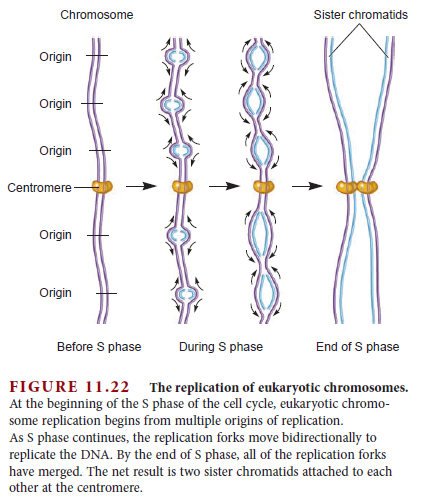
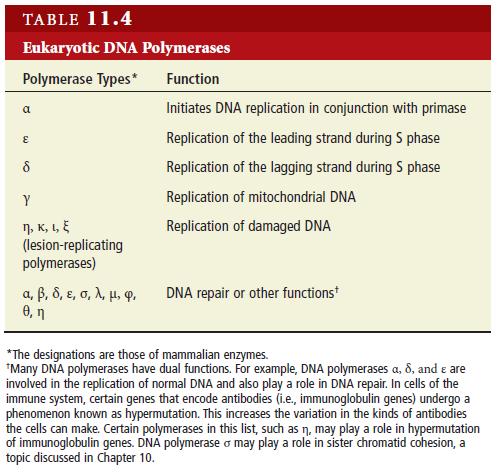
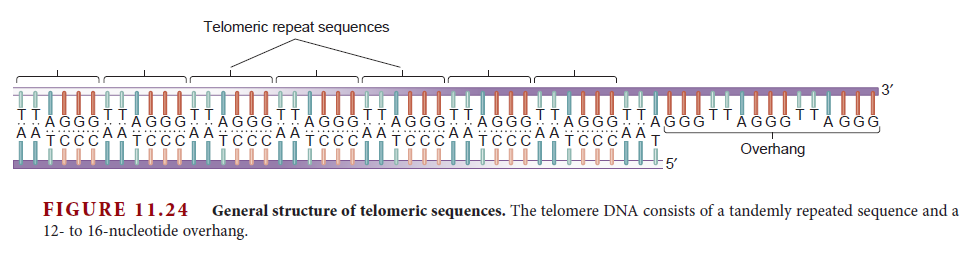
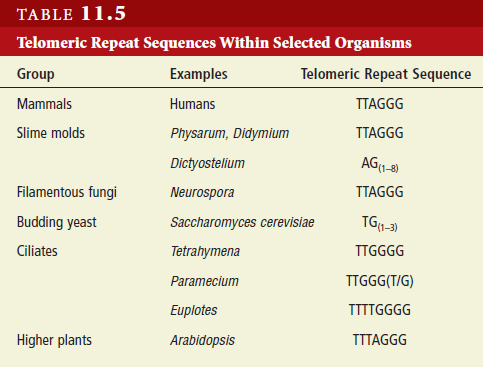
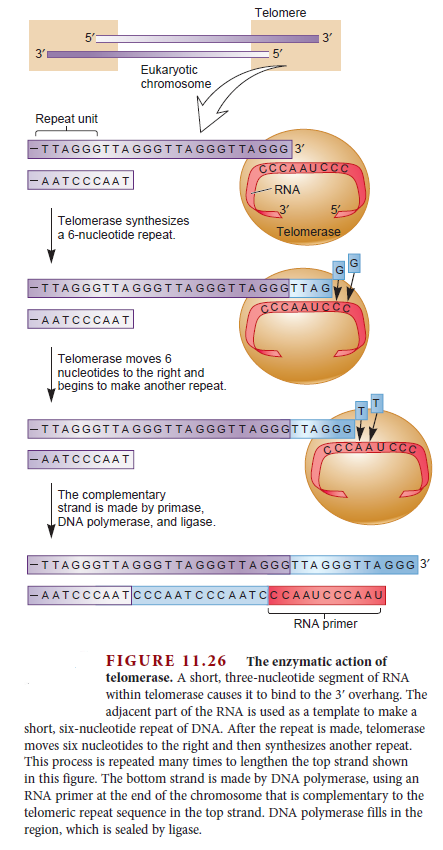
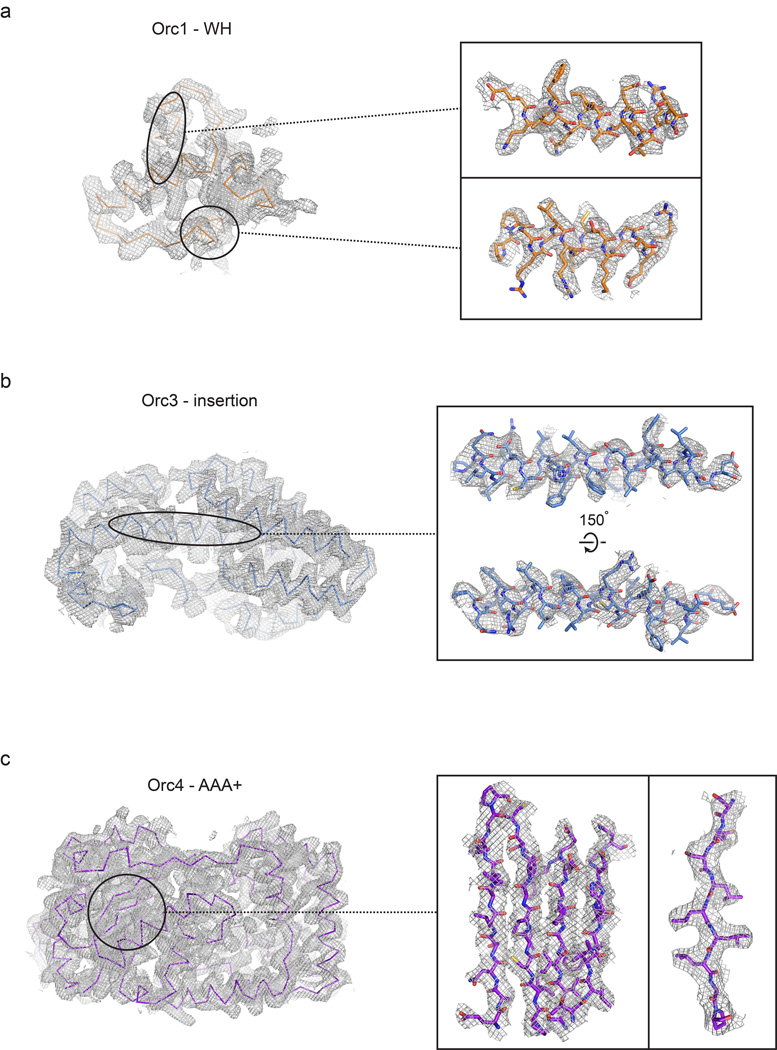
Eukaryotes contain a number of DNA polymerases. Pol � extends primers, the highly processive pol � is the lagging strand polymerase, and pol ε appears to be the leading strand polymerase. Eukaryotic replication has multiple origins and proceeds through nucleosomes. In order to replicate the 5" end of the lagging strand, eukaryotic chromosomes end with repeated telomeric sequences appended by the ribonucleoprotein telomerase. The 3" extension at the end of each chromosome serves as a template for primer synthesis. Somatic cells lack telomerase activity, which may prevent them from transforming to cancer cells.Eukaryotes possess much of the same replication machinery found in prokaryotes. For example, like prokaryotes, a DNA clamp protein acts along with DNA polymerase during DNA synthesis. One such eukaryotic