Chromatin Remodeling in Eukaryotes 1
http://www.nature.com/scitable/topicpage/Chromatin-Remodeling-in-Eukaryotes-1082
chromatin not only serves as a way to condense DNA within the cellular nucleus, but also as a way to control how that DNA is used. In particular, within eukaryotes, specific genes are not expressed unless they can be accessed by RNA polymerase and proteins known as transcription factors. In its default state, the tight coiling that characterizes chromatin structure limits the access of these substances to eukaryotic DNA. Therefore, a cell's chromatin must "open" in order for gene expression to take place. This process of "opening" is called chromatin remodeling, and it is of vital importance to the proper functioning of all eukaryotic cells. In recent years, researchers have discovered a great deal about chromatin remodeling, including the roles that different protein complexes, histone variants, and biochemical modifications play in this process. However, a great deal remains to be learned before chromatin remodeling is fully understood.
Chromatin Remodeling at a Glance
Various molecules called chromatin remodelers provide the mechanism for modifying chromatin and allowing transcription signals to reach their destinations on the DNA strand. Understanding the nature and processes of these cellular construction workers remains an active area of discovery in genetic research.
Currently, investigators know that chromatin remodelers are large, multiprotein complexes that use the energy of ATP hydrolysis to mobilize and restructure nucleosomes. Recall that nucleosomes wrap 146 base pairs of DNA in approximately 1.7 turns around a histone-octamer disk, and the DNA inside each nucleosome is generally inaccessible to DNA-binding factors. Remodelers are thus necessary to provide access to the underlying DNA to enable transcription, chromatin assembly, DNA repair, and other processes. Just how remodelers convert the energy of ATP hydrolysis into mechanical force to mobilize the nucleosome, and how different remodeler complexes select which nucleosomes to move and restructure, remains unknown, however.
Remodelers are partitioned into five families, each with specialized biological roles. Nonetheless, all remodelers contain a subunit with a conserved ATPase domain. In addition to the conserved ATPase, each remodeler complex also possesses unique proteins that specialize it for its unique biological role. However, because all remodelers move nucleosomes and all such movement is ATP dependent, mobilization is most likely a property of the conserved ATPase subunit.
The ATPase domains of remodelers are similar in sequence and structure to known DNA-translocating proteins in viruses and bacteria. Recent evidence from the SWI/SNF and ISWI remodeler families has also revealed that remodeler ATPases are directional DNA translocases that are capable of the directional pumping of DNA. But how is this property applied to nucleosomes? It seems that the ATPase binds approximately 40 base pairs inside the nucleosome, from which location it pumps DNA around the histone-octamer surface. This enables the movement of the nucleosome along the DNA, thus permitting the exposure of the DNA to regulatory factors.
The additional domains and proteins that are attached to the ATPase are important for nucleosome selection, and they also help regulate ATPase activity. These attendant proteins bind to histones and nucleosomal DNA, and their binding to these molecules is affected by the histone modification state. The modification state helps determine whether the nucleosome is an appropriate substrate for a remodeler complex (Saha et al., 2006), as discussed later in this article.
Indeed, canonical histones can themselves be replaced by histone variants or modified by specific enzymes, thereby making the surrounding DNA more or less accessible to the transcriptional machinery.
So far, a number of histone variants have been found and localized to specific areas of chromatin. For instance, H2A.Z is a variant of H2A and is often enriched near relatively inactive gene promoters. Interestingly, H2A.Z does not take its place during replication when the chromatin structure is established. Instead, the chromatin remodeling complex SWR1 catalyzes an ATP-dependent exchange of H2A in the nucleosome for H2A.Z (Wu et al., 2005).
