transcriptional coactivator. Yki was identified via a yeast two-hybrid screen as an interactor of Warts. Over-expression of Yki phenocopies the loss of function of hpo, sav, wts, and mats (all genes of the core kinase cascade) and causes over-growth. Loss of function of yki results in formation of smaller organs due to induction of cell death. Yki activity is regulated by controlling its subcellular localization via phosphorylation-dependent and -independent interactions with the core kinase cascade of the Hippo pathway. Yki associates with Wts, and one mechanism by which the Wts-kinase restricts Yki activity is via phosphorylation at Ser168 that creates a 14-3-3 protein-binding site . Interestingly, only phosphorylated forms ofYki can associate with 14-3-3 proteins. Yki is phosphorylated at multiple sites (e.g., Ser 111 and S250), making it less sensitive to Hpo/Wts-mediated inhibition. These phosphorylation events act in parallel to phosphoYki/14-3-3 mediated mechanisms and inhibit Yki nuclear localization and activity. It is suggested that nuclear export is required for shuttling Yki to the nucleus in response to Hpo signaling, and binding of 14-3-3 proteins is thought to impede nuclear import and/or promote nuclear export thereby facilitating nucleocytoplasmic shuttling of target proteins. Nuclear transport of Yki depends on its binding with cognate transcription factors as Yki does not have an intrinsic nuclear localization signal (NLS). Currently, it is unclear if binding of 14-3-3 proteins to Yki prevents its binding with cognate transcription factors, or masks the NLSs or promotes export from the nucleus. Nevertheless, coactivator Yki/YAP is the critical downstream regulatory target of the Hpo kinase cascade, and regulation of its subcellular localization is the primary mechanism by which the Hpo pathway influences target gene expression. Yki (like Sav) is a WW-domain-containing protein and interacts with the PPxY (where P = Proline; x = any amino acid; Y = Tyrosine) motifs in Wts. Besides Wts, the WW-domains of Yki interact with the PPxY motifs present in other components of Hippo signaling pathway like Expanded (Ex), Hpo, WW-domain-binding protein 2, and Myopic to regulate Hippo signaling via phosphorylation-independent mechanisms. Another protein that acts via its WW-domains is Kibra which associates with the PPxY motifs in Ex . The identification of multiple proteins that act through the interaction between WW-domains and PPxY motifs in the Hippo pathway suggests that these motif-specific interactions are important for regulation of Hippo signaling
1) http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3987945/
Last edited by Admin on Sat Feb 16, 2019 3:20 am; edited 1 time in total