clamp protein, proliferating nuclear cell antigen (PCNA), was originally identified as an antigen that is expressed in the nuclei of dividing cells during S phase. PCNA is a clamp protein for DNA polymerase d. In eukaryotic cells, the enormous length and elaborate folding of the chromosomal DNA molecules pose additional challenges for DNA replication. For example, how are the many replication origins coordinated, and how is their activation linked to other key events in the cell cycle? Answering such questions requires a better understanding of the spatial organization of DNA replication within the nucleus. When cells are briefly incubated with DNA precursors that make the most recently formed DNA fluorescent, microscopic examination reveals that the new,
Parts required in eukaryotes
Origin of replication
The origin of replication (also called the replication origin) is a particular sequence in a genome at which replication is initiated.[1] This can either involve the replication of DNA in living organisms such as prokaryotes and eukaryotes, or that of DNA or RNA in viruses, such as double-stranded RNA viruses.
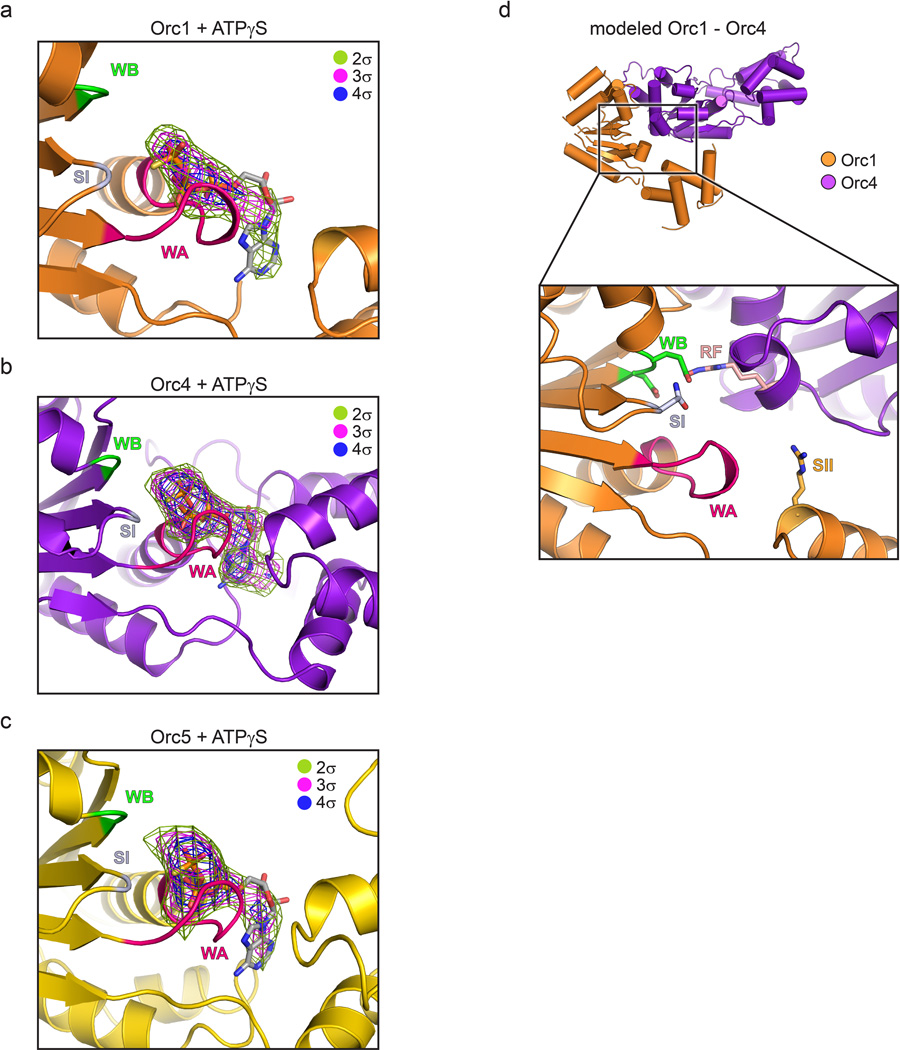
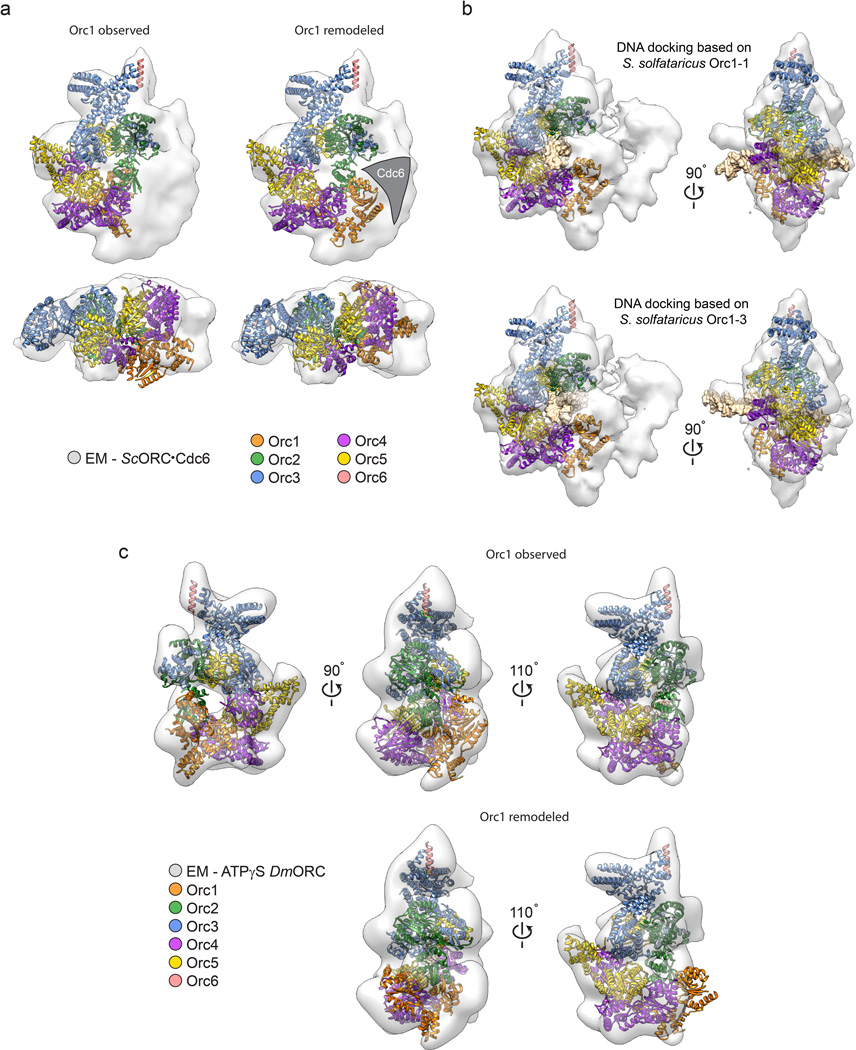
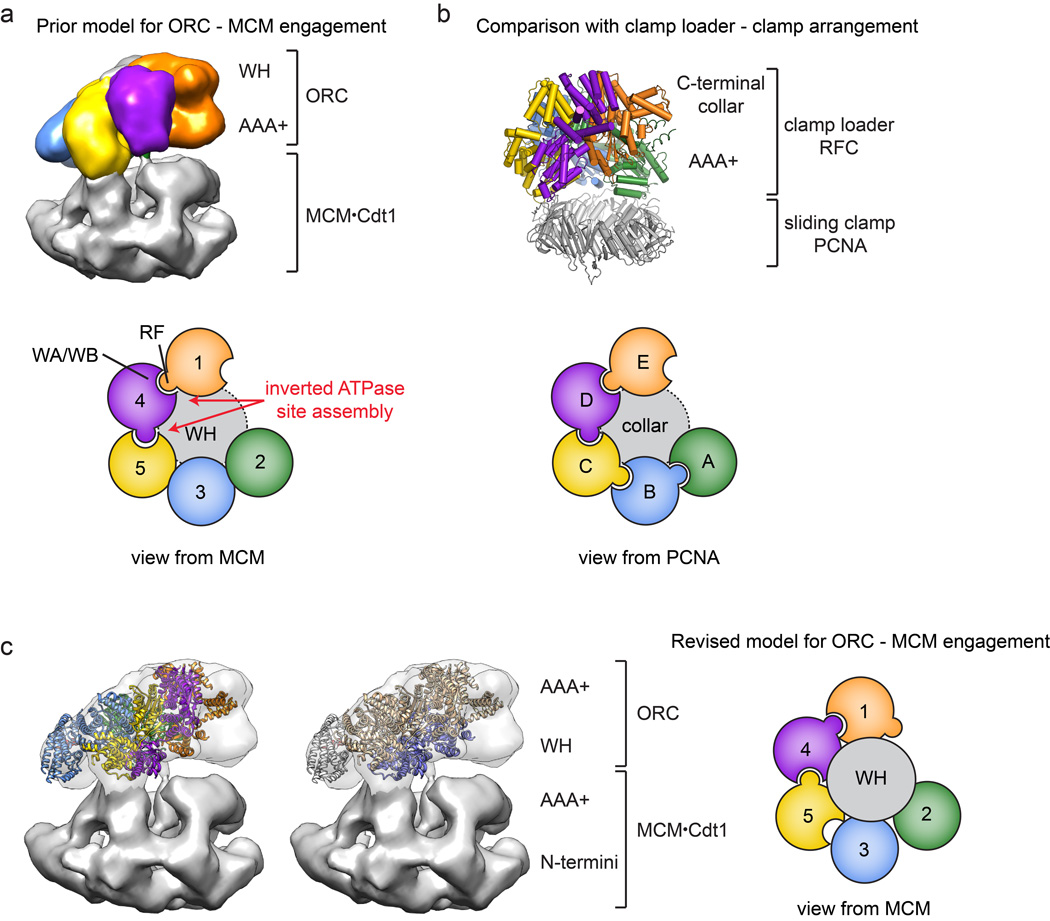
Origin Recognition Complex ( ORC )The origin recognition complex (ORC), a heteromeric six‐subunit protein, is a central component for eukaryotic DNA replication.Primosome ( consists of seven proteins: DnaG primase, DnaB helicase, DnaC helicase assistant, DnaT, PriA, Pri B, and PriC. )helicase-loading proteins Cdc6 and Cdt1a protein called DnaCDNA polymeraseDNA polymerase I for Primer removal after okazaki fragment maturation
In eukaryotes, the budding yeast Saccharomyces cerevisiae has the best characterised replication origins. These origins were first identified by their ability to support the replication of mini-chromosomes or plasmids, giving rise to the name Autonomously replicating sequences or ARS elements. Each budding yeast origin consists of a short (~11 bp) essential DNA sequence (called the ARS consensus sequence or ACS) that recruits replication proteins.