Histone Modification and the Histone Code
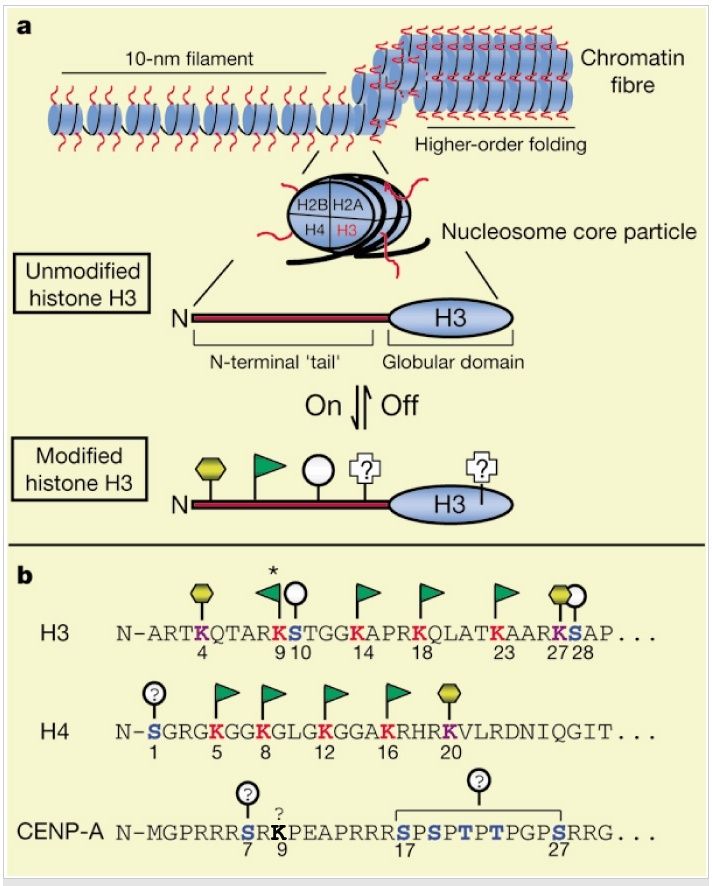
Histone sequences are highly conserved. The diagram below shows a typical chromatin fiber, with the blue cylinders representing histones. Extending from each of the histones is a "tail," called the N-terminal tail because proteins have two ends--an N terminus and C terminus. Here, the C terminus forms a globular domain that is packaged into the nucleosome. The other end of the histone is more flexible and capable of interacting more directly with DNA and the different proteins within the nucleus.
Figure below: (A) General chromatin organization. Like other histone "tails," the N terminus of H3 (red) represents a highly conserved domain that is likely to be exposed or extend outwards from the chromatin fiber. A number of distinct post-translational modifications are known to occur at the N terminus f H3 including acetylation (green flag), phosphorylation (grey circle), and methylation (yellow hexagon). Other modifications are known and may also occur in the globular domain. (B) The N terminus of human H3 is shown in single-letter amino-acid code. For comparison, the N termini of human CENP-A, a centromere-specific H3 variant, and human H4, the nucleosomal partner to H3, are shown. Note the regular spacing of acetylatable lysines (red), and potential phosphorylation (blue) and methylation (purple) sites. The asterisk indicates the lysine residue in H3 that is known to be targeted for acetylation as well as for methylation; lysine 9 in CENP-A (bold) may also be chemically modified (see text). The above depictions of chromatin structure and H3 are schematic; no attempt has been made to accurately portray these structures.
1) http://www.nature.com/scitable/topicpage/Chromatin-Remodeling-in-Eukaryotes-1082
http://www.nature.com/scitable/topicpage/Chromatin-Remodeling-in-Eukaryotes-1082
chromatin not only serves as a way to condense DNA within the cellular nucleus, but also as a way to control how that DNA is used. In particular, within eukaryotes, specific genes are not expressed unless they can be accessed by RNA polymerase and proteins known as transcription factors. In its default state, the tight coiling that characterizes chromatin structure limits the access of these substances to eukaryotic DNA. Therefore, a cell's chromatin must "open" in order for gene expression to take place. This process of "opening" is called chromatin remodeling, and it is of vital importance to the proper functioning of all eukaryotic cells. In recent years, researchers have discovered a great deal about chromatin remodeling, including the roles that different protein complexes, histone variants, and biochemical modifications play in this process. However, a great deal remains to be learned before chromatin remodeling is fully understood.
Chromatin Remodeling at a Glance
Various molecules called chromatin remodelers provide the mechanism for modifying chromatin and allowing transcription signals to reach their destinations on the DNA strand. Understanding the nature and processes of these cellular construction workers remains an active area of discovery in genetic research.