In other eukaryotes, including humans, the DNA sequences at the replication origins vary. Despite this sequence variation, all the origins form a base for assembly of a group of proteins known collectively as the pre-replication complex (pre-RC):
- First, the origin DNA is bound by the origin recognition complex (ORC) which, with help from two further protein factors (Cdc6 and Cdt1), load the mini chromosome maintenance (or MCM) protein complex.
- Once assembled, this complex of proteins indicates that the replication origin is ready for activation. Once the replication origin is activated, the cell's DNA will be replicated.
In metazoans, pre-RC formation is inhibited by the protein geminin, which binds to and inactivates Cdt1. Regulation of replication prevents the DNA from being replicated more than once each cell cycle.
In humans an origin of replication has been originally identified near the Lamin B2 gene on chromosome 19 and the ORC binding to it has extensively been studied.[8]
Question: How did these DNA sequence motifs arise, and how did the proteins " learn " that these specific sequences mean " place to open the DNA strand and start replication ? " trial and error ?
The replisome is a complex molecular machine that carries out replication of DNA. The replisome first unwinds double stranded DNA into two single strands. For each of the resulting single strands, a new complementary sequence of DNA is synthesized. The net result is formation of two new double stranded DNA sequences that are exact copies of the original double stranded DNA sequence.
In terms of structure, the replisome is composed of two replicative polymerase complexes, one of which synthesizes the leading strand, while the other synthesizes the lagging strand. The replisome is composed of a number of proteins including helicase, RFC, PCNA, gyrase/topoisomerase, SSB/RPA, primase, DNA polymerase I, RNAse H, and ligase.
The replisome is a system in which various factors work together to solve the structural and chemical challenges of DNA replication. Chromosomes size and structure varies between organisms, but since DNA molecules are the reservoir of genetic information for all forms of life, many replication challenges and solutions are the same for different organisms. As a result, the replication factors that solve these problems are highly conserved in terms of structure, chemistry, functionality, or sequence. General structural and chemical challenges include the following:
- Efficient replisome assembly at origins of replication (origin recognition complexes or specific replication origin sequences in some organisms)
- Separating the duplex into the leading and lagging template strands (helicases)
- Protecting the leading and lagging strands from damage after duplex separation (SSB and RPA factors)
- Priming of the leading and lagging template strands (primase or DNA polymerase alpha)
- Ensuring processivity (clamp loading factors, ring-shaped clamp proteins, strand binding proteins)
- High-fidelity DNA replication (DNA polymerase III, DNA polymerase delta, DNA polymerase epsilon. All have intrinsically low error rates because of their structure and chemistry.)
- Error correction (replicative polymerase active sites sense errors; 3' to 5' exonuclease domains of replicative polymerases fix errors)
- Synchronised polymerisation of leading and lagging strands despite anti-parallel structure (replication fork structure, dimerisation of replicative polymerases)
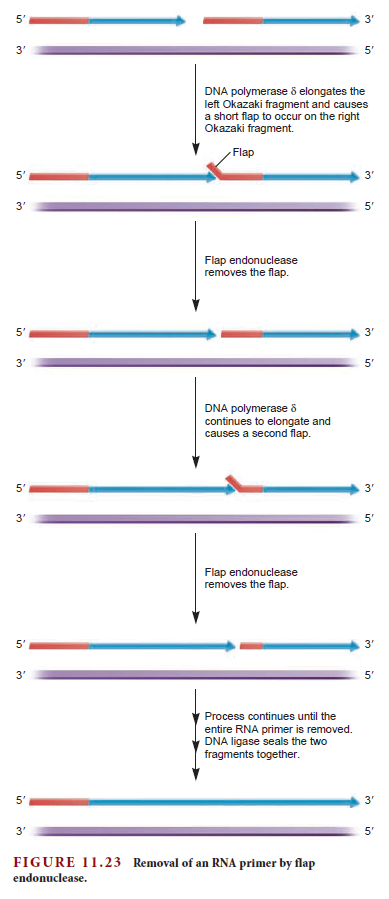
- Primer removal (DNA polymerase I, RNAse H, flap endonucleases such as FEN1, or other DNA repair factors)
- Formation of phosphodiester bonds at gaps between Okazaki fragments (ligase)
In general, the challenges of DNA replication involve the structure of the molecules, the chemistry of the molecules, and, from a systems perspective, the underlying relationships between the structure and the chemistry.Regulated eukaryotic DNA replication origin firing with purified proteins
http://www.nature.com/nature/journal/v519/n7544/full/nature14285.html
DNA codes for the DNA replicating enzyme, DNA polymerase. DNA polymerase reads the chemical code of DNA and faithfully creates another exact duplicate molecule. So for its existence, DNA is dependent on DNA polymerase, the existence of which is dependent on DNA itself. 3
 Replication: Mechanism of Replication
Replication: Mechanism of ReplicationDuring DNA replication, both strands of the double helix act as templates for the formation of new DNA molecules. Copying occurs at a localized region called the replication fork, which is a Y shaped structure where new DNA strands are synthesised by a multi-enzyme complex. Here the DNA to be copied enters the complex from the left. One new strand is leaving at the top of frame and the other new strand is leaving at bottom. The first step in DNA replication is the separation of the two strands by an enzyme called helicase. This spins the incoming DNA to unravel it: at ten thousand RPM in the case of bacterial systems. The separated strands are called three prime and five prime, distinguished by the direction in which their component nucleotides join up. . The 3' DNA strand, also known as the leading strand, is diverted to a DNA polymerase and is used as a continuous template for the synthesis of the first daughter DNA helix. The other half of the DNA double helix, known as the lagging strand, has the opposite 3' to 5' orientation and consequently requires a more complicated copying mechanism. As it emerges from the helicase, the lagging strand is organised into sections called Okazaki fragments. These are then presented to a second DNA polymerase enzyme in the preferred 5' to 3' orientation. These sections are then effectively synthesised backwards. When the copying is complete, the finished section is released and the next loop is drawn back for replication. Intricate as this mechanism appears, numerous components have been deliberately left out to avoid complete confusion. The exposed strands of single DNA are covered by protective binding proteins. And in some systems, multiple Okazaki fragments may be present. The molecular reality is very different from the iconic image of the double helix neatly separating into two DNA copies as so often depicted.
Major Molecular Events of DNA ReplicationArthur Kornberg compared DNA to a tape recording of instructions that can be copied over and over. How do cells make these near-perfect copies, and does the process ever vary?Scientists have devoted decades of effort to understanding how deoxyribonucleic acid () replicates itself. In simple terms, replication involves use of an existing strand of DNA as a template for the synthesis of a new, identical strand. American enzymologist and Nobel Prize winner Arthur Kornberg compared this process to a tape recording of instructions for performing a task: "[E]xact copies can be made from it, as from a tape recording, so that this information can be used again and elsewhere in time and space" (Kornberg, 1960).
In reality, the process of replication is far more complex than suggested by Kornberg's analogy. Researchers typically utilize simple bacterial cells in their experiments, but they still do not have all the answers, particularly when it comes to eukaryotic replication. Nonetheless, scientists are familiar with the basic steps in the replication process, and they continue to rely on this information as the basis for continued research and experimentation.The Molecular Machinery of Bacterial DNA Replication
A typical bacterial has anywhere from about 1 million to 4 million base pairs of DNA, compared to the 3 billion base pairs in the genome of the common house mouse (Mus musculus). Still, even in bacteria, with their smaller genomes, DNA replication involves an incredibly sophisticated, highly coordinated series of molecular events. These events are divided into four major stages: initiation, unwinding, primer synthesis, and elongation. Initiation and Unwinding
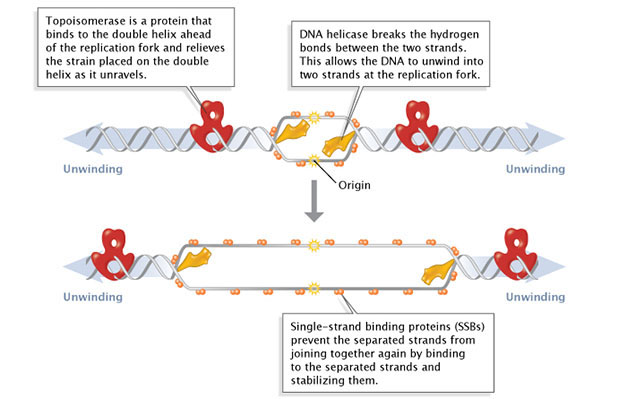
During initiation, so-called initiator proteins bind to the replication origin, a base-pair sequence of nucleotides known as oriC. This binding triggers events that unwind the DNA double helix into two single-stranded DNA molecules. Several groups of proteins are involved in this unwinding (Figure 1). For example, the DNA helicases are responsible for breaking the hydrogen bonds that join the complementary nucleotide bases to each other; these hydrogen bonds are an essential feature of James Watson and Francis Crick's three-dimensional DNA model. Because the newly unwound single strands have a tendency to rejoin, another group of proteins, the single-strand-binding proteins, keep the single strands stable until elongation begins. A third family of proteins, the topoisomerases, reduce some of the torsional strain caused by the unwinding of the double helix.Figure 1: Facilitation of DNA unwinding.During DNA replication, several proteins facilitate the unwinding of the DNA double helix into two single strands. Topoisomerases (red) reduce torsional strain caused by the unwinding of the DNA double helix; DNA helicase (yellow) breaks hydrogen bonds between complementary base-pairs; single-strand binding proteins (SSBs) stabilize the separated strands and prevent them from rejoining. The location at which a DNA strand begins to unwind into two separate single strands is known as the origin of replication. As shown in Figure 1, when the double helix unwinds, replication proceeds along the two single strands at the same time but in opposite directions (i.e., left to right on one strand, and right to left on the other). This forms two replication forks that move along the DNA, replicating as they go. Primer Synthesis
Primer synthesis marks the beginning of the actual synthesis of the new DNA molecule. Primers are short stretches of nucleotides (about 10 to 12 bases in length) synthesized by an RNA polymerase enzyme called primase. Primers are required because DNA polymerases, the enzymes responsible for the actual addition of nucleotides to the new DNA strand, can only add deoxyribonucleotides to the 3'-OH group of an existing chain and cannot begin synthesis
de novo. Primase, on the other hand, can add ribonucleotides
de novo. Later, after elongation is complete, the primer is removed and replaced with DNA nucleotides.