Currently, investigators know that chromatin remodelers are large, multiprotein complexes that use the energy of ATP hydrolysis to mobilize and restructure nucleosomes. Recall that nucleosomes wrap 146 base pairs of DNA in approximately 1.7 turns around a histone-octamer disk, and the DNA inside each nucleosome is generally inaccessible to DNA-binding factors. Remodelers are thus necessary to provide access to the underlying DNA to enable transcription, chromatin assembly, DNA repair, and other processes. Just how remodelers convert the energy of ATP hydrolysis into mechanical force to mobilize the nucleosome, and how different remodeler complexes select which nucleosomes to move and restructure, remains unknown, however.
Remodelers are partitioned into five families, each with specialized biological roles. Nonetheless, all remodelers contain a subunit with a conserved ATPase domain. In addition to the conserved ATPase, each remodeler complex also possesses unique proteins that specialize it for its unique biological role. However, because all remodelers move nucleosomes and all such movement is ATP dependent, mobilization is most likely a property of the conserved ATPase subunit.
The ATPase domains of remodelers are similar in sequence and structure to known DNA-translocating proteins in viruses and bacteria. Recent evidence from the SWI/SNF and ISWI remodeler families has also revealed that remodeler ATPases are directional DNA translocases that are capable of the directional pumping of DNA. But how is this property applied to nucleosomes? It seems that the ATPase binds approximately 40 base pairs inside the nucleosome, from which location it pumps DNA around the histone-octamer surface. This enables the movement of the nucleosome along the DNA, thus permitting the exposure of the DNA to regulatory factors.
The additional domains and proteins that are attached to the ATPase are important for nucleosome selection, and they also help regulate ATPase activity. These attendant proteins bind to histones and nucleosomal DNA, and their binding to these molecules is affected by the histone modification state. The modification state helps determine whether the nucleosome is an appropriate substrate for a remodeler complex (Saha et al., 2006), as discussed later in this article.
Indeed, canonical histones can themselves be replaced by histone variants or modified by specific enzymes, thereby making the surrounding DNA more or less accessible to the transcriptional machinery.
So far, a number of histone variants have been found and localized to specific areas of chromatin. For instance, H2A.Z is a variant of H2A and is often enriched near relatively inactive gene promoters. Interestingly, H2A.Z does not take its place during replication when the chromatin structure is established. Instead, the chromatin remodeling complex SWR1 catalyzes an ATP-dependent exchange of H2A in the nucleosome for H2A.Z (Wu et al., 2005).
Histone Modification and the Histone Code
Histone sequences are highly conserved. The diagram below shows a typical chromatin fiber, with the blue cylinders representing histones. Extending from each of the histones is a "tail," called the N-terminal tail because proteins have two ends--an N terminus and C terminus. Here, the C terminus forms a globular domain that is packaged into the nucleosome. The other end of the histone is more flexible and capable of interacting more directly with DNA and the different proteins within the nucleus.
Figure below: (A) General chromatin organization. Like other histone "tails," the N terminus of H3 (red) represents a highly conserved domain that is likely to be exposed or extend outwards from the chromatin fiber. A number of distinct post-translational modifications are known to occur at the N terminus f H3 including acetylation (green flag), phosphorylation (grey circle), and methylation (yellow hexagon). Other modifications are known and may also occur in the globular domain. (B) The N terminus of human H3 is shown in single-letter amino-acid code. For comparison, the N termini of human CENP-A, a centromere-specific H3 variant, and human H4, the nucleosomal partner to H3, are shown. Note the regular spacing of acetylatable lysines (red), and potential phosphorylation (blue) and methylation (purple) sites. The asterisk indicates the lysine residue in H3 that is known to be targeted for acetylation as well as for methylation; lysine 9 in CENP-A (bold) may also be chemically modified (see text). The above depictions of chromatin structure and H3 are schematic; no attempt has been made to accurately portray these structures.

1) http://www.nature.com/scitable/topicpage/Chromatin-Remodeling-in-Eukaryotes-1082