Elongation
Finally, elongation--the addition of nucleotides to the new DNA strand--begins after the primer has been added. Synthesis of the growing strand involves adding nucleotides, one by one, in the exact order specified by the original (template) strand. Recall that one of the key features of the Watson-Crick DNA model is that adenine is always paired with thymine and cytosine is always paired with guanine. So, for example, if the original strand reads A-G-C-T, the new strand will read T-C-G-A.
DNA is always synthesized in the 5'-to-3' direction, meaning that nucleotides are added only to the 3' end of the growing strand. As shown in Figure 2, the 5'-phosphate group of the new nucleotide binds to the 3'-OH group of the last nucleotide of the growing strand. Scientists have yet to identify a polymerase that can add bases to the 5' ends of DNA strands.
Figure 2: New DNA is synthesized from deoxyribonucleoside triphosphates (dNTPs).
(A) A deoxyribonucleoside triphosphate (dNTP). (B) During DNA replication, the 3'-OH group of the last nucleotide on the new strand attacks the 5'-phosphate group of the incoming dNTP. Two phosphates are cleaved off. (C) A phosphodiester bond forms between the two nucleotides, and phosphate ions are released.
The Discovery of DNA Polymerase
While studying
E. coli bacteria, enzymologist Arthur Kornberg discovered that DNA polymerases catalyze DNA synthesis. Kornberg's experiment involved mixing all of the basic "ingredients" necessary for
E. coli DNA synthesis in a test tube, including nucleotides,
E. coli extract, and , and then purifying and testing the enzymes involved. Using this method, Kornberg not only discovered DNA polymerases, but he also performed some of the initial work demonstrating how enzymes add new nucleotides to growing DNA chains (Kornberg, 1959).
Scientists have since identified a total of five different DNA polymerases in
E. coli, each with a specialized role. For example, DNA polymerase III does most of the elongation work, adding nucleotides one by one to the 3' end of the new and growing single strand. Other enzymes, including DNA polymerase I and RNase H, are responsible for removing the RNA primer after DNA polymerase III has begun its work, replacing it with DNA nucleotides (Ogawa & Okazaki, 1984). When these enzymes finish, they leave a nick between the section of DNA that was formerly the primer and the elongated section of DNA. Another enzyme called DNA ligase then acts to seal the bond between the two adjacent nucleotides.
DNA Polymerase Only Moves in One Direction
After a primer is synthesized on a strand of DNA and the DNA strands unwind, synthesis and elongation can proceed in only one direction. As previously mentioned, DNA polymerase can only add to the 3' end, so the 5' end of the primer remains unaltered. Consequently, synthesis proceeds immediately only along the so-called leading strand. This immediate replication is known as continuous replication. The other strand (in the 5' direction from the primer) is called the lagging strand, and replication along it is called discontinuous replication. The double helix has to unwind a bit before the synthesis of another primer can be initiated further up on the lagging strand. Synthesis can then occur from the 3' end of that new primer. Next, the double helix unwinds a bit more, and another spurt of replication proceeds. As a result, replication along the lagging strand can only proceed in short, discontinuous spurts (Figure 3).

The Challenges of Eukaryotic Replication
Bacterial and eukaryotic cells share many of the same basic features of replication; for instance, initiation requires a primer, elongation is always in the 5'-to-3' direction, and replication is always continuous along the leading strand and discontinuous along the lagging strand. But there are also important differences between bacterial and eukaryotic replication, some of which biologists are still actively researching in an effort to better understand the molecular details. One difference is that eukaryotic replication is characterized by many replication origins (often thousands), not just one, and the sequences of the replication origins vary widely among species. On the other hand, while the replication origins for bacteria, oriC, vary in length (from about 200 to 1,000 base pairs) and sequence, except among closely related organisms, all bacteria nonetheless have just a single replication origin (Mackiewicz
et al., 2004).
Eukaryotic replication also utilizes a different set of DNA polymerase enzymes (e.g., DNA polymerase δ and DNA polymerase ε instead of DNA polymerase III). Scientists are still studying the roles of the 13 eukaryotic polymerases discovered to date. In addition, in eukaryotes, the DNA template is compacted by the way it winds around proteins called histones. This DNA-histone complex, called a nucleosome, poses a unique challenge both for the cell and for scientists investigating the molecular details of eukaryotic replication. What happens to nucleosomes during DNA replication? Scientists know from electron micrograph studies that nucleosome reassembly happens very quickly after replication (the reassembled nucleosomes are visible in the electron micrograph images), but they still do not know how this happens (Annunziato, 2005).
Also, whereas bacterial chromosomes are circular, eukaryotic chromosomes are linear. During circular DNA replication, the excised primer is readily replaced by nucleotides, leaving no gap in the newly synthesized DNA. In contrast, in linear DNA replication, there is always a small gap left at the very end of the chromosome because of the lack of a 3'-OH group for replacement nucleotides to bind. (As mentioned, DNA synthesis can proceed only in the 5'-to-3' direction.) If there were no way to fill this gap, the DNA molecule would get shorter and shorter with every generation. However, the ends of linear chromosomes—thetelomeres—have several properties that prevent this.
DNA replication occurs during the S phase of cell division. In
E. coli, this means that the entire genome is replicated in just 40 minutes, at a pace of approximately 1,000 nucleotides per second. In eukaryotes, the pace is much slower: about 40 nucleotides per second. The coordination of the protein complexes required for the steps of replication and the speed at which replication must occur in order for cells to divide are impressive, especially considering that enzymes are also proofreading, which leaves very few errors behind.
Summary
The study of DNA replication started almost as soon as the structure of DNA was elucidated, and it continues to this day. Currently, the stages of initiation, unwinding, primer synthesis, and elongation are understood in the most basic sense, but many questions remain unanswered, particularly when it comes to replication of the eukaryotic genome. Scientists have devoted decades to the study of replication, and researchers such as Kornberg and Okazaki have made a number of important breakthroughs. Nonetheless, much remains to be learned about replication, including how errors in this process contribute to human disease.
References and Recommended Reading
Annunziato, A. T. Split decision: What happens to nucleosomes during DNA replication?
Journal of Biological Chemistry 280, 12065–12068 (2005)
Bessman, M. J.,
et al. Enzymatic synthesis of deoxyribonucleic acid. II. General properties of the reaction.
Journal of Biological Chemistry 233, 171–177 (1958)
Kornberg, A. The biological synthesis of deoxyribonucleic acid. Nobel Lecture, December 11, 1959. (link to transcript)
———. Biological synthesis of deoxyribonucleic acid.
Science 131, 1503–1508 (1960)
Lehman, I. R.,
et al. Enzymatic synthesis of deoxyribonucleic acid. I. Preparation of substrates and partial purification of an enzyme from
Escherichia coli.
Journal of Biological Chemistry 233, 163–170 (1958)
Losick, R., & Shapiro, L. DNA replication: Bringing the mountain to Mohammed.
Science 282, 1430–1431 (1998)
Mackiewicz, P.,
et al. Where does bacterial replication start? Rules for predicting the oriC region.
Nucleic Acids Research 32, 3781–3791 (2004)
Ogawa, T., & Okazaki, T. Function of RNase H in DNA replication revealed by RNase H defective mutants of
Escherichia coli.
Molecular and General Genetics193, 231–237 (1984)
Okazaki, R.,
et al. Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains.
Proceedings of the National Academy of Sciences 59, 598–605 (1968)
Study reveals the architecture of the molecular machine that copies DNA 4
A series of electron micrographs show the barrel-shaped helicase, which is the enzyme that separates the two DNA strands, along with other components of the replisome, including polymerase-epsilon (green). Credit: Brookhaven National Laboratory
DNA replication is essential to all life, yet many basic mechanisms in that process remain unknown to scientists. The structure of the replisome—a block of proteins responsible for unwinding the DNA helix and then creating duplicate helices for cell division—is one such mystery.Now, for the first time, a team of researchers from The Rockefeller University, Brookhaven National Laboratory, and Stony Brook University has revealed that vital complex's molecular architecture. And to their surprise, it does not look as they had expected."Our finding goes against decades of textbook drawings of what people thought the replisome should look like," says Michael O'Donnell, Anthony and Judith Evnin Professor, head of Rockefeller's Laboratory of DNA Replication and a Howard Hughes Medical Institute investigator. He is the senior author of the study, published November 2 in Nature Structural and Molecular Biology. "However, it's a recurring theme in science that nature does not always turn out to work the way you thought it did."A complete rendering of the eukaryotic replisomeThe findings focus on the replisome found in eukaryotic organisms, a category that includes a broad swath of living things, including humans and other multicellular organisms. O'Donnell and others have long studied replisomes in simpler, single-celled bacteria, but the more complex version has over 30 different gears, or proteins, and required about 15 years for O'Donnell's lab to obtain. Through these previous studies, his lab has learned how to assemble the more complex replisome from its components.But until now, no pictures existed to show just how everything fit together in the eukaryotic replisome. To create them, the team began building the complete structure piece by piece, and examining its shape in the electron microscope—a powerful device used to study protein structures, and a specialty of co-author Huilin Li, a molecular biologist at Brookhaven National Laboratory and Stony Brook University. The pictures Li and members of his lab captured were the first ever made of a complete replisome from any type of cell.
Previously (left), the replisome's two polymerases (green) were assumed to be below the helicase (tan), the enzyme that splits the DNA strands. The new images reveal one polymerase is located at the front of the helicase, causing one strand …moreThe DNA helix has two DNA strands, and each is duplicated by a separate DNA polymerase, an enzyme that creates DNA molecules by pairing single nucleotides, the basic units of DNA, with their matching partners. Another enzyme in the replisome is the helicase that, like a zipper, is responsible for separating DNA into two single strands in preparation for replication. For years, the two polymerases were thought to follow along behind the helicase, or below it, as it unzips the strands. But the new pictures of the replisome showed that one polymerase sits above the helicase.A link to epigeneticsTo identify which polymerase was at the top of the helicase, the team enlisted the help of co-authors postdoc Yi Shi and Brian Chait, the Camille and Henry Dreyfus Professor at Rockefeller and head of the Laboratory of Mass Spectrometry and Gaseous Ion Chemistry. They identified the top polymerase as Pol-ε.Three-dimensional structure of the active DNA helicase, which unzips DNA strands, is bound to the polymerase called Pol epsilon, which copies them. The DNA polymerase epsilon (green) sits on top rather than the bottom of the helicase. Credit: Brookhaven National LaboratoryWhy the eukaryotic replisome developed such a structure is not known. O'Donnell and his colleagues suspect that it may have something to do with the evolution of multicellularity. As the helicase unzips two strands of DNA, it encounters nucleosomes, particles that tightly bundle DNA to fit it into a cell's nucleus. These must be dislodged and then distributed to one new strand or the other. Previous work has shown that Pol-ε binds nucleosomes, and it may be that while riding atop the helicase, Pol-ε is in charge of distributing nucleosomes to the two strands, O'Donnell suggests."Changes to nucleosomes carry epigenetic information that instructs different cells to become the different tissues of the body, such as heart, brain, and other organs during embryonic development," O'Donnell says. "So we can speculate that Pol-ε's interaction with nucleosomes could play a role in assigning different epigenetic identities to the two new daughter cells after cell division, that instruct them to form different organs during development of a multicellular animal."Genesis of the enzyme that divides the DNA double helix during cell replication 5
Averaged electron microscope images of two intermediate helicase structures. The top shows an ORC binding the two ring structures together, and the bottom shows the completed double-ring structure with the ORC detached.The proteins that drive DNA replication—the force behind cellular growth and reproduction—are some of the most complex machines on Earth. The multistep replication process involves hundreds of atomic-scale moving parts that rapidly interact and transform. Mapping that dense molecular machinery is one of the most promising and challenging frontiers in medicine and biology.Now, scientists have pinpointed crucial steps in the beginning of the replication process, including surprising structural details about the enzyme that "unzips" and splits the DNA double helix so the two halves can serve as templates for DNA duplication.The research combined electron microscopy, perfectly distilled proteins, and a method of chemical freezing to isolate specific moments at the start of replication. The study—authored by scientists from the U.S. Department of Energy's Brookhaven National Laboratory, Stony Brook University, Cold Spring Harbor Laboratory, and Imperial College, London—published on Oct. 15, 2014, in the journal Genes and Development."The genesis of the DNA-unwinding machinery is wonderfully complex and surprising," said study coauthor Huilin Li, a biologist at Brookhaven Lab and Stony Brook University. "Seeing this helicase enzyme prepare to surround and unwind the DNA at the molecular level helps us understand the most fundamental process of life and how that process might go wrong. Errors in copying DNA are found in certain cancers, and this work could one day help develop new treatment methods that stall or break dangerous runaway machinery."The research picks up where two previous studies by Li and colleagues left off. They first determined the structure of the "Origin Recognition Complex" (ORC), a protein that identifies and attaches to specific DNA sites to initiate the entire replication process. The second study revealed how the ORC recruits, cracks open, and installs a crucial ring-shaped protein structure (Mcm2-7) that lies at the core of the helicase enzyme.But DNA replication is a bi-directional process with two helicases moving in opposite directions. The key question, then, was how does a second helicase core get recruited and loaded onto the DNA in the opposite orientation of the first?"To our surprise, we found an intermediate structure with one ORC binding two rings," said Brookhaven Lab biologist and lead author Jingchuan Sun. "This discovery suggests that a single ORC, rather than the commonly believed two-ORC system, loads both helicase rings."
Three-dimensional model (based on electron microscopy data) of the double-ring structure loaded onto a DNA helix.One step further along, the researchers also determined the molecular architecture of the final double-ring structure left behind after the ORC leaves the system, offering a number of key biological insights."We now have clues to how that double-ring structure stably lingers until the cell enters the DNA-synthesis phase much later on in replication," said study coauthor Christian Speck of Imperial College, London. "This study revealed key regulatory principles that explain how the helicase activity is initially suppressed and then becomes reactivated to begin its work splitting the DNA."Precision methods, close collaborationExamining these fleeting molecular structures required mastery of biology, chemistry, and electron microscopy techniques."This three-way collaboration took advantage of each lab's long standing collaboration and expertise," said study coauthor Bruce Stillman of Cold Spring Harbor. "Imperial College and Cold Spring Harbor handled the challenging material preparation and functional characterization, while Brookhaven and Stony Brook led the sophisticated molecular imaging and three-dimensional image reconstruction."The researchers used proteins from baker's yeast—a model organism for the more complex systems found in animals. The scientists isolated the protein mechanisms involved in replication and removed structures that might otherwise complicate the images.
Collaborating scientists and study coauthors Zuanning Yuan of Stony Brook University (standing), Huilin Li of Stony Brook and Brookhaven Lab (seated, back), and Jingchuan Sun of Brookhaven Lab (seated, front) examining protein structures.Once the isolated proteins were mixed with DNA, the scientists injected chemicals to "freeze" the binding and recruitment process at intervals of 2, 7, and 30 minutes.They then used an electron microscope at Brookhaven to pin down the exact structures at each targeted moment in a kind of molecular time-lapse. Rather than the light used in a traditional microscope, this technique uses focused beams of electrons to illuminate a sample and form images with atomic resolution. The instrument produces a large number of two-dimensional electron beam images, which a computer then reconstructs into three-dimensional structure."This technique is ideal because we're imaging relatively massive proteins here," Li said. "A typical protein contains three hundred amino acids, but these DNA replication mechanisms consist of tens of thousands of amino acids. The entire structure is about 20-nanometers across, compared to 4 nanometers for an average protein."Unraveling the DNA processes at the most fundamental level, the focus of this team's work, could have far-reaching implications."The structural knowledge may help others engineer small molecules that inhibit DNA replication at specific moments, leading to new disease prevention or treatment techniques," Li said.1) https://www.dnalc.org/resources/3d/04-mechanism-of-replication-advanced.html
2) http://www.nature.com/scitable/topicpage/major-molecular-events-of-dna-replication-413
3) http://dialogue.adventist.org/articles/15_2_ward_e.htm
4) http://phys.org/news/2015-11-reveals-architecture-molecular-machine-dna.html
5) http://phys.org/news/2014-10-genesis-enzyme-dna-helix-cell.html